Miichthys miiuy selenoproteins have been studied by comparing Danio rerio (Zebrafish) and Homo sapiens (Human) selenoproteins. We have focused our analysis on Danio rerio because it is an animal model commonly used in some research fields such as Developmental Biology, and therefore this fish's genome is the most studied and well characterised in comparison with other fishes. Regarding Homo sapiens, its proteome has also been considered because its genome has been completely sequenced.
In order to compare Zebrafish's and Human's selenoproteins with Miichthys miiuy's genome, the queries have had to be obtained from SelenoDB. In this database not all the proteins do start with Methionine, and therefore, in this cases, we can only conclude that the obtained predictions are just a part of the protein, but not the whole protein.
Fishes have a larger selenoproteome than mammals. In fact, fish selenoproteomes are among the largest known. Even though there are some differences between mammals and fishes, the same core selenoprotein families are found. One important difference between fishes and mammals is that fishes have some additional selenoproteins such as SELENOE, SELENOJ and SELENOL6. It has also been reported that fishes have some duplicated segments that result from a genome-wide duplication, and therefore, fishes often contain multiple copies of a gene that is present as a single copy in mammals. For that reason, it is expected to find some duplicated selenoproteins in our results.24
In order to conclude whether the predicted proteins are actually selenoproteins, it has been considered if after the Sec residue the rest of the sequence is conserved. On the contrary, if there was not conservation after the supposed Sec residue, that would mean that it is not a Sec residue, but a codon stop.
From that point on, the results of our analysis are going to be discussed by comparing them with the literature.
Selenoproteins:
15kDa selenoprotein (Sel15)
Sel15 was identified in 1998 by experimental procedures. It contains a thioredoxin-like domain and an N-terminal region consistent with its ER-location.
Once the analysis of the results has been completed, we have been able to predict this protein in Miichthys miiuy's genome. This selenoprotein contains the Sec residue and a SECIS element in 3'UTR of the gene that corresponds with the SECIS element predicted by Seblastian. Since this protein is not fully annotated in SelenoDB, the first exon is missing in our prediction. Therefore, the prediction made by Seblastian is, in this case, more complete and informative.
Fish selenoprotein 15 (SELENOE)
This protein can be also called Fep15 and it is an ER-resident selenoprotein of unknown function.10 It is related to members of the family of 15 kDa selenoproteins (Sel15) and is absent in mammals, so it can be detected only in fishes. While other members of the family contain both Sec and Cys residues, SELENOE only has Sec.6 This assumption coincides with our results because as it can be seen in our prediction, it only contains a Sec residue.
After analysing our results we have been able to predict this selenoprotein in Miichthys miiuy. Seblastian has also predicted this selenoprotein but it has taken SELENOM as the query. We hypothesize that Seblastian is doing this because SELENOM and SELENOE are really similar and Seblastian may not have the query corresponding to SELENOE. However, when we have compared the results made by Seblastian with the ours, we have observed that the results are really similar. Nevertheless, since we have used the SELENOE query from SelenoDB, we think that our results are more accurate and reliable than the Seblastian ones. According to SECIS elements, one of grade A has been identified and it coincides with the one predicted by Seblastian. All in all, it can be confirmed that Miichthys miiuy has SELENOE protein in its genome.
Glutathione peroxidase (GPx)
Glutathione peroxidases (GPx) are the largest selenoproteins family which are spread in all three domains of life. They play a wide range of physiological functions and are involved in hydrogen peroxide signaling, detoxifications of hydroperoxidases and maintenance of cellular redox homeostasis. 9,10.
In Zebrafish GPx1a, GPx1b, GPx2, GPx3a, GPx3b, GPx4a, GPx4b, GPx7 and GPx8 have been described, while GPx5 and GPx6 are absent. For this reason, these two queries have been obtained from Homo sapiens SelenoDB in order to see if Miichthys miiuy have them in its genome.
Since GPx is a really big family and a lot of scaffolds with a good E-value appear when these selenoproteins are blasted, we have had to decide which scaffold has to be used to analyze each query. First of all, we took into consideration in which scaffold the alignment length was the largest, but we also considered not to repeat scaffold for the different proteins of the same family.
Considering all this requirements, every query has been analyzed in the scaffold presented in the table, except GPx4a that was firstly analyzed in scaffold JXSJ01000006.1, and GPx4b that was firstly analyzed in scaffold JXSJ01000014.1. Then, we wanted to check if that was correctly done, and we created a phylogenetic tree with phylogeny.fr in order to see if every predicted protein was close to its query. The phylogenetic tree is shown below:
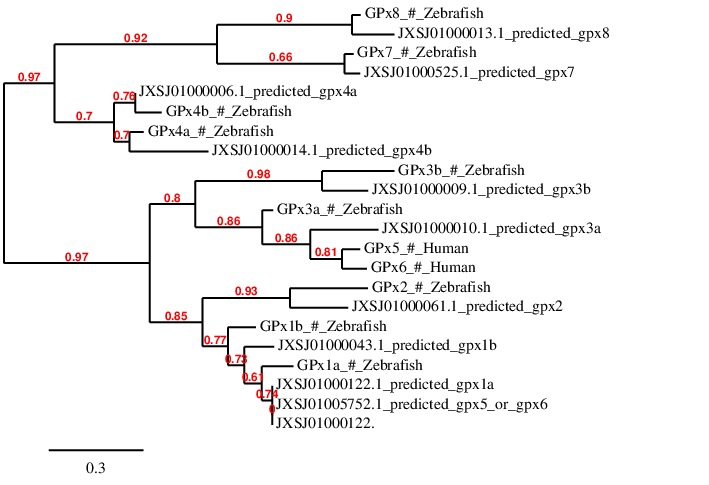
As it can be observed, the query corresponding to GPx4a is closer to scaffold JXSJ01000014.1 than the query corresponding to GPx4b, while GPx4b is closer to scaffold JXSJ01000006.1 than GPx4a. That is the reason why we have finally analyzed Gpx4a in scaffold JXSJ01000014.1 and GPx4b in scaffold JXSJ01000006.1.
Regarding GPx4a, Seblastian has not predicted the same selenoprotein than our program: the exons positions are not the same ones than ours. We hypothesize that the differences in the gene structure of both predictions are due to the fact that the query we have chosen does not start by Methionine, and therefore a fragment of the protein is missing. The predicted SECIS element is the same one that has been predicted by Seblastian. Taking into account all this information, we can conclude that Miichthys miiuy contains GPx4a in its genome.
Concerning GPx4b, Seblastian has predicted two additional exons, one in 5' and the other one in 3'. We hypothesize that the reasons are the same ones described for GPx4a, because the GPx4b query does not start by Methionine. The predicted SECIS element is the same one that has been predicted by Seblastian. Therefore, we can conclude that GPx4b is present in Miichthys miiuy's genome.
Another limitation that has had to be faced is how to identify the scaffold corresponding to the queries of GPx5 and GPx6. We found that GPx5 and GPx6 have the same two scaffolds (JXSJ01000122.1 and JXSJ01005752.1) as the best ones (as it can be observed in the blast files in the results table), and the gene is located in the same positions of both scaffolds. Moreover, the positions of the scaffold JXSJ01000122.1 in which we find these two proteins have already been used for query GPx1a. Thus, the only scaffolds that we could use are JXSJ01005752.1 for GPx5 and JXSJ01005752.1 or JXSJ01002762.1 for GPx6.
Then, we have proceeded to predict GPx5 by running our program in scaffold JXSJ01005752.1, and as it can be seen in the alignment below, no selenoprotein is found in this scaffold. We have also ran our program to align the query GPx6 in the scaffolds JXSJ01005752.1 and JXSJ01002762.1. In the first one, we have obtained the same alignment than with GPx5, and in the second scaffold no selenoprotein could be predicted due to a computational error (the Exonerate does not work for this protein, even though we have tried to align the protein step by step, without using our program).
In the phylogenetic tree we can observe that GPx5 and GPx6 are very close one another and both of them are closer to the prediction in scaffold JXSJ01000010.1, where we have predicted GPx3a, than to the scaffold in which GPx5 and GPx6 have been predicted. Taking into account all the information mentioned above and the phylogenetic tree, it is suggested that, since GPx5 and GPx6 are always predicted in the same positions of the same scaffolds, Miichthys miiuy has just one of them, GPx5 or GPx6. One of them, whichever Miichthys miiuy has, is probably located in scaffold JXSJ01000010.1. The problem is that GPx5 or GPx6 is aligned in the same positions that we had previously predicted GPx3a. That would mean that GPx3a is not present in Miichthys miiuy>'s genome. This idea is supported by the phylogenetic tree, in which GPx3a is not close to any of the scaffolds included in the tree.
The GPx5 query from Human, is a Cys-containing homologous protein, while GPx6 contains a Sec residue. As we can observe in the results table, Miichthys miiuy has a Sec in the prediction from scaffold JXSJ01000010.1. That means that, if Miichthys miiuy had GPx5, this would have evolved from a Sec (in Miichthys miiuy) to a Cys (in Homo sapiens). However if Miichthys miiuy had GPx6, the Sec would have conserved in humans.
Since our results are confusing and the phylogenetic tree does not help us to solve these confusions, we cannot confirm whether Miichthys miiuy has GPx5 or GPx6. However, in the results table we have uploaded the files corresponding to Gpx5 prediction because this prediction is a bit larger than Gpx6 prediction in scaffold JXSJ01000010.1, and therefore, it is more informative.
Even though selenoprotein GPx3b has been correctly aligned in Miichthys miiuy's genome, Seblastian has not predicted any selenoprotein and SECIS element. Even though in the T_coffee alignment a Sec appears in Miichthys miiuy's genome, the fact that Seblastian has not been able to predict any selenoprotein and that no SECIS element has been found, makes us doubt about the reliability of our results. Therefore, we cannot definitely conclude that Miichthys miiuy has GPx3b in its genome.
Regarding GPx1a it has been predicted in scaffold JXSJ01000122.1 but as we can observe in the phylogenetic tree, it is as close to this scaffold as to scaffold JXSJ01005752.1. That means that even though we have determined the prediction in scaffold JXSJ01000122.1 as the best one, it is important to consider that GPx1a could also be found in scaffold JXSJ01005752.1.
Seblastian has predicted this selenoprotein with the same exons positions, and the SECIS element is the same one than the one predicted by Seblastian. This information allows us to conclude that Miichthys miiuy has GPx1a in its genome.
Concerning Gpx1b, it has been found in Miichthys miiuy's genome containing a Sec residue. Seblastian has also correctly predicted this protein and the SECIS element. Therefore, we can conclude that Miichthys miiuy has GPx1b in its genome.
GPx2 has been found in Miichthys miiuy's genome. It contains a Sec residue, and Seblastian has been able to predict this selenoprotein with the same gene structure than our program. The predicted SECIS element is the same one than the one predicted by Seblastian. Therefore, we can conclude that Miichthys miiuy has Gpx2 in its genome.
Regarding GPx7 and GPx8 they are Cys-containing homologous because they do not contain a Sec in their sequence. For that reason, Seblastian has not predicted any selenoprotein because both of them are not selenoproteins. No SECIS elements have been predicted neither. Since we have obtained correct alignments of both proteins, we can conclude that both of them are present in Miichthys miiuy.
In order to definitely confirm that the proteins we have only predicted a fragment of them are truly found in the genome of Miichthys miiuy, we suggest that a complete query should be taken from another database in order to compare the new prediction results with the Seblastian ones.
Iodothyronine deiodinase (DIO)
This family regulates the activation and inactivation of thyroid hormones. There are 3 different DIO subfamilies that contain selenocysteine residues in the N-terminal region: DIO1, DIO2 and DIO3.
It is known that DIO2 has a second Sec residue located in the C-terminal region but its function is unknown. This residue does not participate in the catalytic mechanism and it is indispensable for the DIO2 functional activity. The reaction that converts T4 in T3, the active thyroid hormone isoform, is catalyzed by DIO1 and DIO2. T3 and T4 can be inactivated by DIO3 and under specific conditions DIO1 will convert T3 in reverse T3 or T2 (the inactive isoform). Therefore, Deiodinases play an important role in maintaining levels of thyroid hormones and its activity.9,10
Since in Zebrafish DIO family is a family formed by 4 different subfamilies, we have considered the same things than in Gpx family: the alignment length of each scaffold and the fact of not repeating any scaffold in the whole family. Then, we have also checked whether we have chosen the correct scaffold for each query by doing a phylogenetic tree that can be seen below:
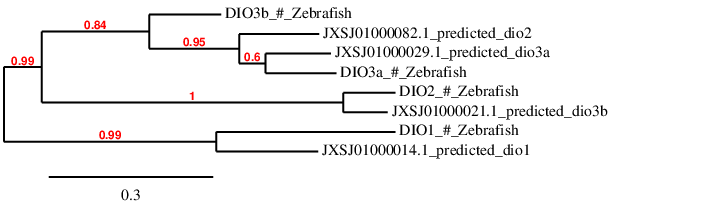
As it can be observed, according to the phylogenetic tree, the prediction of DIO1 in scaffold JXSJ01000014.1 is similar to its query DIO1, and the prediction of DIO3a in scaffold JXSJ01000029.1 is also similar to its query DIO3a. Therefore, we can consider that these two protein subfamilies have been correctly assigned to their scaffold.
Regarding DIO1, we have found it in Miichthys miiuy's genome with a Sec resdiue. Seblastian has predicted an additional exon in 3' and moreover, it has predicted an additional fragment of the first exon, including Metionine as the first amino acid. Our prediction does not contain this amino acid because the query from SelenoDB is not complete. Seblastian has also predicted two SECIS elements of grade A, as well as SECISearch3, that has predicted these two and one more. Even though we have predicted a shorter form of this selenoprotein, we can conclude that Miichthys miiuy has part of DIO1 in its genome.
Concerning DIO3a, it has been found in Miichthys miiuy's genome with a Sec residue. Seblastian has predicted the same selenoprotein than our program, but with an additional part of the exon. We hypothesize that this is due to the missing part of the query from SelenoDB. The predicted SECIS element is the same one than the one predicted by Seblastian. Therefore, we can conclude that Miichthys miiuy contains DIO3a in its genome.
However, when we look at query DIO3b in the phylogenetic tree it is closer to the prediction in the scaffold JXSJ01000082.1 than to the scaffold JXSJ01000021.1, which is the one we have used to predict DIO3b. The same happens to query DIO2, which is closer to the prediction obtained in the scaffold JXSJ01000021.1 than to the prediction we obtained in the scaffold JXSJ01000082.1. That made us think that query DIO3b should be predicted in scaffold JXSJ01000082.1 and the query DIO2 should be predicted in the scaffold JXSJ01000021.1. We have proceed to do so and we have obtained the alignment of DIO2 in scaffold JXSJ01000021.1 and the alignment of DIO3b in scaffold JXSJ01000082.1.
As it can be observed in the T_coffee alignment, the prediction of DIO2 in scaffold JXSJ010000021.1 is much worse than in scaffold JXSJ01000082.1, and the prediction of DIO3b is also worse in scaffold JXSJ01000082.1 than in scaffold JXSJ01000021.1. For that reason, we have decided to trust more in our alignments than in the phylogenetic tree, and therefore, we have continued our analysis considering that DIO2 is found in scaffold JXSJ01000082.1 and that DIO3b is found in scaffold JXSJ01000021.1.
Moreover, if we look at the blast files, in the one corresponding to DIO2, the scaffold JXSJ01000082.1 has a better E-value than scaffold JXSJ01000021.1, and in the blast corresponding to DIO3b the scaffold JXSJ01000021.1 has a better E-value than JXSJ01000082.1
Therefore, in order to decide which scaffold to choose for each query, we have taken into consideration these two things: The quality of the T_coffee alignment and the E-values of the scaffolds. This two aspects have been the ones that have made us take the decision of choosing scaffold JXSJ01000082.1 for DIO2 and the scaffold JXSJ01000021.1 for DIO3b.
Seblastian has not predicted DIO2, therefore the fact of not having the Seblastian evidences that Miichthys miiuy contains DIO2, we cannot definitely conclude that Miichthys miiuy has DIO2 in its genome.
Regarding DIO3b, Seblastian has predicted it, and therefore that supports our hypothesis that DIO3b is definitely found in Miichthys miiuy's genome. As it has been reported in literature, DIO3 is duplicated in all bony fishes and this duplication has been named as DIO3b.9,10
Thus, our results totally correlate with this assumption.
Methionine sulfoxide reductase A (MsrA)
MsrA can catalyze the reduction of free methionine residues, but it can also reduce methionines that are present in proteins. In some organisms such as unicellular eukaryotes and anaerobic bacterium, MsrA has a Sec residue located in the active site while in other organisms, such as vertebrates, this residue is a Cys. It has been shown that Sec residue provides catalytic advantages in the redox-active enzymes.10
MsrA (1) is a Cys-containing homologous protein which has been found in Miichthys miiuy. Since it does not have a Sec residue, Seblastian has not predicted any selenoprotein and any SECIS element.
MsrA (2) is also a Cys-containing homologous protein that has also been found in Miichthys miiuy's genome. In this case, Seblastian has not predicted any selenoprotein, but a SECIS element of grade B has been identified. We hypothesize that this SECIS element, which does not have a really good score, does not correspond to this protein and it may have been a software error.
In order to check whether the predicted selenoprotein is similar to the query, we have done a phylogenetic tree and we have obtained the following results:
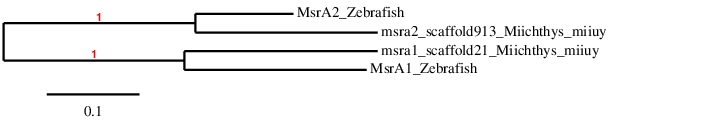
As we can observe, each query has been correctly assigned to its scaffold. For that reason and also for the fact that we have obtained two good alignments for both queries, we can conclude that Miichthys miiuy has MsrA (1) and MsrA(2) in its genome. Moreover, our results agree with the literature because, as it has been previously reported, all vertebrates have a Cys residue in this protein.
Selenophosphate synthase (SEPHS)
This family is also known as selenophosphatase synthetase (SPS1) and (SPS2). Sec synthesis requires this protein and that is the reason why it is conserved in all prokaryotic and eukaryotic genomes encoding selenoproteins. SEPHS is found in many species although some functional homologs that replace the Sec site with cysteine (Cys) are common. Sec and Cys homologs are expected to have the same molecular function.
In eukaryotes, SEPHS, apart from being a machinery protein, it is also a selenoprotein by itself. In vertebrates two paralogous SEPHS have been reported: SEPHS2, which is a selenoprotein, and SEPHS 1, which is a machinery gene since carries a threonine instead of Sec. The conversion of these residue seems to abolish the selenophosphate synthase function. Since Miichthys miiuy is a vertebrate it is expected to find SEPHS2 with a Sec residue, and SEPHS (1) with a threonine instead of a Sec.9,11
SEPHS(1) is a Cys-containing homologous. Since it is not a selenoprotein per se Seblastian has not predicted it. However, one SECIS element has been identified. We hypothesize that since SEPHS(1) and SEPHS2 are paralogous proteins, SEPHS(1) may have evolved losing the Sec residue but it may have conserved the SECIS element. We have obtained a really good alignment in Miichthys miiuy's genome, and this is a proof that Miichthys miiuy contains SEPHS(1) in its genome.
Regarding SEPHS2, it contains a Sec residue. We have correctly predicted this protein in the genome of Miichthys miiuy but when we have started to analyse its results, we have realized that in the Exonerate file we have obtained two results. The first result contains all the exons except the first one, that is in the second result. We think that in the Exonerate file appears as different results because the region between these two results cannot be detected by Exonerate, and therefore, this program consider them as different genes, instead of different exons of the same gene.
For that reason, we have used the Genewise program to obtain a predicted protein with more information. We have compared the predicted protein by the Exonerate program and by the Genewise making a T_coffee and we observe that the missed region in the Exonerate file is present in the Genewise file, so we have decided to use the Genewise file to do the rest of the analysis.
Once we have assigned a scaffold to each query, in order to check whether the predicted proteins are similar to the queries we have done a phylogenetic tree and we have obtained the following results:

It has to be considered that the name of the query corresponding to SEPHS(1) has appeared cut in the phylogenetic tree. In this phylogenetic tree we observe that our predicted proteins are crossed over between them, that is to say, SEPHS2 query is aligned with the prediction made in the scaffold JXSJ01000007.1 and our SEPHS(1) query is better aligned with the prediction made in scaffold JXSJ01000927.1.
Observing this result, we have tried to predict the SEPHS(1) protein in the scaffold JXSJ01000927.1 and the SEPHS2 protein in the scaffold JXSJ01000007.1. As it can be observed, these two alignments are worse than the ones we had obtained before. Moreover, when we blast Miichthys miiuy>'s genome against the SEPHS(1) protein of Danio rerio the scaffold JXSJ01000007.1 has a better E-value and T_coffee score than the other scaffold, and vice versa in SEPHS2. Taking this into account, we confirm that our results, in this case, are more reliable than the phylogenetic tree information.
To correlate our results with the literature, we have aligned our predictions corresponding to SEPHS(1) and SEPHS2 in order to check whether the Sec residue from SEPHS2 aligns with a Thr from SEPHS(1). As it can be observed, our results totally agree with literature, and therefore it confirms that our predictions are, indeed, SEPHS(1) and SEPHS2.
One SECIS element has been predicted by SECISearch3 but Seblastian has not been able to predict SEPHS2, and this was not expected because SEPHS2 is a selenoprotein. In this particular case, these results do not make us doubt about the presence of this protein in Miichthys miiuy because we have already confirmed that Miichthys miiuy contains SEPHS2.
All in all, these results confirm that the SEPHS(1) and SEPHS2 proteins are present in the Miichthys miiuy's genome.
Selenoprotein H (SELENOH)
SELENOH is a 14 kDa selenoprotein that contains a Sec residue within the Cys-xx-Sec motif. The expression of this protein is relatively low in adult tissues but higher at embryonic development. It has a AT-hook motif which is present in the DNA binding proteins of AT-hook family. It has been described that this protein binds to sequences containing heat shock and stress response elements. Moreover, SELENOH has glutathione peroxidase activity.10
After analysing our results, we have been able to predict SELENOH in Miichthys miiuy's genome. Seblastian has also predicted this protein but with one exon less in 3'. Therefore, in this case, our result is more informative.
The predicted SECIS element, which has grade A, is the same one than the predicted by Seblastian. Moreover, as it can be seen in the T_coffee alignment, the predicted protein contains the Sec residue within the Cys-xx-Sec motif, as it has been previously described in the literature. This totally confirms that Miichthys miiuy has SELENOH in its genome.
Selenoprotein I (SELENOI)
SELENOI is one of the less studied selenoproteins. It has 7 transmembrane domains that correspond to the predicted topologies of CHP1 (choline phosphotransferase) and CEPT1 (choline/ethanolamine phosphotransferase).10
Observing our results, we have been able to predict SELENOI in Miichthys miiuy's genome. Seblastian has predicted the same selenoprotein and SECIS element even though the query we have used is not complete. That could be explained by the fact that this selenoprotein is not fully studied and therefore, its sequence may not be complete in any database. Considering this results, we can conclude that Miichthys miiuy has SELENOI in its genome.
Selenoprotein J (SELENOJ)
It is a selenoprotein that appears restricted in actinopterygian fishes and sea urchin. Since Miichthys miiuy is an actinopterygian, this protein is expected to be found in this organism. It has been showed that SELENOJ and J1-crystallins have been derived from ADP-ribosylation enzymes. Therefore, in contrast with all other selenoproteins that have an enzymatic function, SELENOJ has a structural function. It has preferential expression in the eye lens in early stages of zebrafish development.14
As it has been described in the results, SELENOJ has been found duplicated in different regions of the same scaffold JXSJ01000123.1. To reject that this observed duplication is an assembly problem, a T_coffee with the two predicted proteins, and another T_coffee with the nucleotides codifying for both predictions have been done. As it can be seen, the nucleotides as well as the amino acids are similar but not identical. If they were identical, the probability that it was a duplication would be minimal because when a gene is duplicated it normally changes some nucleotides. These results show that they are actually a duplication and not an assembly problem. Therefore, we can conclude that this protein is duplicated in Miichthys miiuy's genome.
The first and the second copy of SELENOJ have been predicted by Seblastian with the same number and positions of the exons, and also the SECIS elements predicted by Seblastian are the same ones than the ones predicted by SECISearch3. This helps us to conclude that Miichthys miiuy contains both copies of SELENOJ in its genome.
Selenoprotein K (SELENOK)
SELENOS and SELENOK can be assigned to the Selk/SelS family of related selenoproteins based on their topology. This family is involved in processing and removing misfolded proteins from the ER to the cytosol where they can be polyubiquitinated and degraded through proteasome complexes.13 Both homologs contain Sec in the third or second position from the COOH terminal. So, Selenoprotein K has the same characteristics as selenoprotein S.9,10
SELENOK has been predicted in Miichthys miiuy genome with a Sec residue. However, Seblastian has not been able to predict it, but one SECIS element has been identified as the best one. We suggest that Seblastian may not have this query in its database and that is the reason why it has not predicted it. A very good alignment has been obtained and moreover, in correlation with the literature, it contains the Sec residue in the third position from the COOH terminal. Even though Seblastian has not predicted it, all these evidences show that Miichthys miiuy contains SELENOK in its genome.
Selenoprotein L (SELENOL)
It is a selenoprotein that is found among aquatic eukaryotes such as fishes. It contains two Sec residues organized in a UxxU domine. Some proteins distantly related to SELENOL are present in some organisms, but both Sec residues are replaced by Cys. It has been experimentally confirmed that between both Sec residues of this protein a diselenide bond is formed.15
Seleno L has been predicted in the scaffold JXSJ01000513.1 between the positions 156452 and 150743 in the reverse strand. This gene is composed by 6 exons, without any selenocysteine.The structure of this gene, predicted by Exonerate, is shown below:
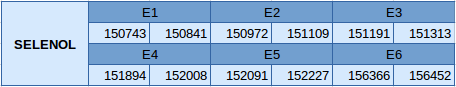
In this protein no SECIS elements were predicted and no Seblastian results have been obtained neither.
As it can be observed in the T_coffee alignment, Sec residues from the query are not correctly aligned with the Sec residues from our prediction, and moreover, the two Sec residues found in the predicted protein are not organized in a UxxU domain. Therefore, we cannot definitely conclude that SELENOL is found in Miichthys miiuy's genome.
Selenoprotein M (SELENOM)
selenoprotein M is a thioredoxin-like fold endoplasmic reticulum (ER) resident protein. SELENOM was identified as an homolog of the Sel15, but is mainly localized in the brain. Both share a common thioredoxin-like domain and contain a N-terminal consistent with their ER-location.10,12
SELENOM has been predicted in Miichthys miiuy's genome with a Sec residue. Seblastian has been able to predict it and one SECIS element has been identified as the best one. Since the alignment is good, we can conclude that Miichthys miiuy contains SELENOM in its genome.
Selenoprotein N (SELENON)
It is an ER-resident transmembrane glycoprotein which is highly expressed in the embryonic development and in a lot of adult tissues, but the function in these tissues are still unknown.9,10
SELENON has been predicted in the genome of Miichthys miiuy with a Sec residue. Seblastian has been able to predict it but with one additional exon in 5' region. The SECIS element predicted by Seblastian is the same that the SECISearch3 predicted one. This protein has a Sec residue and it is correctly aligned with the queries' Sec residue. Therefore, we can conclude that Miichthys miiuy has SELENON in its genome.
Selenoprotein O (SELENOO)
It is one of the most characterized human selenoproteins but no structural or biochemical characterisation has been reported. Homologs of human Selenoprotein O have been detected in a wide variety of species. SELENOO contains a single Sec residue located in the antepenultimate region in C-terminal end. However, the majority of homologs contains a Cys residue in place of a Sec. 9,10.
SELENOO1 has been correctly aligned in Miichthys miiuy's genome. Seblastian has predicted the selenoprotein as well as the SECIS element. In order to correlate our results with the literature, we have checked the position of the Sec residue, and we have observed that it is indeed in the antepenultimate position. Therefore, we can confirm that Miichthys miiuy contains SELENOO1 in its genome.
The SELENOO2 protein is located in the scaffold JXSJ01000232.1 in the reverse strand between positions 334116 and 339104. This gene contains 10 exons, without any selenocysteine.
The structure of the gene has been analyzed in the Exonerate file and the results are described below:
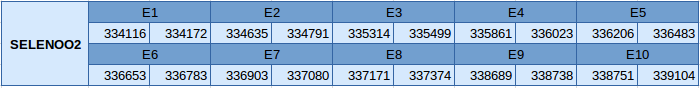
When we have analyzed the results from SELENOO2 we have realized that the Exonerate file contains two different results. However, we know it is only one protein, and that is the reason why we hypothesize that Exonerate is not able to recognise these two results as the same gene. We think the reason is that there is a big region between these two parts of the protein that prevents it from being recognised only as a single one. Moreover, as it can be seen in the T_coffee alignment, SELENOO2 is very well aligned but Miichthys miiuy does not contain the part of the protein where the Sec residue should be. That suggests that Miichthys miiuy contains SELENOO2 but it has lost the region that contains Sec residue. Therefore, since this prediction does not contain a Sec residue, we cannot conclude that Miichthys miiuy has this selenoprotein in a complete form.
In this protein one SECIS element of grade A has been predicted. This SECIS element is in the reverse strand and is located in the 3'UTR. No selenoprotein has been predicted by Seblastian because it is a Cys-containing protein.
In order to check whether the predicted selenoproteins are similar to the query, we have done a phylogenetic tree and we have obtained the following results:

As it can be observed, the queries have been correctly assigned to their scaffold. Therefore, we can conclude that SELENOO1 is found in Miichthys miiuy but SELENOO2, even though it is found in Miichthys miiuy, it does not contain the Sec residue, so it is not a selenoprotein.
Selenoprotein P (SELENOP)
Some SELENOP homologs are found predominantly in vertebrates. This protein has multiple Sec residues, however the number of these residues vary a lot between different species of vertebrates. 9,10In fishes, SELENOP has two different isoforms, SelPa and SelPb with one Sec residue.6
In order to check that the predicted protein is similar to the query, we have done a phylogenetic tree and we have obtained the following:
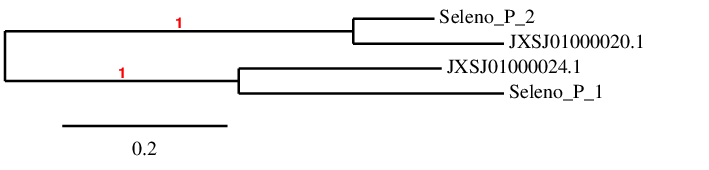
As we can see, the predicted proteins are correctly assigned to their scaffold. So we can conclude that SELENOP(1) and SELENOP(2) have been correctly assigned to their scaffolds, and therefore our results are reliable.
SELENOP(1) has been predicted in Miichthys miiuy with a Sec residue. Seblastian has predicted this selenoprotein but with an additional exon in 3' region and we hypothesize that Seblastian may have a larger query than ours from SelenoDB. Seblastian and SECISearch3 have predicted the same number of SECIS elements, but only one of them is the best one. We can observe in the alignment that only the first half of the protein is correctly aligned. Therefore, we can only conclude that, at least one part of the SELENOP(1), that contains the Sec residue, is present in Miichthys miiuy genome.
SELENOP(2) has also been predicted in Miichthys miiuy with a Sec residue. Seblastian has not predicted this selenoprotein, but it has predicted many SECIS elements. The fact that Seblastian has not been able to predict it make us doubt about the reliability of our results. Therefore, we cannot definitely conclude that SELENOP(2) is in Miichthys miiuy's.
Selenoprotein R (MSRB)
It is a zinc-containing selenoprotein that was initially identified as selenoprotein R (MSRB1) and selenoprotein X (SelX). Even though these proteins have structural differences, they have complementary functions. The Sec-containing MSRB1 is the most abundant MSRB in mammals and it is mainly localized in the cytosol and nucleus. Two additional MSRB homologous (MSRB2 and MSRB3) contain a Cys residue instead of the Sec residue in the active site.
Since it is also a big family, in order to check whether the analyzed scaffolds have been correctly assigned to each query, a phylogenetic tree has been performed.
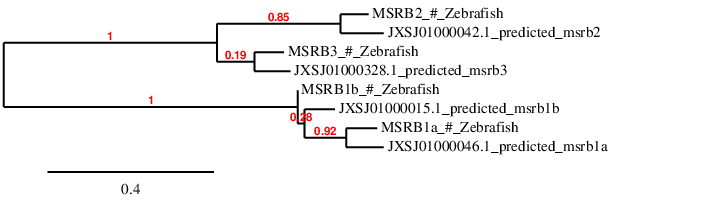
As it can be observed, each prediction is close to the query we have used to predict the protein, therefore it can be assumed that the results and the conclusions below are reliable.
MSRB1a from Danio rerio has been predicted in Miichthys miiuy's genome with a Sec residue. Seblastian has also predicted this selenoprotein, but two different SECIS has been identified. With these results, we can conclude that Miichthys miiuy contains MSRB1a in its genome.
In some organisms an additional MSRB was discovered and named as MSRB1b which reduces free methionine-R-sulfoxide.10 MSRB1b is a Sec containing protein that has been predicted in Miichthys miiuy. Seblastian has obtained the same prediction for MSRB1b than for MSRB1a, and therefore the Seblastian exon structure does not correlate with the one we have obtained for MSRB1b. We hypothesize that Seblastian has used the same query to predict MSRB1a than for MSRB1b. For that reason, we suggest that our prediction, in this case, is more reliable and informative. Since MSRB1b has been predicted in Miichthys miiuy's genome, we can conclude that this organism contains MSRB1b.
MSRB2 from Danio rerio is a Cys-containing homologous. However, we have predicted this protein, and, interestingly, our prediction contains a Sec residue, what differs from the literature. We suggest that Miichthys miiuy may have evolved changing the Cys residue for a Sec one, but it is also possible that the Sec residue has not been changed for a Cys residue yet. Since the alignment we have obtained in this scaffold is good, we can conclude that Miichthys miiuy contains MSRB2 in its genome.
MSRB3 from Danio rerio is a Cys-containing homologous, and we have predicted it with this Cys residue in Miichthys miiuy's genome. Since it does not has a Sec residue, Seblastian has not predicted any selenoprotein. No SECIS elements have been predicted neither. Since the alignment of the query with the genome is good, we can conclude that Miichthys miiuy contains MSRB3 in its genome.
Selenoprotein S (SELENOS)
As it has been said before, SELENOS and SELENOK can be assigned to the Selk/SelS family of related selenoproteins based on their topology. Therefore this prediction should contain Sec in the third or second position from the COOH terminal.
SELENOS has been predicted in Miichthys miiuy's genome with a Sec residue. Seblastian has been able to correctly predict the selenoprotein and the SECIS element. Therefore, since the alignment is good, we can conclude that SELENOS is found in Miichthys miiuy's genome.
Selenoprotein T (SELENOT)
It is a selenoprotein predominantly localized to the ER and Golgi and is expressed both during embryonic development and adult tissues. SELENOT belongs to the Rdx family and possesses a thioredoxin-like fold that is characterized of a conserved Cys-XX-Sec motif. It contains a conserved sequence of amino acids in the c-terminal region of the protein. 10
We have created a multiblast with the three subfamilies, SELENOT1, SELENOT1b and SELENOT2, in order to choose a different scaffold for each query, but also to check that these proteins are not overlapped in the same positions of the same scaffold. What we observed is that only two scaffolds appeared in the blast for the three subfamilies, and the three proteins appeared almost in the same positions of the scaffolds. We performed a phylogenetic tree with the three queries and the predictions obtained in both scaffolds, and the results are shown below:
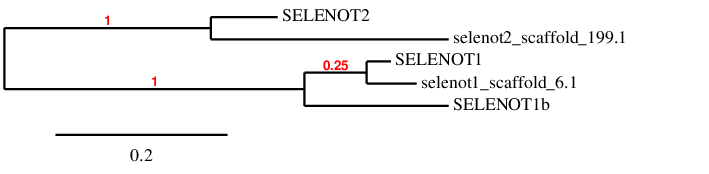
It can be observed that scaffold JXSJ000199.1 is the closest one to SELENOT2, and the scaffold JXSJ01000006.1 is the closest one to SELENOT1. Considering all this information, it is suggested that SELENOT1 is found in scaffold JXSJ01000006.1, that SELENOT2 is found in scaffold JXSJ000199.1 and that SELENOT1b is not found in Miichthys miiuy's genome.
SELENOT1 has been predicted in Miichthys miiuy's genome with a Sec residue in it. Seblastian has predicted this selenoprotein and also the best SECIS element. Our results do correlate with the literature because our prediction also contains this conserved motif Cys-xx-Sec, so we can conclude that SELENOT1 is present in Miichthys miiuy's genome.
Regarding SELENOT2, it has also been predicted in Miichthys miiuy's genome with a Sec residue in it. Seblastian has predicted this protein but with an additional exon in 3' region, because it may have a larger query, and the best SECIS element has also been predicted. The results of this prediction also correlate with the literature because it also contains the conserved motif Cys-xx-Sec, so we can conclude that SELENOT2 is present in Miichthys miiuy's genome.
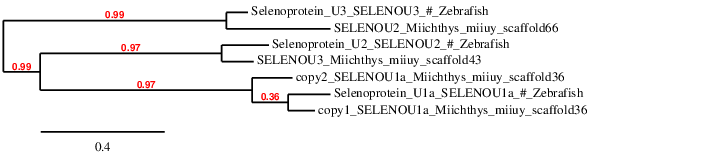
Selenoprotein U (SELENOU)
It has been shown that this family of proteins appear duplicated in bony fishes. Two proteins of this family are found in some fishes and they are called SELENOU1a and SELENOU1b. In Medaka this gene has a Sec residue while in Stickleback the Sec residue is replaced by a Cys. The function of this family is still unknown.9
In order to check whether the predicted proteins have been correctly assigned to their scaffold we have done a phylogenetic tree and we have obtained the following: In this phylogenetic tree the prediction obtained from scaffold JXSJ01000043.1 is aligned with the SELENOU2 query and the SELENOU3 query is aligned with the prediction obtained from the scaffold JXSJ01000066.1. When we observed the blast file, our chosen scaffold for each prediction was the only one possible, because no more possibles scaffolds have been obtained. For that reason, we assigned the scaffold JXSJ01000043.1 for query SELENOU3 and scaffold JXSJ01000066.1 for the query SELENOU2. We suggest that even though the phylogenetic tree does not correlate with our results, we have been forced to choose these scaffolds because no more options have been provided by the blast file.
SELENOU1a has been predicted twice in the same scaffold of Miichthys miiuy's genome with a Sec residue in both cases. To reject that this observed duplication is an assembly problem a T_coffee with the two predicted nucleotide sequences has been done. The results show that these two proteins are similar but not identical, so we can conclude that these protein is duplicated in the Miichthys miiuy's genome. Since it has been described that some fishes have SELENOU1a and SELENOU1b, we hypothesize that the duplication of SELENOU1a we have found is actually SELENOU1b.
Seblastian has also predicted this selenoprotein but with an additional exon in 3' region. Since our query does not start by Methionine, our prediction is not complete. Therefore, we can only conclude that a part of SELENOU1a has been predicted, and it is not sure (because we do not have enough evidences) that we have also predicted SELENOU1b.
Since our fish is phylogenetically closer to Stickleback than to Medaka, we expected to find the selenoprotein SELENOU1a with a Cys residue. However, it has been found a Sec residue.
SELENOU2 and SELENOU3 are Cys-containing homologous proteins. No SECIS elements have been predicted and Seblastian has not predicted any selenoprotein neither. Since both alignments are good, we can definitely conclude that Miichthys miiuy also contains SELENOU2 and SELENOU3.
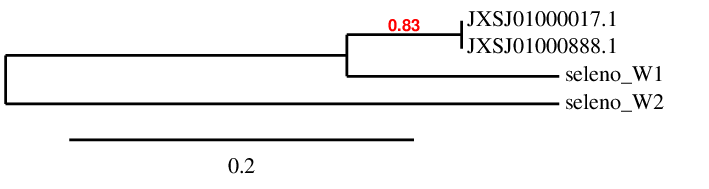
It is a small 9-kDa selenoprotein and it is sensitive to dietary Selenium intake. It has been proposed that Selenoprotein W could be involved in redox regulation, but the exact function remains unknown. Several W homologs have been observed across non-mammalian vertebrates. A phylogenetic analysis distincted a new group of selenoproteins, SELENOW2. This selenoprotein was found in bony fishes, frog and elephant shark what suggested that this protein is part of the ancestral vertebrate selenoproteome. Therefore, since Miichthys miiuy is a bony fish, we expect to find multiple copies of SELENOW2. This family possesses a thioredoxin-like fold that is characterized by a conserved Cys-XX-Sec motif, and they contain a conserved sequence of amino acids in the C-terminal region of the protein.
In the blast results we have observed that SELENOW(1) and SELENOW(2) could be aligned in the Miichthys miiuy genome in the same positions of two different scaffolds, so they were overlapping in each scaffold. To clarify which protein was situated in each scaffold a phylogenetic tree, showed below : In this tree, we can observe that both predicted proteins in the scaffolds JXSJ01000017.1 and JXSJ01000888.1 appear together aligned with the query of SELENOW1 and the query of SELENOW2 appear distant from the others. This information suggests that in the Miichthys miiuy do not exist both proteins, just SELENOW1 is present and this one is duplicated in both scaffolds. In order to check whether this supposed duplication is an assembly problem we aligned the sequence of nucleotides of both predictions, that are in different scaffolds. As we can observe in the T_coffee both sequences are identical. Therefore, we can conclude that is an assembly problem and SELENOW1 is only in one of these two scaffolds. The scaffold JXSJ01000017.1 has a better E-value in the blast file, so we could assume that SELENOW1 is only in this scaffold. Since in the blast file of SELENOW2 the scaffold JXSJ01000888 is the one with best E-value, this scaffold has been assigned to SELENOW2.
SELENOW(1) has been found in Miichthys miiuy's genome with a Sec residue. Seblastian has also predicted this selenoprotein and the SECIS element. In order to correlate our results with the literature, we have observed if our prediction contains the conserved Cys-xx-Sec motif. We have observed that it contains Ser-xx-Sec. Therefore Miichthys miiuy does not have this motif conserved. However, since we have predicted this selenoprotein and Seblastian too, we can conclude that Miichthys miiuy has SELENOW1 in its genome.
SELENOW(2) has also been found in Miichthys miiuy's genome with a Sec residue. Seblastian has also predicted the SECIS element and this selenoprotein but with an additional exon, suggesting that our query may not be complete. We have also checked whether it contains the Cys-xx-Sec motif but it has the same one than SELENOW1: Ser-xx-Sec. It is known that bony fishes contain multiple copies of SELENOW29,10. However, we have only found one copy of this gene but we suggest that there may be other copies of SELENOW2 but we have not found them. Even though we have just found one copy, we can conclude that, at least, we have SELENOW2 in Miichthys miiuy's genome.
When we blasted SELENOW3 from Danio Rerio with the Miichthys miiuy's genome no hits of a valid E-value were found so we have concluded that the M.miiuy does not have this protein.
These proteins are oxidoreductases that, together with thioredoxin, constitute the disulfide reduction system of the cell. It has been shown that TXNRD1 is also implicated in DNA repair, maintaining redox homeostasis and regulation of cell signaling. Multiple TXNRD1 isoforms have been described, at least 6 have been found in mammals. Multiple TXNRD3 isoforms have been described too. Since TXNRD1 and TXNRD3 are present in all vertebrates, Miichthys miiuy is expected to have them. 9,10
In order to check whether the queries have been correctly assigned to their scaffold, we have done a phylogenetic tree and the results obtained are the following ones: The results obtained in the phylogenetic tree suggest that the proteins TXNRD2 and TXNRD3 are correctly predicted in Miichthys miiuy genome. However, the TXNRD1 protein from Homo sapiens is not aligned with any prediction, and that suggests that TXNRD1 is not present in Miichthys miiuy
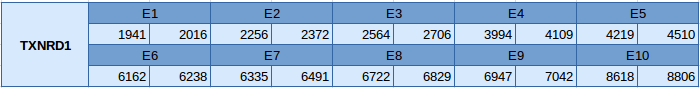
TXNRD1 was predicted blasting Miichthys miiuy's genome against the Homo sapiens TXNRD1 protein found in SelenoDB. TXNRD1 has been predicted in the scaffold JXSJ01000182.1 between positions 1941 and 8806 in the forward strand. This gene contains 10 exons and the structure of this gene is the following: After analysing our results we have realised that we have obtained two results in the Genewise file, so we have used the Genewise program to obtain a better prediction of the protein. Moreover, we have also observed that the predicted protein has almost the same length than the query but it does not contain the region where the Sec residue should be. Therefore we cannot conclude that this selenoprotein is present in Miichthys miiuy's genome. This idea is also supported by the phylogenetic tree, in which we can see that TXNRD1 is not close to any scaffold.
In this protein two SECIS elements of grade A have been obtained. The SECIS element of grade A with the best score is located in the middle of the exon 9. The second SECIS element of grade A is in the forward strand in the 3'UTR region. Since the predicted protein does not contain any Sec, Seblastian has not predicted any selenoprotein.
We hypothesize that Miichthys miiuy has evolved losing the region where the Sec residue should be found, but it has maintained the SECIS element.
TXNRD2 has been predicted in Miichthys miiuy with a Sec residue. However, Seblastian has not predicted it, but only one suitable SECIS has been identified. The fact that Seblastian has not predicted this selenoprotein make us doubt about the reliability of our results. Therefore, we cannot definitely conclude that TXNRD2 is present in Miichthys miiuy's genome.
TXNRD3 has been found in Miichthys miiuy's genome in two different scaffolds. To reject that this duplication observed is an assembly problem, a T_coffee with the nucleotides codifying for the two predicted proteins has been done. The results show that these two proteins are identical. Therefore, we can conclude that this protein is not duplicated in the Miichthys miiuy genome but it is an assembly problem. In order to decide which scaffold use to predict this selenoprotein, we have taken into consideration which one is more informative. We have observed that in scaffold JXSJ01000302.1 the alignment of the query with Miichthys miiuy genome is larger.
Seblastian has also predicted this protein with an additional exon in 5' region. We hypothesize that this is due to the fact that Seblastian may take a different query from the one we have taken.Therefore, we can conclude that Miichthys miiuy contains TXNRD3 in its genome.
eEFsec, PSTK and SBP2 are three proteins of the group selenoproteins machinery. All of them have been predicted in the Miichthys miiuy's genome.
SECIS binding protein 2 (SBP2) has its binding site in the stem region of the SECIS element.16
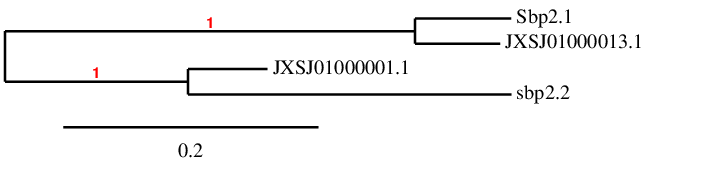
When we have decided in which scaffold each protein was situated we have performed a phylogenetic tree comparing both queries and both predicted proteins to check if our prediction agreed with those results. The figure of the phylogenetic tree is the following: SBP2(1) has been correctly predicted in Miichthys miiuy's genome. Seblastian has not predicted it because it does not contain a Sec residue, and no SECIS elements have been predicted neither. Since the alignment is good, we can conclude that Miichthys miiuy contains SBP2(1) in its genome.
SBP2(2) has been predicted in the genome of Miichthys miiuy. In the Exonerate file we obtained 13 different Exonerates results, so we think that the program did not recognize that the different exons were part of the same protein. In order to solve this problem, a Genewise was performed and the result was used to predict the selenoprotein and final alignment. The results of Genewise obtained showed that this protein is divided in 13 exons.
The alignment between the predicted protein and the SBP2(2) query is not good and the predicted protein has three selenocysteines that are not aligned with any Sec or Cys residue. Since it is not expected to find Sec residues in this protein, we cannot conclude that Miichthys miiuy contains SBP2(2).
Selenocystein synthase (SecS) dephosphorilates phosphoseryl-tRNA[Ser]Sec, and incorporates monoselenophosphate, which is the active form of selenium. Since it is a Cys containing protein, it is not a selenoprotein.
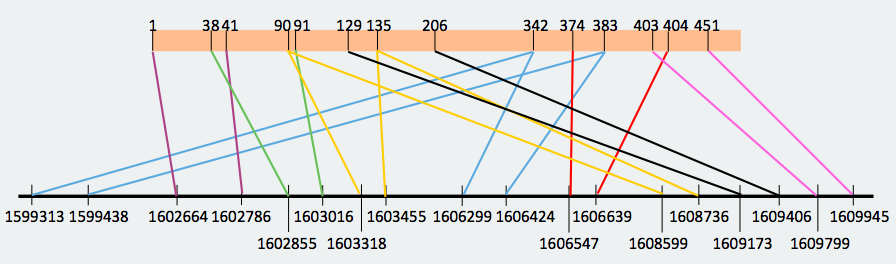
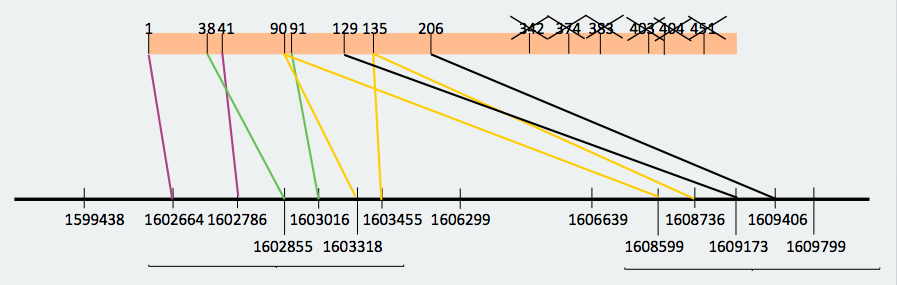
SECS protein is found in scaffold JXSJ01000055.1 in the forward strand. We observed in the blast file that there were a lot of overlapped hits. For that reason, we tried to reconstruct the distribution of the different fragments of the protein in the genome, and the result is the following one: As we can observe, there are a lot of overlapped hits. For example, we can see that the hit from amino acid 342 to 383 appears in the beginning of the sequence as well as before the hit 129-206, and it should go after it. We decided that we could omit these overlapped hits in order to identify the protein fragments that are entirely found in the Miichthys miiuy's genome . For that reason, we made a reconstruction taking into consideration the omitted hits that did not follow a logic order, and these results were obtained:
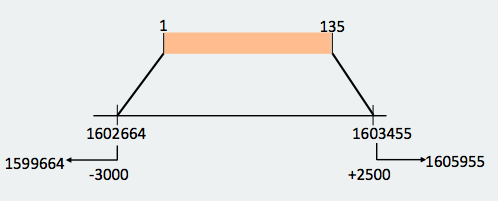
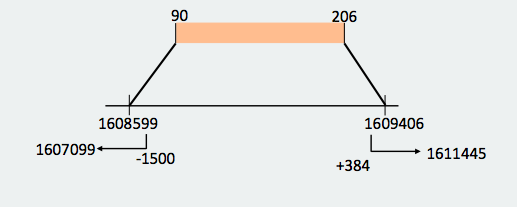
With this new reconstruction, we divided the protein in two big fragments that are found in different parts of the genome but in a logic order: one fragment is from amino acid 1 to 135 and the other one is from amino acid 90 to 206. When we made the prediction we took into account where the omitted hits started and finished in order not to include the genomic region corresponding to these hits when lengthening the prediction. In the end, the start and end coordinates in the genome were the following ones: Looking at the T_coffees from both fragments, we can observe that, in effect, there is a high conservation between the SecS from Danio rerio and both fragments of the protein that we have decided to use. If we observe both predictions, we can see that are consecutive between them, what reaffirms that they are different fragments from the same protein that are spread in different parts of the genome.
Seblastian has not predicted the selenoprotein and the SECIS element because, as it has been said, it does not contain a Sec residue, and moreover we have not provided to Seblastian the whole genome sequence.
We reject the hypothesis that there has been something that has been "introduced" in the middle of the protein resulting in the fragmentation of it, because there are some fragments (from amino acid 90 to amino acid 135) that are found in both fragments predicted by us. Therefore, the only conclusion we can make is that has these two fragments of the protein in different parts from the same scaffold. Even though the scaffold analyzed is the only one possible and we have not been able to find it there, we hypothesize that this organism may contain the whole protein because, since it is a machinery protein, it is indispensable for selenoproteins biosynthesis.
All in all, our results do not allow us to conclude that Miichthys miiuy contains SecS in its genome.
All these proteins have been predicted in Miichthys miiuy. Any of them contains a Sec residue, except SARS2 that has been predicted with one Sec residue. However, Seblastian has not predicted any of them, but SECIS elements have been identified by SECISearch3. That would mean that these proteins may have evolved from a Sec-containing protein and may have lost the Sec residue but still conserving the SECIS element.
All these proteins have been predicted in Miichthys miiuy with any Sec residue in any of them. For that reason, Seblastian has not predicted them, and no SECIS element have been identified. They have been correctly aligned with their respective queries, and therefore we can conclude that G6Pd, RPL30, SCLY and TTPA are present in Miichthys miiuy's genome.
CELF1 has been found in Miichthys miiuy without any Sec residue. Seblastian has not predicted it but one SECIS element has been identified as the best one. Even though our program has only predicted the first and the last part of the protein, we can conclude that Miichthys miiuy contains CELF1.
LRP8 is a cell-surface receptor involved in the reelin signaling pathway. It has been seen that mutations in LRP8 do no affect Selenium metabolism but it constitutes a Se-uptake and delivery system essential for supplying Se to target organs such as brain, bone and testes.
Selenoprotein W (SELENOW)
Thioredoxin reductase (TXNRD)

Selenoproteins machinery:
eEFsec, PSTK and SECp43
SECp43 and eEFsec have a SECIS element predicted by SECISearch3, but any SECIS element has been predicted for PSTK. Furthermore, Seblastian has not been able to predict any selenoprotein because they do not contain any Sec residue. Good alignments have been obtained in the SECp43 and eEFsec analysis, so we can conclude that Miichthys miiuy contains this two proteins in its genome.
However, the reliability of the PSTK prediction is uncertain, because even though many Cysteines are aligned, the score of the t_coffee is not good enough. Therefore, we cannot conclude that the PSTK is present in the Miichthys miiuy's genome. In future studies, a PSTK of another species closer to Miichthys miiuy should be analysed to confirm the presence or the absence of this protein in its genome.
SECIS binding protein 2 (SBP2)
Selenocysteine synthase (SecS)
Proteins related to selenium metabolism:
ELAVL1, EIF4A3, XPO1, SELENBP1 and SARS2
Since all of them are good aligned with their respective queries, we can conclude that Miichthys miiuy contains ELAVL1, EIF4A3, XPO1, SELENBP1 and SARS2 in its genome.
G6PD, RPL30, SCLY and TTPA
Elav-like family member 1 (CELF1)
Low density lipoprotein receptor-related protein 8 (LRP8)
When the query of LRP8 was blasted, it was found that it seemed to be duplicated in the same scaffold, JXSJ01000652.1, because it appeared twice in different positions. Then, we have ran our program for these two parts of the genome, and then we have aligned the nucleotides codifying for the predictions found in order to check whether it was a duplication or an assembly problem. What we can see in the T_coffee alignment, the nucleotides are completely identical, and therefore this is an assembly problem.
Seblastian has not predicted this protein because it does not contain a Sec residue, and no SECIS element has been found neither. Since at least one part of this protein has been aligned in the genome
