In the next tables we show the prediction of the selenoproteins we have found in the genome of Miichthys miiuy. In order to correctly interprete the table we have used some symbols:
 |
Danio rerio (Zebrafish) |  |
Homo sapiens |  |
Found in Miichthys miiuy |  |
Not found in Miichthys miiuy |
Selenoproteins:
GENEWISE |
Photo |
Info |
|||||||||
| Sel15 |  |
Sec |  |
 |
JXSJ01000006.1 | 2280275-2283892(-) |  |
 |
 |
 |
|
|---|---|---|---|---|---|---|---|---|---|---|---|
| SELENOE |  |
Sec |  |
 |
JXSJ01000024.1 | 1414823-1416749(+) |  |
 |
 |
 |
|
| GPx1a |  |
Sec |  |
 |
JXSJ01000122.1 | 469000-470574(-) |  |
 |
 |
 |
|
| GPx1b |  |
Sec |  |
 |
JXSJ01000043.1 | 1230876-1231894(-) |  |
 |
 |
 |
|
| GPx2 |  |
Sec |  |
 |
JXSJ01000061.1 | 670956-672041(+) |  |
 |
 |
 |
|
| GPx3b |  |
Sec |  |
 |
JXSJ01000009.1 | 4882795-4884192(-) |  |
 |
 |
 |
|
| GPx4a |  |
Sec |  |
 |
JXSJ010000014.1 | 7520-8330(-) |  |
 |
 |
 |
|
| GPx4b |  |
Sec |  |
 |
JXSJ01000006.1 | 3977178-3978784(-) |  |
 |
 |
 |
|
| GPx5/GPx6 |  |
Sec |  
|  |
JXSJ01000010.1 | 2819476-2821100(+) |  |
 |
 |
 |
GPx7 |  |
Cys |  |
 |
JXSJ01000525.1 | 179948-181053(+) |  |
 |
 |
 |
| GPx8 |  |
Cys |  |
 |
JXSJ01000013.1 | 1301222-1302748(-) |  |
 |
 |
 |
|
| DIO1 |  |
Sec |  |
 |
JXSJ01000014.1 | 2396940-2398257(+) |  |
 |
 |
 |
|
| DIO2 |  |
Sec |  |
 |
JXSJ01000082.1 | 1146249-1148716(+) |  |
 |
 |
 |
|
| DIO3a |  |
Sec |  |
 |
JXSJ01000029.1 | 1651922-1652713(+) |  |
 |
 |
 |
|
| DIO3b |  |
Sec |  |
 |
JXSJ01000021.1 | 1252836-1253576(-) |  |
 |
 |
 |
|
| MsrA(1) |  |
Cys |  |
 |
JXSJ01000021.1 | 792956-823482(+) |  |
 |
 |
 |
|
| MsrA(2) |  |
Cys |  |
 |
JXSJ01000913.1 | 69102-86182(-) |  |
 |
 |
 |
|
| SEPHS(1) |
 |
Cys |  |
 |
JXSJ01000007.1 | 6480332-6483728(-) |  |
 |
 |
 |
|
| SEPHS2 |
 |
Sec |  |
 |
JXSJ01000927.1 | 74199-78967(+) |  |
 |
 |
 |
|
| SELENOH |  |
Sec |  |
 |
JXSJ01000003.1 | 9698935-9700841(-) |  |
 |
 |
 |
|
| SELENOI |  |
Sec |  |
 |
JXSJ01000029.1 | 3504276-3512380(-) |  |
 |
 |
 |
|
| SELENOJ |  |
Sec Sec |
 |
 
| JXSJ01000123.1 JXSJ01000123.1 |
355390-357844(-) 36133-364072(-) |
 
|  
|   |
  |
|
| SELENOK |  |
Sec |  |
 |
JXSJ01000005.1 | 6790241-6791962(-) |  |
 |
 |
 |
|
| SELENOM |  |
Sec |  |
 |
JXSJ01000037.1 | 1022323-1024868(-) |  |
 |
 |
 |
|
| SELENON |  |
Sec |  |
 |
JXSJ01000021.1 | 1515343-1519319(+) |  |
 |
 |
 |
|
| SELENOO1 |  |
Sec |  |
 |
JXSJ01000027.1 | 201128-205951(+) |  |
 |
 |
 |
|
| SELENOP(1) |  |
Sec |  |
 |
JXSJ01000024.1 | 2362763-2363619(+) |  |
 |
 |
 |
|
| SELENOP(2) |  |
Sec |  |
 |
JXSJ01000020.1 | 4078401-4080381(+) |  |
 |
 |
 |
|
| MSRB1a |  |
Sec |  |
 |
JXSJ01000046.1 | 2417768-2418712(-) |  |
 |
 |
 |
|
| MSRB1b |  |
Sec |  |
 |
JXSJ01000015.1 | 2035871-2037319(-) |  |
 |
 |
 |
|
| MSRB2 |  |
Sec |  |
 |
JXSJ01000042.1 | 365418-399496(+) |  |
 |
 |
 |
|
| MSRB3 |  |
Cys |  |
 |
JXSJ01000328.1 | 132096-137920(-) |  |
 |
 |
 |
|
| SELENOS |  |
Sec |  |
 |
JXSJ01001008.1 | 75534-77926(-) |  |
 |
 |
 |
|
| SELENOT1 |  |
Sec |  |
 |
JXSJ01000006.1 | 6154489-6158134(-) |  |
 |
 |
 |
|
| SELENOT2 |  |
Sec |  |
 |
JXSJ01000199.1 | 220221-221833(+) |  |
 |
 |
 |
|
| SELENOU1a |  |
Sec Sec |
 |
  |
JXSJ01000036.1 JXSJ01000036.1 |
1859825-1861770(-) 1865654-1866392(-) |
 
|  
|  
|  
| |
| SELENOU2 |  |
Cys |  |
 |
JXSJ01000066.1 | 443031-444802(-) |  |
 |
 |
 |
|
| SELENOU3 |  |
Cys |  |
 |
JXSJ01000043.1 | 74604-82871(+) |  |
 |
 |
 |
|
| SELENOW(1) |  |
Sec |  |
 |
JXSJ01000017.1 | 774172-774663(+) |  |
 |
 |
 |
|
| SELENOW(2) |  |
Sec |  |
 |
JXSJ01000888.1 | 2038-2369(+) |  |
 |
 |
 |
|
| TXNRD2 |  |
Sec |  |
 |
JXSJ01000073.1 | 512657-531121(-) |  |
 |
 |
 |
|
| TXNRD3 |  |
Sec Sec |
 |
  |
JXSJ01000302.1 JXSJ01000728.1 |
316058-326975(+) 133825-143097(-) |
  |
  |
  |
  |
|
Selenoproteins machinery:
GENEWISE |
Photo |
Info |
|||||||||
| eEFsec |  |
Cys |  |
 |
JXSJ01000351.1 | 76191-85452(-) |  |
 |
 |
 |
|
|---|---|---|---|---|---|---|---|---|---|---|---|
| PSTK |  |
Cys |  |
 |
JXSJ01000009.1 | 5173702-5174952(+) |  |
 |
 |
 |
|
| SBP2(1) |  |
Cys |  |
 |
JXSJ01000013.1 | 206114-208113(-) |  |
 |
 |
 |
|
| SBP2(2) |  |
Cys |  |
 |
JXSJ01000001.1 | 182184-191334(+) |  |
 |
 |
 |
|
| SecS(1) |  |
Cys |  |
 |
JXSJ01000055.1 | 1602664-1603455(+) |  |
 |
 |
 |
|
| SecS(2) |  |
Cys |  |
 |
JXSJ01000055.1 | 1608599-1609403(+) |  |
 |
 |
 |
|
| SECp43(1) |  |
Cys |  |
 |
JXSJ010005029.1 | 237740-239453(-) |  |
 |
 |
 |
|
| SECp43(2) |  |
Cys |  |
 |
JXSJ01000502.1 | 79222-84336(+) |  |
 |
 |
 |
|
Proteins related to selenium metabolism:
GENEWISE |
Photo |
Info |
|||||||||
| ELAVL1 |  |
Cys |  |
 |
JXSJ01000369.1 | 274913-277728(-) |  |
 |
 |
 |
|
|---|---|---|---|---|---|---|---|---|---|---|---|
| CELF1 |  |
Cys |  |
 |
JXSJ01000100.1 | 86195-107585(+) |  |
 |
 |
 |
|
| EIF4A3 |  |
Cys |  |
 |
JXSJ01000207.1 | 410532-414259(-) |  |
 |
 |
 |
|
| XPO1 |  |
Cys |  |
 |
JXSJ01000048.1 | 532824-546124(+) |  |
 |
 |
 |
|
| G6PD |  |
Cys |  |
 |
JXSJ01000005.1 | 2740552-2744079(-) |  |
 |
 |
 |
|
| LRP8 |  |
Cys Cys |
 |
 
| JXSJ01000652.1 JXSJ01000652.1 |
11081-15068(+) 31374-35820(+) |
  |
  |
  |
  |
|
| RPL30 |  |
Cys |  |
 |
JXSJ01000353.1 | 48544-50822(-) |  |
 |
 |
 |
|
| SCLY |  |
Cys |  |
 |
JXSJ01000121.1 | 196657-200005(-) |  |
 |
 |
 |
|
| SELENBP1 |  |
Cys |  |
 |
JXSJ01000048.1 | 387018-393128(-) |  |
 |
 |
 |
|
| SARS2 |  |
Sec |  |
 |
JXSJ01000068.1 | 356430-363543(+) |  |
 |
 |
 |
|
| TTPA |  |
Cys |  |
 |
JXSJ01000006.1 | 61548-66016(-) |  |
 |
 |
 |
|
Selenoproteins analysis:
15kDa selenoprotein (Sel15)
The protein Sel15 is located in the scaffold JXSJ01000006.1 between the positions 2283892 and 2280275 in the reverse strand. This gene is formed by 4 exons with a selenocysteine located in the second exon.
The structure of the gene, analyzed with the Exonerate file, is described below:

In this protein two Secis elements, one of grade A and another of grade B have been predicted. The best secis element is the one with grade A which is located in the reverse strand and in the 3'UTR. The result that has been predicted with Seblastian correlates with our result but one additional exon in 5' has been found by Seblastian.
Fish selenoprotein 15 (SELENOE)
SELENOE is located in the scaffold JXSJ01000024.1 between positions 1414823 and 1416749 in the forward strand. The gene contains 4 exons, with a selenocysteine in the second one.
We have analyzed the gene structure with the Exonerate file and the results are described below:

In this protein three Secis elements, one of grade A and two of grade B have been predicted. The grade A SECIS element is located in the 3'UTR in the forward strand. Seblastian has been able to predict a selenoprotein with more exons than our prediction.
Glutathione peroxidase (GPx)
The query corresponding to GPx1a from Zebrafish has been aligned in the scaffold JXSJ01000122.1 from Miichthys miiuy's genome. Gpx1a is concretely found from position 469000 to 470574 in the reverse strand. This protein only contains two exons, and the selenocysteine is located in the first one.
The structure of the gene, analyzed with the Exonerate file, is described below:

Seblastian has been able to correctly predict the selenoprotein and two SECIS elements have been found: one of grade A in the reverse strand and one of grade B in the forward strand. Therefore, since this gene is located in the reverse strand and the SECIS element of grade A is in 3'UTR, this one has been chosen as the best one.
GPx1b has been found in the scaffold JXSJ01000043.1 from Miichthys miiuy's genome. This protein contains only two exons, and the selenocysteine is located in the fist one. The positions in the genome that codify for this protein go from 1230876 to 1231894 in the reverse strand.
The structure of the gene, analyzed with the exonerate file, is described below:
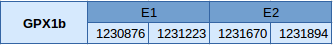
Seblastian has correctly predicted this selenoprotein, and only one SECIS element located in 3'UTR has been predicted.
GPx2 is located between the positions 670956 and 672041 of the genome in the forward strand, in the scaffold JXSJ01000061.1. This protein has two exons and the selenocysteine is located in the first one.
The structure of the gene, analyzed with the Exonerate file, is described below:
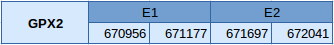
Three SECIS elements have been predicted, but only two of them are found in the forward strand and in the region 3'UTR of the gene. Even though both of them may be correctly predicted, there is one of them that has better score than the other one, and that is the reason we have chosen it, as it can be seen in the table.This protein has also been correctly predicted by Seblastian.
GPx3b is located between the positions 4882795 and 4884192 of the reverse strand, corresponding to scaffold JXSJ01000009.1. This protein is formed by four different exons, and the selenocysteine is found in the fist one.
The structure of the gene, analyzed with the Exonerate file, is described below:

This protein could not be predicted by Seblastian, and no SECIS elements have been identified by SECISearch3.
Gpx4a has been found in scaffold JXSJ01000014.1 between the positions 7520 and 8330 in the reverse strand. This gene contains three exons, with a selenocysteine in the fist exon.
The structure of the gene, analyzed with the Exonerate file, is described below:

Two SECIS elements have been identified in this protein, one of grade A and one of grade B. Since the one of grade A is the only one which is located in the 3'UTR of the gene, this one has been selected as the most suitable one.
Seblastian has been able to predict this selenoprotein, and even though it has predicted the same number of exons than our program (3 exons), they have been predicted in quite different positions. Therefore, the selenocysteine is found in the first exon according to Exonerate but in the second one according to Seblastian.
Gpx4b has been found in scaffold JXSJ01000006.1 between the positions 3977178 and 3978784 in the reverse strand. This gene contains 4 exons, with a selenocysteine in the first one.
The structure of the gene, analyzed with the Exonerate file, is described below:

SECISearch3 has predicted 2 different SECIS elements, one of grade A and one of grade B. Even though they both are found in 3'UTR of the gene, the one with best score has been chosen.
Seblastian has predicted the selenoprotein but it has predicted two additional exons, one in the 5' and other in the 3'.
The prediction made in scaffold JXSJ01000010.1, either it corresponds to Gpx5 or Gpx6, is found between the positions 2819476 and 2821100 in the forward strand. This gene contain 4 exons, with a selenocysteine in the first one.
The structure of the gene, analyzed with the exonerate file, is described below:

One SECIS element, with grade A, has been identified in the 3'UTR of the gene. Seblastian has also predicted this protein but with an additional exon.
Gpx7 is a cysteine containing homologue, and has been aligned in scaffold JXSJ01000525.1 between the positions 179948 and 181053 in the forward strand. This protein has 2 exons, without any selenocysteine.
The structure of the gene, analyzed with the Exonerate file, is described below:
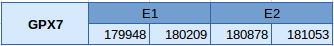
Seblastian has not predicted any selenoprotein because, as mentioned above, it is a cysteine homologue. For the same reason, no SECIS elements have been identified.
Gpx8 is also a cysteine containing homologue, and has been found in scaffold JXSJ01000013.1 between the positions 1301222 and 1302748 in the reverse strand. This protein contains 3 exons.
The structure of the gene, analyzed with the Exonerate file, is described below:

Seblastian has not predicted any selenoprotein because, as mentioned above, it is a cysteine homologue. For the same reason, no SECIS elements have been identified.
Iodothyronine deiodinase (DIO)
DIO1 has been found in scaffold JXSJ01000014.1 from position 2396940 to 2398257, in the forward strand. This protein has 3 exons, and the selenocysteine is located in the second one.
The structure of the gene, analyzed with the Exonerate file, is described below:

Seblastian has predicted one additional exon, but it still predicts the selenocysteine in the second one. Three SECIS elements have been predicted, and all of them are located in 3'UTR. However, we have selected the one with better score.
DIO2 has been predicted in scaffold JXSJ01000082.1 between positions 1146249 and 1148716 in the forward strand. This protein has 3 exons, and the selenocysteine is located in the third one.
The structure of the gene, analyzed with the Exonerate file, is described below:

Seblastian has not been able to predict any selenoprotein, and no SECIS elements have been predicted.
DIO3a is located between the position 1651922 and 1652713 in the forward strand, in scaffold JXSJ01000029.1. This protein has only one exon.
The structure of the gene, analyzed with the Exonerate file, is described below:

Seblastian has also correctly predicted this selenoprotein, and two SECIS elements have been identified. Only one of them is in the forward strand, so we have chosen this one as the best one.
DIO3b is located in scaffold JXSJ01000021.1 between the positions 1252836 and 1253576 in the reverse strand. This protein has only one exon.
The structure of the gene, analyzed with the Exonerate file, is described below:

Seblastian has also predicted this selenoprotein with only one exon, and 3 SECIS elements have been identified in 3'UTR. However, we have only selected the one with best score.
Methionine sulfoxide reductase A (MsrA)
There are two different MsrA subfamilies in SelenoDB whose names are not specified. For this reason, we have called them MsrA (1) and MsrA (2), following the order they appeared in the zebrafish SelenoDB.
The MsrA (1) protein is located in the scaffold JXSJ01000021.1 between positions 792956 and 823482 in the forward strand. It is a cysteine homologue since the gene contains 6 exons, without any selenocysteine.
The structure of the gene was analyzed with the Exonerate and it is described below:
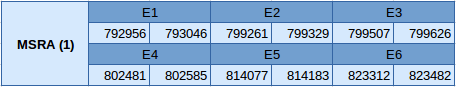
Eventhough Seblastian has not predicted any selenoprotein because it is not a selenoprotein, one SECIS element of grade B has been predicted in the in the 3'UTR region in the forward strand.
The MsrA (2) protein is in the scaffold JXSJ01000913.1. The gene is located between positions 69102 and 86182 in the reverse strand. This gene contains 5 exons, without any selenocysteine.
The structure of the gene, analyzed with the Exonerate file, is described below:

No SECIS element has been predicted and because it is not a selenoprotein, Seblastian has not predicted it.
Selenophosphate synthase (SEPHS)
There are two different SEPHS subfamilies in SelenoDB, SEPHS2 and another one whose name is not specified (appears in this database as "none"). For that reason, we have called it SEPHS (1) following the order they appeared in the zebrafish SelenoDB.
The SEPHS (1) protein is located in the scaffold JXSJ01000007.1 between positions 6480332 and 6483728 in the reverse strand. This gene contains 9 exons, without any selenocysteine.
We have analyzed the structure of the gene with the Exonerate and the results obtained are the following ones:
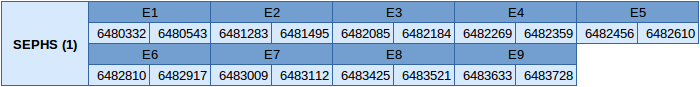
In this protein one SECIS element of grade B has been predicted. This SECIS element is in the reverse strand in the 3'UTR. Since this is not a selenoprotein, Seblastian has not been able to predict any selenoprotein.
The SEPHS2 protein is in the scaffold JXSJ01000927.1 between positions 74199 and 78967 in the forward strand. This gene contains 8 exons, with a selenocysteine in the first exon.
We have analyzed the structure of the gene with the Exonerate program and the results obtained are the following:
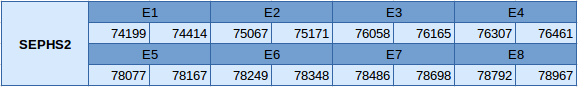
In this protein one SECIS element of grade B has been predicted. This SECIS element is located in the forward strand but is in the 5'UTR region. No Seblastian prediction has been obtained.
Selenoprotein H (SELENOH)
The protein SELENOH is found in the scaffold JXSJ0100003.1. The gene is located between positions 9698935 and 9700841 in the reverse strand. This gene contains 3 exons, with a selenocysteine in the second one.
The structure of the gene was analyzed with the Exonerate and it is described below:

In this protein two SECIS elements have been predicted, one of grade A and another of grade B. The grade A SECIS element predicted is in the reverse strand and is located in the 3'UTR.
Selenoprotein I (SELENOI)
The protein SELENOI is located in the scaffold JXSJ01000029.1 between positions 3504276 and 3512380 in the reverse strand. The gene contains 10 exons, with a selenocysteine in the last exon.
The structure of this gene was analyzed with the Exonerate and is described below:
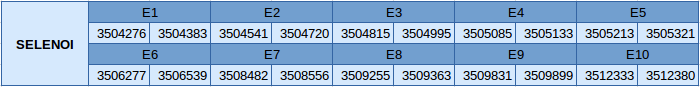
In this protein one SECIS element of grade A has been predicted. This SECIS element is in the reverse strand and is located in the 3'UTR. The selenoprotein predicted with Seblastian is very similar to our predicted protein.
Selenoprotein J (SELENOJ)
The protein SELENOJ is duplicated in the reverse strand of scaffold JXSJ01000123.1. The first copy is between positions 355390 and 357844 and the second copy is between positions 361337 and 364072. Both copies of the gene contain 9 exons, with a selenocysteine in the seventh one.
The structure of the gene, analyzed in the Exonerate file, is described below:
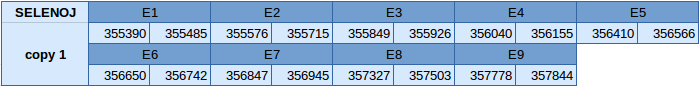
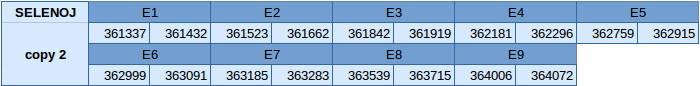
In the first copy of the gene, two SECIS elements have been obtained, one of grade A and another of grade B. The grade A SECIS element is in the 3'UTR region in the reverse strand. Seblastian has been able to predict a selenoprotein and this is very similar to our prediction.
In the second copy of the gene, only one SECIS element of grade A has been predicted in the reverse strand and is in the 3'UTR. Seblastian has been able to predict a selenoprotein and this is very similar to our prediction.
Selenoprotein K (SELENOK)
SelenoK is located in the reverse strand between the positions 791962 and 6790241. It is found in the scaffold JXSJ01000005.1 and the gene is composed by 4 exons. The selenocysteine is located in the last one.
The structure of this gene, which is described in the Exonerate file, is the following:

No Seblastian results have been predicted. However, two secis elements have been found, but just the one of grade A was located in the reverse strand and in 3'UTR.
Selenoprotein M (SELENOM)
The protein Seleno M is codified in the scaffold JXSJ01000037.1 between the positions 1024868 and 1022323 in the reverse strand. This protein has 5 exons, and the selenocysteine is located in the second one. Its gene structure is described below:

Four Secis elements have been predicted but just two of them are in the reverse strand and are located in the 3'UTR. one of them has grade A and the other grade B. Also, the Seblastian results we have obtained are similar to the ones we have predicted.
Selenoprotein N (SELENON)
The protein SELENON is located in the scaffold JXSJ01000021.1 between the positions 15153432 and 15193119 in the forward strand. This gene has 11 exons, and the selenocysteine is found in the 8th one.
The structure of the gene, analyzed with the Exonerate file, is described below:
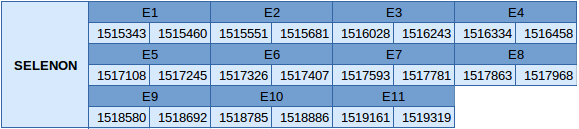
Three SECIS elements have been predicted but just one of them, which is of grade A, is in the forward strand and in 3'UTR. Also, it has been obtained a Seblastian prediction similar to the prediction we have made, but an extra exon was described by Seblastian. Therefore the selenocysteine is found in the 8th exon according to Exonerate but in the 9th according to Seblastian prediction.
Selenoprotein O (SELENOO)
There are two different SELENOO subfamilies in SelenoDB, SELENOO1 and SELENOO2.
The SELENOO1 protein is in the scaffold JXSJ01000027.1 in the forward strand. The gene location is between positions 201128 and 205951 and contains 9 exons, with a selenocysteine in the last exon.
The structure of the gene was analyzed in the Exonerate file and the results are described below:
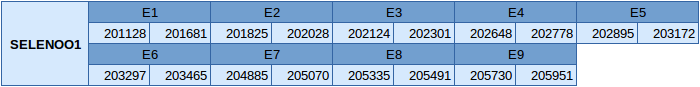
In this protein two SECIS elements have been predicted, one of grade A and another of grade B. The predicted SECIS element of grade A is in the forward strand and is located in the 3'UTR. Seblastian has been able to predict a selenoprotein and the results obtained agree with our results.
Selenoprotein P (SELENOP)
This family of selenoproteins was incompletely annotated because it is formed by two different proteins which did not have the subfamily classification annotated (it appeared as "NONE"). In order to make our work easier we have named them Seleno P(1) and Seleno P(2) following the order they appeared these queries from the Danio rerio in SelenoDB.
SelenoP1 is located in the forward strand of the scaffold JXSJ01000024.1 between the positions 2362763 and 2363619. This protein is composed by 3 exons with the selenocysteine located in the first one. The structure of this protein is described below:

Two different SECIS elements have been found in the forward strand and in 3'UTR, one of grade A and another one of grade B. Therefore, we have chosen the one with the grade A as the best. A Seblastian prediction with an extra exon was found.
The protein SelenoP2 is located in the scaffold JXSJ01000020.1 between the positions 4078401 and 4080381 in the forward strand. This gene is composed by 3 exons described in the Exonerate with a selenocysteine located in the first exon.
The structure of the gene, analyzed with the Exonerate file, is described below:

Six SECIS elements have been predicted in this protein but just two of them were found in the forward strand and in 3'UTR: one of grade A, and another of grade B. So the one with grade A was chosen as the optimal. Though, no Seblastian results have been obtained.
Selenoprotein R (MSRB)
MSRB family is formed by 4 different subfamilies: MSRB1a, MSRB1b, MSRB2 and MSRB3.
MSRB1a has been found in scaffold JXSJ01000046.1 between the positions 2417768 and 2418712 in the reverse strand. This protein has 3 exons and the selenocysteine is found in the last one. The location of the three exons are shown below:

Seblastian has also predicted this selenoprotein with the same number of exons. Three SECIS elements have been predicted but only two of them are found in the reverse strand and in 3'UTR. Since there is one with best score than the other, we have selected the first one as the best one.
MSRB1b has been predicted in scaffold JXSJ01000015.1, concretely between the position 2035871 and 2037319 in the reverse strand. This protein has also 3 exons whose positions are shown below:

Seblastian has also been able to correctly predict the selenoprotein, with the selenocysteine in the first exon. Five different SECIS elements have been predicted in this selenoprotein but only three of them are found in the reverse strand and in 3'UTR. However, only the one with best score has been considered as the best one.
MSRB2 is located between the position 365418 and 399496 in the forward strand and in the scaffold JXSJ01000042.1. This protein contains 6 exons, and the selenocysteine is located in the first one. The location of these exons is shown here:
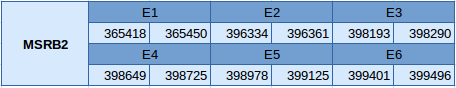
Seblastian has not been able to predict this selenoprotein, but SECISearch3 has found two different SECIS elements. Both of them are in the forward strand and in 3'UTR, but only the one with best score has been selected.
MSRB3 is a Cys-containing homologue that has been predicted in scaffold JXSJ01000328.1 between positions 132096 and 137920 in the reverse strand. This protein is formed by 6 exons, which can be observed below:
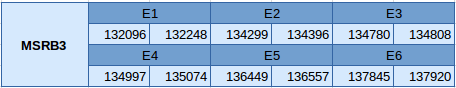
Seblastian and SECISearch3 have not been able to predict the selenoprotein or the SECIS elements.
Selenoprotein S (SELENOS)
Seleno S is located between the position 75534 and 77926 in the reverse strand. It belongs to the scaffold JXSJ01001008.1 and the gene is composed by 5 exons. The selenocysteine is located in the last one.
The structure of this gene, that is described in the Exonerate file, is the following one:

The Seblastian prediction we have obtained correlates with the final results predicted with our program. Also, a SECIS element, which is of grade A and it is located in 3'UTR of the reverse strand, has been obtained.
Selenoprotein T (SELENOT)
There are three different SELENOT proteins that belong to two different subfamilies: SELENOT1, SELENOT1b and SELENOT2.
The SELENOT1 protein is located in the scaffold JXSJ01000006.1 between positions 6154489 and 6158134 in the reverse strand. This gene contains 5 exons, with a selenocysteine in the second exon.
The structure of the gene was analyzed in the Exonerate file and the results are described below:

In this protein four SECIS elements have been predicted, one of grade A and the other three of grade B. The SECIS element of grade A predicted is located in the 3'UTR and in the reverse strand. Seblastian has been able to predict the selenoprotein and the results obtained agree with our results.
The SELENOT2 protein is in the scaffold JXSJ01000199.1 between positions 220221 and 221833. The gene is located in the forward strand and contains 6 exons with a selenocysteine in the third one.
We have analyzed the structure of the gene in the Exonerate file and the results obtained are described below:
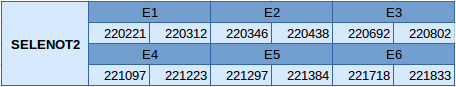
In this protein two SECIS elements have been predicted, one of grade A and another of grade B. The predicted SECIS element of grade A is in the 3'UTR and in the forward strand. The selenoprotein predicted with Seblastian is correlated with our prediction but one additional exon in the 3'UTR region is missing in the Seblastian results.
Selenoprotein U (SELENOU)
There are three different SelenoU subfamilies in SelenoDB database, SELENOU1a, SELENOU2 and SELENOU3.
The SELENOU1a protein is duplicated in the scaffold JXSJ01000036.1 in the reverse strand. The first copy of the gene is between positions 1859825 and 1861770 and contains 5 exons, with a selenocysteine in the second one. The second copy is between positions 1865654 and 1866392 and contains 4 exons, with a selenocysteine in the first exon.
We have analyzed the structure of the gene with the Exonerate program and the results are described below:


In the first copy of the gene, one SECIS element of grade A has been predicted. This SECIS element of grade A is located in the 3'UTR region in the reverse strand. The predicted selenoprotein with Seblastian is correlated with our predicted selenoprotein but one additional exon in the 5' appears in the Seblastian results.
In the second copy of the gene, two SECIS elements have been predicted, one of grade A and another of grade C. The SECIS element of grade A is in the reverse strand in the 3'UTR. The selenoprotein predicted with Seblastian is similar to our prediction, but one additional exon in 5' is present in the selenoprotein predicted by Seblastian.
The SELENOU2 protein is located in the scaffold JXSJ01000066.1 between positions 443031 and 444802 in the reverse strand. This gene contains 6 exons, without any selenocysteine.
We have analyzed the structure of the gene with the Exonerate program and the results are described below:
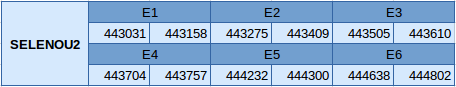
In this protein any SECIS element has been predicted and, since it is not a selenoprotein, Seblastian has not been able to predict any selenoprotein.
The SELENOU3 protein is in the scaffold JXSJ01000043.1 between positions 74604 and 82871 in the forward strand. This gene contains 6 exons without any selenocysteine.
The structure of the gene, analyzed with the Exonerate program, is described below:
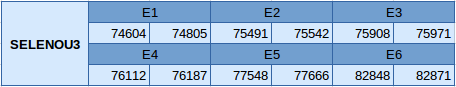
In this protein no SECIS element has been predicted and, since it is not a selenoprotein, Seblastian has not been able to predict any selenoprotein.
Selenoprotein W (SELENOW)
There are three subfamilies from Danio Rerio in SelenoDB whose names are not specified (“NONE” appeared as subfamily). In order to make our work easier we have named them as selenoprotein W1, W2 and W3 following the order in which they appeared in the Danio rerio SelenoDB web page.
Seleno W1 is a protein situated in the scaffold JXSJ01000017.1 between the positions 774172 and 774663 in the forward strand. This protein has 3 exons with the selenocysteine located in the first one. The structure of this protein is described below:
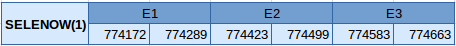
One SECIS element of grade A has been predicted in the forward strand and in 3'UTR and a Seblastian result has been obtained that correlates with the results we have obtained in the Exonerate.
SELENOW(2) is in the scaffold JXSJ01000888.1 between the positions 2038 and 253 in the forward strand. This protein has 2 exons, with the selenocysteine located in the first one. The structure of the protein is described below:
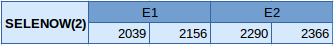
Two SECIS elements have been predicted but just one of grade A was located in the 3'UTR and forward strand, so this one was chosen as the best one. It has been also obtained a Seblastian prediction that coincide with our results.
Thioredoxin reductase (TXNRD)
There are three different TXNRD families in SelenoDB: TXNRD1 from Homo sapiens and TXNRD2 and TXNRD3 from Danio rerio.
The TXNRD2 protein is located in the scaffold JXSJ01000073.1 between positions 512657 and 531121 in the reverse strand. This gene contains 14 exons, with a selenocysteine in the last exon.
We have analyzed the gene structure with the Exonerate program, and the results obtained are the following:
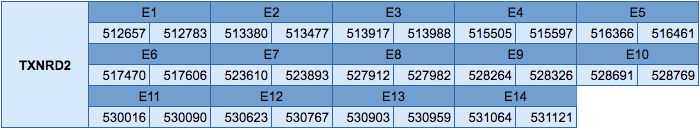
In this protein two SECIS elements have been predicted, one of grade A and another of grade B. The SECIS element of grade A is in the 3'UTR in the reverse strand. No Seblastian prediction has been obtained.
The TXNRD3 protein is located in two different scaffolds: in the scaffold JXSJ01000302.1 between positions 316058 and 326975 in the forward strand and in the scaffold JXSJ01000728.1 between positions 133825 and 143097 in the reverse strand. The gene in the first scaffold contains 15 exons, with a selenocysteine in the last exon. The gene of the second scaffold contains 14 exons, with a selenocysteine in the last exon.
We have analyzed the structure of the gene with the Exonerate and the results obtained are the following:
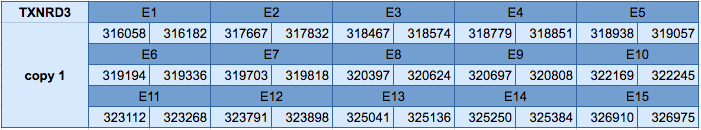
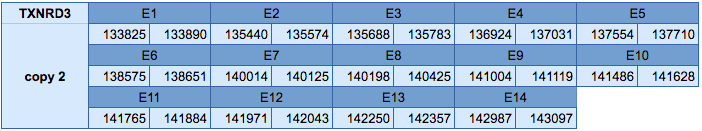
In the protein predicted in the scaffold JXSJ01000302.1 three SECIS elements have been identified, one of grade A and two of grade B. The grade A SECIS element is predicted in the forward strand in the 3'UTR. Seblastian has been able to predict a selenoprotein that is very similar to our predicted selenoprotein.
Moreover, in the protein predicted in the scaffold JXSJ01000728.1 two SECIS elements have been predicted, one of grade A and another of grade B. The SECIS element of grade A is in the reverse strand in the 3'UTR. Seblastian has been able to predict a selenoprotein that is very similar to our predicted selenoprotein.
Selenoproteins machinery analysis:
Eukaryotic elongation factor (eEFsec)
The protein eEFsec is located in the reverse strand in the scaffold JXSJ01000351.1. It is situated between the positions 76191 and 85452 in the Miichthys miiuy genome. This gene is composed by 7 exons distributed as shown below:
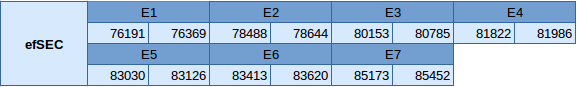
No Secis elements or Seblastian results have been predicted in this protein because it is not a selenoprotein.
Phosphoseryl-tRNA kinase (PSTK)
PTSK is situated between the positions 5173702 and 5174952 in the scaffold JXSJ01000009.1 in the forward strand.
This protein contains 3 exons. The scheme of its structure is shown below:

It has been described a SECIS element in the forward strand and in 3'UTR of these protein. Though, no Seblastian results have been obtained.
SECIS binding protein 2 (SBP2)
This family of machinery genes was not properly described in the database SelenoDB, because the subfamily queries of Danio rerio were described as "NONE". In order to make our predictions easier we have named the queries in order of appearance in the selenoDB: SBP2 (1) and SBP2 (2).
SBP2 (1) is a machinery protein situated in the scaffold JXSJ01000013.1 in the Miichthys miiuy genome. It is located between the positions 206114 and 208113 in the reverse strand. This protein is divided in 7 exons whose structure is showed below:
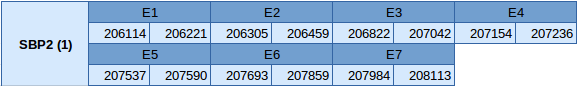
No SECIS elements or Seblastian results have been predicted in this protein.
SBP2 (2) is a protein of the group of the machinery. It is situated in the forward strand between the positions 182184 and 191334, in the scaffold JXSJ01000001.1.
The scheme of the distribution of this protein is shown below:
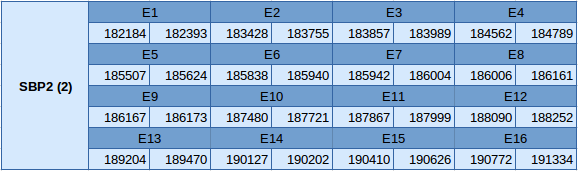
In this protein no SECIS elements or Seblastian results have been predicted.
Selenocysteine synthase (SecS)
The SECS protein is in the scaffold JXJ01000055.1 in the forward strand but is truncated. The first part of the protein contains 3 exons and the second one contains 2 exons. Any of them contains a selenocysteine.
We have analyzed the structure of the gene with the Exonerate file and we have obtained the following results:

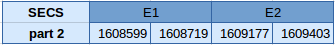
Since it is not a selenoprotein, in any of the two fragments of the protein Seblastian has been able to predict any selenoprotein and no SECIS elements have been obtained.
tRNA Sec1 associated protein 1 (SECp43)
SECp43 family contains two subfamilies whose names are not specified.
The first one (SECp43 (1)) is located in the scaffold JXSJ010005029.1 between the positions 237740 and 239453 in the reverse strand. This gene contains three exons without any selenocysteine.
The structure of the gene, analyzed with the Exonerate file, is described below:

In this protein two SECIS elements have been predicted, one of grade A in the forward strand and one of grade B in the reverse strand which correspond with the selenoprotein predicted. Seblastian result was not found.
The second one (SECp43 (2)) is located in the scaffold JXSJ01000502.1 between the positions 79222 and 84336 positive strand. This gene has eight exons, which are shown below:
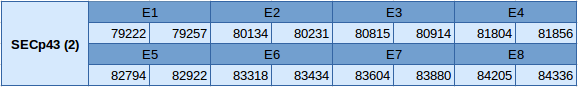
In this protein no Seblastian and SECISearch3 results were found.
Proteins related to selenium metabolism analysis:
All the proteins explained below have been predicted blasting Miichthys miiuy genome against the different protein families of Homo sapiens found in SelenoDB.
ELAV like RNA binding protein 1 (ELAVL1)
This protein is located in the scaffold JXSJ01000369.1 between the positions 274913 and 277728 in the reverse strand. This gene contains 5 exons without any selenocysteine. The structure is described below:

In this protein no SECIS elements have been predicted neither Seblastian results were found.
Elav-like family member 1 (CELF1)
The CELF1 protein is located in the scaffold JXSJ01000100.1 between positions 86195 and 107585 in the reverse strand. This gene contains 7 exons, with any selenocysteine.
The structure of the gene, analyzed with the Exonerate file, is described below:
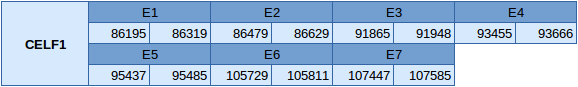
In this protein one SECIS element of grade B has been predicted. This SECIS element is in the forward strand in the 3'UTR. Seblastian has not been able to predict any selenoprotein.
Eukaryotic translation initation factor 4A3 (EIF4A3)
The EIF4A3 protein is in the scaffold JXSJ01000207.1 between positions 410532 and 414259 in the reverse strand. This gene contains 11 exons without any selenocysteine.
We have analyzed the gene structure with the Exonerate program, and the results obtained are described below:
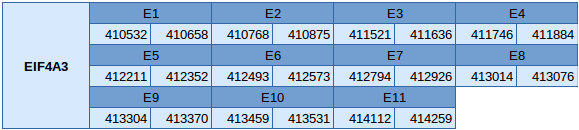
In this protein one SECIS element of grade B has been predicted. This SECIS element is in the reverse strand but is in the 5'UTR, so any suitable SECIS element can be predicted for this protein. Seblastian has not been able to predict any selenoprotein.
Exporin 1 (XPO1)
The XPO1 protein is in the scaffold JXSJ01000048.1 between positions 532824 and 546124 in the forward strand.
We have analyzed the gene structure with the Exonerate file and the results are the following ones:
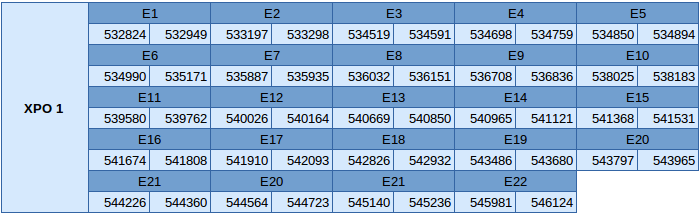
In this protein only one SECIS element of grade B has been predicted. However, this SECIS element is in the reverse strand, so any suitable SECIS element can be predicted. Seblastian has not been able to predict any selenoprotein.
Glucose-6-phophate deshydrogenase (G6PD)
The G6PD protein is located in the scaffold JXSJ01000005.1 between positions 2740552 and 2740552 in the reverse strand. This gene contains 12 exons without any selenocysteine.
The structure of the gene has been analyzed with the Exonerate file and is described below:
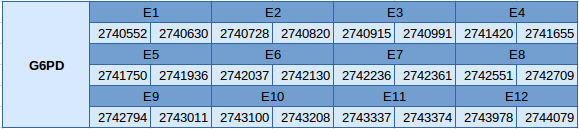
In this protein no SECIS element has been predicted and Seblastian has not predicted any selenoprotein.
Low density lipoprotein receptor-related protein 8 (LRP8)
This protein is duplicated in the same scaffold JXSJ01000652.1. The first copy of this gene is located between the position 11081 and 15068 in the forward strand. This protein has 9 exons and the selenocysteine is found in the last one. The positions of the exons are shown below:
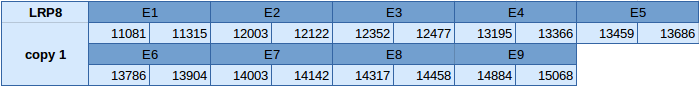
Seblastian has not predicted this selenoprotein and no SECIS element has been predicted neither.

The second copy of this protein is between the positions 31374 and 32820. This duplication is much shorter than the first copy, and it has been only predicted from amino acid 83 to 264. Therefore, this protein contains only 3 exons and the selenocysteine is also in the last one. The structure of the exons is shown below:
Seblastian has not predicted any selenoprotein, and no SECIS elements have been predicted neither.
Ribosomal protein L30 (RPL30)
This protein has been predicted in scaffold JXSJ010000353.1 between the positions 48544 and 50822 in the reverse strand. This protein has 4 exons, whose positions are shown below:

Since it is not a selenoprotein, Seblastian has not been able to predict it. Two different SECIS elements have been predicted but only one is in the reverse strand. However, this one is not found in the 3’UTR, therefore it cannot be concluded that a SECIS element has been found.
Selenocysteine lyase (SCLY)
This protein has been found in scaffold JXSJ01000121.1 between the position 196657 and 200005 in the reverse strand. This protein has 11 exons, whose positions can be observed below:
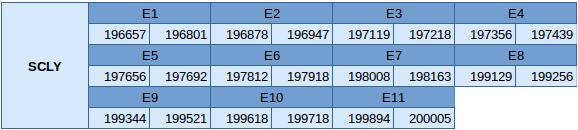
This is not a selenoprotein, therefore Seblastian has not predicted anything. Two different SECIS elements have been predicted but only one is in the reverse strand. However this one is not in the 3'UTR, so it cannot be concluded that this protein has a SECIS element.
Selenium binding protein 1(SELENBP1)
This protein has been predicted in scaffold JXSJ01000048.1 between the positions 48544 and 50822 in the reverse strand. It has 9 exons whose positions are shown below:
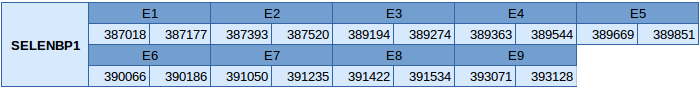
Since it is not a selenoprotein, Seblastian has not been able to predict it. Two different SECIS elements of grade B, but only one is in the reverse strand and in the 3'UTR.
Seryl-tRNA sinthetase 2 mitochondrial (SARS2)
The SARS2 protein is in the scaffold JXSJ01000068.1 in the forward strand between positions 356430 and 363543. This gene contains 14 exons.
The gene structure, analyzed with the exonerate file, is described below:
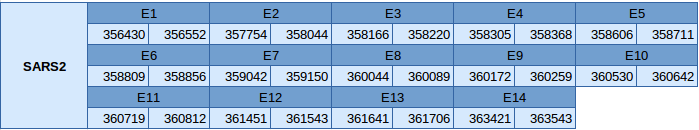
In this protein one SECIS element of grade C has been predicted. This SECIS element is in the forward strand but is in the 5'UTR region, so any suitable SECIS element has been predicted. Seblastian has not been able to predict any selenoprotein.
Tocopherol (alpha) transfer proteins (TTPA)
This protein is located in scaffold JXSJ01001106.1 between the position 61548 and 66016 in the reverse strand. This protein has 5 different exons, which are shown below:

This protein does not have any selenocysteine, therefore Seblastian has not predicted any selenoprotein. One SECIS element has been predicted but it is in the forward strand. Therefore, it cannot be concluded that this protein contains a SECIS element.
