Selenium:
Selenium is an essential element that modulates important biological processes such as immunological and oxidative stress responses, redox signalling, cellular differentiation, protein folding and modulation of the aging process1,4. It has also been studied that selenium deficit increases the risk of having severe diseases such as stroke, sepsis, autoimmune diseases or even cancer. However there is a narrow window between Selenium deficiency and toxicity, meaning that its concentration in the living organism has to be strictly regulated to guarantee the organism well being2. For this reason, studying Selenium containing proteins is crucial for preventing and treating diseases.
Selenoproteins:
The way selenium is able to perform all the functions mentioned above is by means of selenocysteine (Sec), the 21st amino acid, which is a cysteine that contains selenium instead of sulphur. Proteins having at least one selenocysteine are called selenoproteins, and the set of selenoproteins is known as the selenoproteome.
It is known that selenoproteins are present in all three the domains: bacteria, archae and eukaryote. While 45 selenoprotein subfamilies have been identified in vertebrates, humans only contain 25 selenoproteins. Aquatic organisms, whether they are fishes or plants, are the only ones containing the highest number of selenoproteins6. For example, bonny fishes have been reported to have 41 selenoprotein subfamilies4.
There are some organisms that do not contain selenoproteins because they have evolved to convert Sec into Cysteine (Cys). This event can be explained by the challenges aquatic animals had to face when they changed the aquatic habitat for the terrestrial one, in which availability of some elements, such as Selenium, was diminished6.
One of the most important features of selenoproteins is that they are encoded by a UGA codon, which normally codifies for a stop codon. However, there is a variety of cis- and trans- elements that control and regulate selenoproteins biosynthesis, without which it would be difficult to differentiate between a normal stop codon. Therefore, strict regulatory elements are needed to ensure a correct incorporation of Sec instead of a stopping protein traduction.
Selenoproteins biosynthesis:
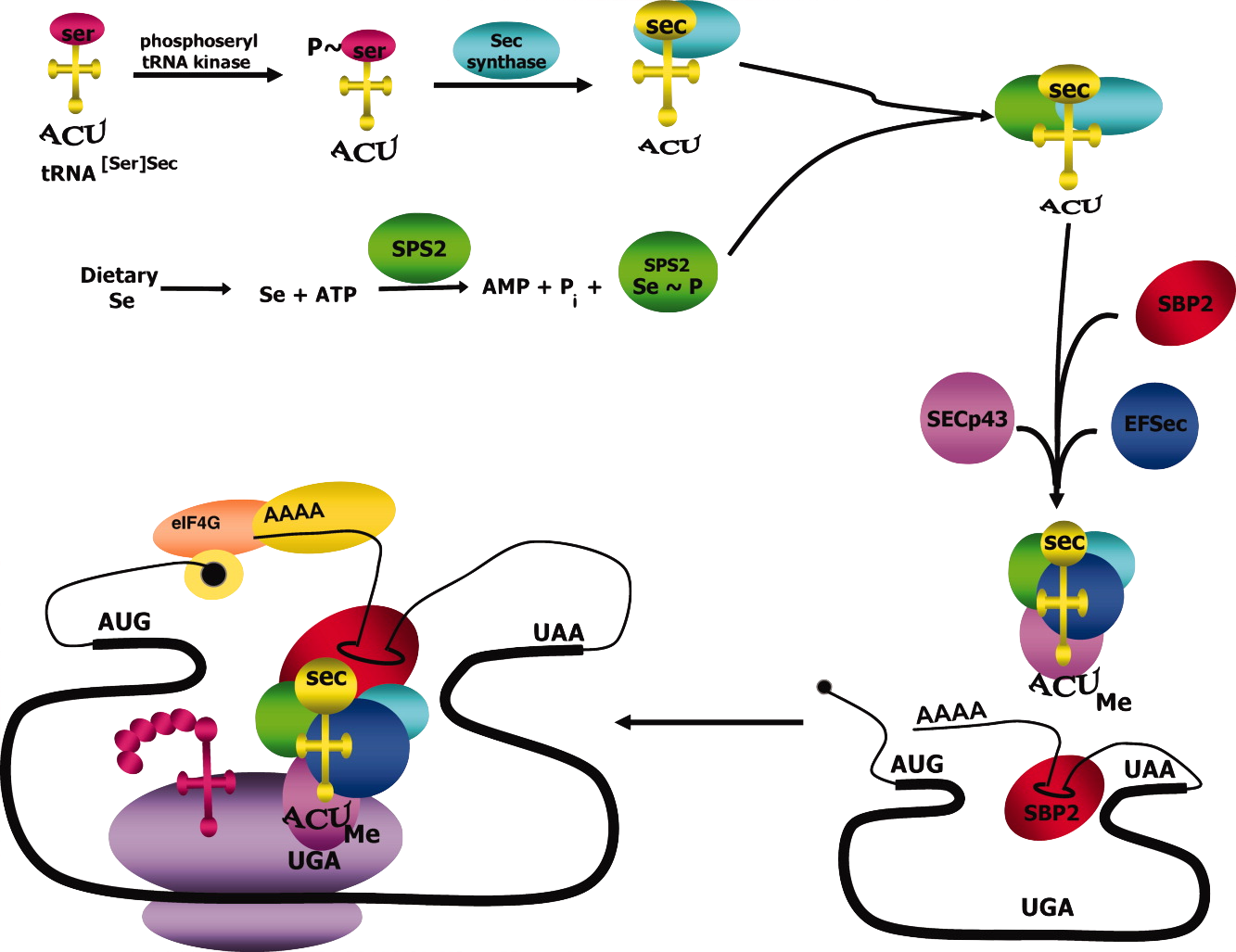
As it has been mentioned, selenoproteins byosinthesis needs to be strictly controlled in order to decode the stop codon as a Sec. There is a variety of cis- and trans- elements that control this process, without which it would be difficult to differentiate between a normal stop codon and a Sec.
Regarding cis-acting elements, which are the ones located in the mRNA sequence, the UGA codon and the Selenocystein Insertion Sequence (SECIS) element are the most important ones. SECIS is a stem-loop structure, and it is essential because it allows Sec incorporation. It is located in the 3'- untranslated region (3'-UTR) of the selenoprotein-encoding mRNA, meaning that even though it is found in a non-codifying region, it is transcripted into RNA5.
On the other hand, trans-acting elements are the ones that are not found directly in the mRNA sequence. The essential ones are SBP2, eEFsec, PSTK, SecS, Secp43, SPS1 and SPS2.
SECIS binding protein 2 (SBP2) has its binding site in the stem region of the SECIS element. When SBP2 binds to the SECIS element, SBP2 recruits and interacts with eEFsec, an elongation factor that allows incorporation of the specific Sec tRNA into the ribosome5. tRNA, suffers quite many modifications until it becomes selenocysteyl-tRNA[Ser]Sec. In eukarya domain, tRNA is aminoacylated with serine, leading to the formation of seryl-tRNA[Ser]Sec. PSTK is a kinase that phosphorilates the seryl part of the tRNA, resulting in phosphoseryl-tRNA[Ser]Sec. Later on, Selenocysteine synthase (SecS) dephosphorilates phosphoseryl-tRNA[Ser]Sec, and incorporates monoselenophosphate, which is the active form of selenium. This process leads to the generation of selenocysteyl-tRNA[Ser]Sec. After that, SecP43 methylates selenocysteyl-tRNA[Ser]Sec resulting in the shuttling of the tRNA complex between the nucleus and the cytoplasm, and therefore, enhancing selenoprotein expression7,8.
Selenophosphate synthetase 1 and 2 (SEPHS1 and SEPHS2) are the proteins in charge of synthesizing monoselenophosphate, which is the active form of selenium. SEPHS1 has been speculated to play a role in selenocysteine recycling and selenium salvage. However, SEPHS2, which is a selenoprotein, is involved in generating the active selenium donor to synthesise selenocysteine8.
Selenoproteins families:
Sel15 and Selenoprotein M (SELENOM):
The 15-kDa selenoprotein (sel15) and selenoprotein M are thioredoxin-like fold endoplasmic reticulum (ER) resident proteins.
Sel15 was identified in 1998 by experimental procedures and SELENOM was identified as an homolog of the first one, but mainly localized in the brain. Both share a common thioredoxin-like domain and contain a N-terminal consistent with their ER-location.10,12
Selenoprotein E (SELENOE):
This protein can also be called Fep15 and it is related to the other members of the selenoprotein family of 15 kDa. It is an ER-resident selenoprotein of unknown function found only in fish.10
Glutathione peroxidases (GPx):
This is the largest selenoprotein family which is spread in all three domains of life. GPxs play a wide range of physiological functions and are involved in hydrogen peroxide signaling, detoxifications of hydroperoxidases and maintenance of cellular redox homeostasis.
GPx1 is a cytosolic enzyme that catalyzes glutathione (GSH)-dependent reduction of hydrogen peroxide into water. It is an intracellular antioxidant, so its overexpression may lead to reductive stress and may disrupt H2O2 signalling. By degrading H2O2, high GPx1 levels have been implicated in promoting insulin secretion.
GPx2 and GPx3 are expressed just in certain tissues. GPx2 is found in the epithelium of the gastrointestinal tract and the GPx3 is secreted primarily from kidney and is the major GPx form in plasma.
GPx4 is expressed in a wide range of cell types and tissues. It is involved in the reduction of membrane-bound phospholipid and cholesterol hydroperoxides and also in the inhibition of lipid peroxidation which is considered it is primary role in most tissues. It has also been shown that it is implicated in the regulation of protein tyrosine phosphatases.
GPx6 is only found in the olfactory epithelium and during embryonic development.
GPx7 and GPx8 evolved from a GPx4-like selenoprotein prior to the separation of mammals and fishes.
All the tetrameric vertebrate GPx have specificity for hydrogen peroxide and other soluble low-molecular weight peroxidases.9,10
Iodothyronine deiodinases (DIO):
This family regulates the activation and inactivation of thyroid hormones. There are 3 different DIO subfamilies that contain selenocysteine residues in the N-terminal region: DIO1, DIO2 and DIO3. They have distinct subcellular location and tissue expression: DIO1 and DIO3 are located on the plasma membrane, but DIO2 is found in the endoplasmic reticulum.
The deiodinases possess a thioredoxin-fold and show significant intrafamily homology. All three deiodinases are selenoenzymes containing a single transmembrane domain and they form a homodimer structure.
The protein DIO3 is duplicated in all bony fishes and has been called DIO3b, which irreversibly inactivates the thyroid hormone by deiodination of the inner tyrosyl ring. All detected DIO3 genes are ironless.
DIO2 is an ER-resident protein which activates the thyroid hormone by deiodination of the outer tyrosyl ring. It has a second Sec residue located in the C-terminal region whose function in unknown. This residue does not participate in the catalytic mechanism but it is indispensable for the DIO2 functional activity.
The thyroid hormone is mainly produced in the inactive form (T4). The reaction that converts T4 in T3 (active hormone isoform) is catalyzed by DIO1 and DIO2. T3 and T4 can be inactivated by DIO3 and under specific conditions DIO1 will convert T3 in reverse T3 or T2 (the inactive isoform). Therefore, deiodinases play an important role in maintaining levels of thyroid hormones and its activity.9,10
Methionine-R-sulfoxide reductase (MSRB) and methionine-S-sulfoxide reductase (MsrA):
MSRB1 is a zinc-containing selenoprotein that was initially identified as selenoprotein R and selenoprotein X (SelX). This protein was found to work as a stereospecific methionine-R-sulfoxide reductase, which catalyzes the reparation of the R enantiomer of oxidized methionine residues in proteins. Based on it is similarity to MsrA, which catalyzes the reduction of the other isomer (R), this selenoprotein was renamed as MSRB. But this two proteins have structural differences although they have complementary functions.
In some organisms such as unicellular eukaryotes and anaerobic bacterium, MsrA has a Sec residue located in the active site while in other organisms, such as vertebrates, this residue is a Cys. It has been shown that Sec residue provides catalytic advantages in the redox-active enzymes.
The Sec-containing MSRB1 is the major MSRB in mammals and it is mainly located in the cytosol and nucleus. Two additional MSRB homologous (MSRB2 and MSRB3) contain a Cys residue instead of the Sec residue in the active site and have different cellular distributions. MSRB2 is located in the mitochondria whereas MSRB3 is found in the ER.
MsrA can catalyze the reduction of free methionine residues and also methionine when it is compressed in a protein, whereas MSRB can reduce methionine-R-sulfoxide only in proteins. In some organisms an additional MSRB was discovered, MSRB1b, which reduce free methionine-R-sulfoxide.10
Selenophosphate synthase (SEPHS):
This family of proteins is also known as SPS (1 and 2). Sec synthesis requires the enzyme selenophosphatase synthetase that is conserved in all prokaryotic and eukaryotic genomes encoding selenoproteins. SEPHS is found in many species although some functional homologs that replace Sec with Cys are common.
SEPHS is unique among the selenoproteins because it is considered a biosynthesis machinery protein that is also known as selenoprotein by itself. It catalyzes the synthesis of monoselenophosphate from selenide, ATP and water. The monoselenophosphate is the selenium donor for the synthesis of Sec. As it has been mentioned before, this family of proteins is conserved in the evolution from bacteria to human. In eukaryotes, SEPHS is generally found as selenoprotein whereas in prokaryotes some homologs which contain a Cys aligned with Sec are commonly found. Sec and Cys homologs are expected to have the same molecular function.
In vertebrates two paralogous SEPHS have been reported: SEPHS2, which is a selenoprotein, and SEPHS, which is a machinery gene since it contains a threonine instead of Sec. The conversion of these residue seems to abolish the selenophosphate synthase function.9,11
Selenoproteins W, T, H and V belong to the Rdx family and possess a thioredoxin-like fold that is characterized by a conserved Cys-XX-Sec motif. They contain a conserved amino acid sequence in the c-terminal region. The Rdx family are thiol-based selenoproteins but the exact function of these protein remains unknown.
Selenoprotein H (SELENOH):
It is a 14 kDa selenoprotein that contains a Sec residue within the cys-xx-SEc motif and has a conserved nuclear targeting RKRK motif in the N-terminal sequence. It is specifically localized in the nucleoli.
The expression of this protein is relatively low in adult tissues but higher in embryonic development. It is sensitive to dietary Se intake, as selenoW. It has a AT-hook motif which is present in the DNA binding proteins of AT-hook family. It has been described that this protein binds to sequences containing heat shock and stress response elements. Moreover, SELENOH has glutathione peroxidase activity.10
Selenoprotein I (SELENOI):
SELENOI is one of the least studied selenoproteins. It contains a CDP-alcohol phosphatidyltransferase domain that it is shown to be highly conserved. This domain is found in choline phosphotransferase (CHPT1), that catalyzes the transfer of choline to diacylglycerol from CDP-choline, and also choline/ethanolamine phosphotransferase (CEPT1) which catalyzes an analogous reaction with both choline and ethanol. SELENOI has 7 transmembrane domains that correspond to the predicted topologies of CHP1 and CEPT1.10
Selenoprotein J (SELENOJ):
It is a selenoprotein that appears restricted in actinopterygian fishes and sea urchin, and there are some Cys homologues found only in cnidarians. This protein has a potential role as a crystallin since it has significant similarity to the jellyfish J1-crystallins. It has been showed that SELENOJ and J1-crystallins have been derived from ADP-ribosylation enzymes. Therefore, in contrast with all other selenoproteins that have an enzymatic function, SELENOJ has a structural function. It has preferential expression in the eye lens in early stages of zebrafish development.14
Selenoprotein S (SELENOS) and Selenoprotein K (SELENOK):
SELENOS is a resident protein of the ER membrane. It is involved in processing and removing missfolded proteins from the ER to the cytosol where they can be polyubiquitinated and degraded through proteasome complexes.13
SELENOS and SELENOK can be assigned to the Selk/SelS family of related selenoproteins based on their topology. Both homologs contain SEc in the third or second position from the COOH terminal. So, Selenoprotein K has the same characteristics as selenoprotein S.9,10
Selenoprotein L (SELENOL):
This selenoprotein is manifested among aquatic eukaryotes such as fish, invertebrates and marine bacteria. It contains two Sec residues organized in a UxxU domine. Some proteins distantly related to SELENOL are present in some organisms, but both Sec residues are replaced by Cys. It has been experimentally confirmed that between both Sec residues of this protein a diselenide bond is formed.15
Selenoprotein N (SELENON):
It is an ER-resident transmembrane glycoprotein which is highly expressed in the embryonic development and in a lot of adult tissues, but the function in these tissues is still unknown. SELENON is required for muscle development and differentiation and it plays an important role in the maintenance of satellite cells and it is needed for regeneration of skeletal muscle tissue after stress or injury.
The ryanodine receptor (RyR) has been identified as a binding partner of Selenoprotein N. RyR forms a calcium release channel that allows the release of calcium from the sarcoplasmic reticulum during muscle contraction.9,10
Selenoprotein O (SELENOO):
It is one of the most characterized human selenoproteins but no structural or biochemical characterisation has been reported. Homologs of human Selenoprotein O have been detected in a wide variety of species.
SELENOO contains a single Sec residue located in the antepenultime region in C-terminal end. However, the majority of homologs contain a Cys residue in place of a Sec. The function of selenoprotein O and any of it is homologs is yet unknown.9,10
Selenoprotein P (SELENOP):
This selenoprotein is a secreted protein that contains about 50% of the selenium in human plasma and it is very important for transporting selenium to the brain.
It is mainly synthesized in the liver but some SELENOP mRNA expression has been found in most of tissues. Some SELENOP homologs are found predominantly in vertebrates.
This protein has multiple Sec residues, however the number of these residues variate a lot between different species of vertebrates. A part of the full protein, 3 other isoforms were found that appeared to be produced by prematuretermination at the UGA codons in Selenoprotein P mRNA.9,10
Selenoprotein T (SELENOT):
It is a selenoprotein predominantly located in the ER and Golgi and is ubiquitously expressed during embryonic development and adult tissues. It has been identified as the target of the neuropeptide pituitary adenylate cyclase activation polypeptide (PACAP). SELENOT has a role in the regulation of calcium homeostasis and neuroendocrine function. Moreover, it has been found that it is implicated in the regulation of pancreatic beta-cell function and glucose homeostasis.10
Selenoprotein U (SELENOU)
It has been shown that this family of proteins appear duplicated in bony fishes. Two proteins of this family are found in some fishes and they are called SELENOU1a and SELENOU1b. In Medaka this gene has a Sec residue while in Stickleback the Sec residue is replaced by a Cys. The function of this family is still unknown.9
Selenoprotein V (SELENOV):
It is the least conserved mammalian selenoprotein that probably arose from a duplication of the SELENOW in the placental stem. The function of SELENOV and SELENOW are not known. These two proteins have the same gene structure (6 exons with intron locations and phases conserved). It has been reported that SELENOV was lost by deletion specifically in gorilla.9,10
Selenoprotein W (SELENOW):
This familly contains small 9-kDa selenoproteins that are located in the cytosol and expressed at high levels in muscles and brain. They belong to the stress-related group of selenoproteins. They are sensitive to dietary Se intake. It has been proposed that Selenoprotein W could be involved in redox regulation of 14-3-3 protein, but the exact function remains unknown.
Several W homologs have been observed across non-mammalian vertebrates. A phylogenetic analysis distincted a new group of selenoproteins, SelenoproteinW2. This selenoprotein was found in bony fishes, frog and elephant shark what suggested that this protein is part of the ancestral vertebrate selenoproteome. In bony fishes multiple copies of SELENOEW2 were found.9,10
Thioredoxin reductases (TXNRD):
They are oxidoreductases that, together with thioredoxin, constitute the disulfide reduction system of the cell.
TXNRD1 is located in the cytosol and nucleus. The major role of TXNRD1 is the NADPH-dependent reduction of Trx1. The cytosolic thioredoxin (Trx1) is the major substrate for TXNRD1 which can also reduce a variety of low-molecular-weight compounds. Trx1 is involved in the control of many physiological processes such as antioxidant defense or apoptosis. It has been shown that TXNRD1 is also implicated in DNA repair, maintaining redox homeostasis and regulation of cell signaling. Multiple TXNRD1 isoforms have been described, at least 6 have been found in mammals.
TXNRD3 is localized into the mitochondria where is involved in reduction of mitochondrial thioredoxin (Trx2) and glutaredoxin 2 (Grx2). Multiple TXNRD3 isoforms have been described.
TXNRD1 and TXNRD3 are present in all vertebrates.9,10
Specific vertebrate selenoproteins:
Among the ancestral 28 selenoproteins, 6 are detected uniquely in vertebrates: SELENOE, GPx2, DIO2, DIO3, SELENOI, and SELENOP. Most of them (SELENOE, GPx2, SELENOI and SELENOPb) showed a less partial conservation of intron structure with their closest homologs what suggest that were generated during the whole genome duplication occurred at the root of vertebrates.9
Proteins related to selenium metabolism:
In the SelenoDB website, selenoproteins, cysteine homologues and genes related to the selenoproteins machinery are annoted. Moreover, in the Homo sapiens SelenoDB entry there are some genes that do not match any of the categories mentioned above. These genes correspond to genes related to selenium metabolism, so we decided to predict them in Miichthys miiuy. We may say that the information concerning their role in the selenium metabolism is limited.
ELAV like RNA binding protein 1 (ELAVL1)
ELAVL1 regulates the translation of SBP2 protein. It binds the 3' end of SBP2 mRNA competing with the mRNA degradation machinery for mRNA binding preventing the degradation process.16
Elav-like family member 1 (CELF1)
CELF1 perform like ELAVL1 protein as it has been said before.16
Eukaryotic translation initiation factor 4A3 (EIF4A3)
EIF4a3 is a non-essential SECIS binding protein that is not fully understood. It is an RNA dependent ATPase, ATP dependent RNA helicase, and a DEAD-box protein family protein family member that regulates Sec incorporation. This protein binds to the GPX1 SECIS element preventing SBP2 binding and inhibiting Sec incorporation in vitro. It seems to be inversly regulated by Se levels in order to regulate a cohort of non-essential selenoproteins.16
Low density lipoprotein receptor-related protein 8 (LRP8)
LRP8 is is highly homologous to the low density lipoprotein receptor (LDL-R) and to the very density lipoprotein receptor (VLDL-R). This receptors participate in signal transduction and, more specifically, LRP8 is a cell-surface receptor involved in the reelin signaling pathway. Reelin is a large neuronal signaling molecule that guide neuronal cell migration during central nervous system and interact with Lrp8 and VLDL-R.
It has been seen that mutations in LRP8 do no affect Se metabolism but it constitute a Se-uptake and delivery system essential for supplying Se to target organs such as brain, bone and testes.16
Ribosomal protein L30(RPL30)
RPL30 is another SECIS binding protein implicated in Sec incorporation, it is a part of the large ribosomal subunit. It was shown to simulate Sec incorporation in transfected cells but it is still unknown whether it is essential for Sec incorporation or whether it can bind to all SECIS elements.
Its current model activity is in the promotion of the dissociation of the SBP/SECIS complex by binding to SECIS and allowing the ribosomal elongation to continue.16
Selenocysteine lyase (SCLY)
Selenocysteine lyase (SCLY; EC 4.4.1.16) catalyzes the pyridoxal 5-prime phosphate-dependent conversion of L-selenocysteine to L-alanine and elemental selenium.17
Selenium binding protein 1 (SELENBP)
This gene encodes a member of the selenium-binding protein family. This protein may play a selenium-dependent role in ubiquitination/deubiquitination-mediated protein degradation. Alternatively spliced transcript variants encoding multiple isoforms have been observed for this gene.18
Seryl-tRNA synthetase 2 mitochondrial (SARS2)
SARS2 gene encodes the mitochondrial seryl-tRNA synthethase precursor. This enzyme catalyzes the ligation of Serine to tRNA(ser) and participates in the biosynthesis of selenocysteinyl-tRNA(sec) in mitochondria. It contains, in the N-terminal region, a tRNA binding and a core catalytic domain. It works as a homodimer which is stabilized by tRNA binding.19
Tocopherol (alpha) transfer proteins (TTPA)
TTPA is essential for early development of the vertebrate central nervous system. It has been demostrated that a Zebrafish TTPA-/- results in severe malformations.20
Miichthys miiuy:
The cientific classification of Miichthys miiuy is the next one presented:
Kingdom: Animalia
Filum: Chordata
Class: Actinopterygii
Ordrer: Perciformes
Suborder: Percoidei
Family: Sciaenidae
Genus: Miichthys
Species: M. miiuy
It is distributed from western Japan to the East China Sea.21 It lives mainly in Zhoushan Fisheries located at the estuary of the Yangtze River, a muddy areas. Therefore it is a carnivorous benthic organism.22 In China, this species represents an important aquaculture fish since 1990. However, its aquaculture has been hampered by diseases caused by pathogens and parasites to which revolutionized the study of the immune system of these fish as they are considered a good model system for both immune and sensory adaptations.
It lives in 15-100m of depth in temperate and demersal climates.22 It avoids clear waters and prefers to live in estuaries, bays and muddy shores of rivers.
According to its morphology, males can reach a total length of 70 cm.23 Like the rest of Sciaenidae fishes, M. miiuy is known for having some exceptionally large otolith that provide a highly developed auditory system. These fishes are often called croakers or drums fishes because of the sounds they produce with their swim bladder.
For more information you can visit: Viquipèdia

