Abstract
Selenoproteins represent a portion of the proteome present in the three domains of life. These ones are characterized by presenting at least one residue of selenocysteine (U or Sec), the 21st amino acid which is encoded by the UGA stop codon. That’s why a complex enzymatic machinery is required to recognize and insert this residue in a protein sequence. Moreover, the presence of a SECIS element in the 3’-UTR region of the mRNAs that code for selenoproteins is essential to achieve its recognition.
The aim of this project has been to predict the selenoproteins and the associated machinery of Ammotragus lervia by bioinformatic methods (such as tBlastn, Exonerate, Genewise, Seblastian and T-Coffee) that have allowed to compare the genome of this mammal with the selenoproteome of Homo sapiens and Bos taurus. The sequence alignments obtained against these reference species and the characterization of SECIS candidates (by SECISearch3 online program), have provided a notable approach about the conservation and composition of Ammotragus lervia’s selenoproteome.
The analysis performed has lead to the identification of 23 selenoproteins, 9 Cys-containing homologs and 7 synthesis associated machinery proteins in Ammotragus lervia. These findings represent a notable contribution to the characterization of the selenoproteome across mammalian and vertebrates organisms.
Introduction
Selenoproteins
1. Definition
Selenoproteins are a unique group of proteins containing at least one selenocysteine (Sec), which is a selenium-containing amino acid. In fact, this residue is the 21st “naturally occurring” amino acid in the genetic code and quite similar to cysteine (Cys), which contains a sulfur (S) atom instead of Sec.
Sec is encoded by the UGA codon, whose main function is to act as a stop codon. However, thanks to the intervention of a complex enzymatic machinery and some other regulator elements, premature termination of translation is prevented and a Sec residue is sometimes incorporated to a nascent protein at this point [1].
With a few exceptions, Sec is located in the enzyme active sites in order to perform mainly catalytic redox reactions[1, 2].
Since selenoproteins genes contain UGA codons, which are normally used as stop signals during protein translation, selenoproteins are often misannotated in sequence data-bases.
2. Biosynthesis of selenoproteins
Taking into account the aim of this project, the following explanations will mainly regard to the biosynthesis process in eukaryotes.
2.1. Enzyme machinery involved
Fig1. Model of the Human tRNA[Ser]Sec
The key component of selenoprotein biosynthesis is tRNA[Ser]Sec (Fig.1), whose corresponding gene is Trsp [3]. In order to incorporate the Sec residue to tRNA[Ser]Sec different steps take place:
- tRNA[Ser]Sec is initially aminoacylated with serine (Ser) by seryl-tRNA synthetase (SerS).
- Phosphoseryl-tRNA kinase (PSTK) gives place to the phosphorylated intermediate PSer-tRNA[Ser]Sec
- In parallel, Selenophosphate synthetases SPS1 and SPS2 catalyze the production of selenophosphate (H2SePO-3) from selenite and ATP. In fact, SPS2 is also a selenoprotein itself and has been described to participate in Cys biosynthesis. Moreover, it possibly serves as an autoregulator of selenoprotein synthesis and that is why is essential for this process [4].
- H2SePO-3 acts as a donor of Se for Sec synthase (SecS), which catalyzes the conversion of the serine moiety on tRNA[Ser]Sec to selenocysteyl-tRNA[Ser]Sec (Sec-tRNA[Ser]Sec). This way, Sec is incorporated to its corresponding tRNA [1] (Fig.2).
2.2. SECIS elements
Sec insertion sequence (SECIS) elements are cis stem-loop RNA structures located in the 3’-untranslated regions (3’-UTR) of all eukaryotic and archaeal selenoprotein mRNAs [5]. In bacteria, SECIS elements are found immediately downstream of the UGA encoding Sec, within the coding region of selenoprotein genes [6]. The main function of SECIS is to lead recoding of UGA codon as Sec.
When a ribosome encounters the UGA codon (Fig.3), SECIS elements and trans protein factors interact with the translation machinery to augment the coding potential of that codon. SECIS binding protein 2 (SBP2) binds to ribosomes and to SECIS elements with high affinity. It also interacts with the eukaryotic Sec-specific translation elongation factor (eEFSec), which recruits Sec-tRNA[Ser]Sec and facilitates incorporation of Sec into the nascent protein giving rise to a selenoprotein this way [1].
Additional SECIS-binding proteins like ribosomal protein L30, eukaryotic initiation factor 4a3 (eIF4a3), and nucleolin have been described as regulatory factors in the modulation of selenoproteins synthesis [1, 7].
Fig2. Pathway of Sec and Cys biosynthesis in eukaryotes
3. Evolution of selenoproteins
Selenoproteins are present in eukarya, archaea and eubacteria domains and have been also observed in viruses. The distribution is widespread among these domains and varies greatly among species. The number of proteins in selenoproteomes (a full set of selenoproteins in an organism) can range from one, as in C elegans, to as many as 59 found in the pelagophyte Aureococcus anophagefferens [1].
Fungi, plants, some animal species and several insects don’t present selenoproteins. Furthermore, comparative analysis revealed that several groups of terrestrial organisms reduced their utilization of Sec by replacing selenoproteins with Cys homologs or completely losing some selenoproteins. In contrast, most aquatic organisms have large selenoproteomes, what suggests that environment plays a role in the evolution of selenoproteins [8].
On the other hand, analyses of vertebrate and mammalian selenoproteomes also demonstrated a trend toward reduced selenoprotein usage in mammals. Indeed, all Cys/Sec replacements found in mammals were from Sec to Cys [9].
Fig3. Mechanism of Sec insertion in eukaryote
4. Families of selenoproteins
Approximately, there are more than 50 selenoprotein families known, the majority of which have been identified by bioinformatics methods. Currently, 21 families of selenoproteins have been found in vertebrates which are: Fep15, SelI, SelJ, SelN, SelP, SelS, SelV, SPS, Sep15, DIOs, GPxs, MsrB, SelH, SelK, SelL, SelM, SelO, SelT, SelU, SelW, TRs ( or TrxRs, Txnrds) [10]. Nevertheless, the quantity varies among the different species and some duplications events of selenoprotein genes have been detected. Specifically, 37 families have been identified in Homo sapiens.
Within the whole set of selenoprotein families, three of them are some of the most remarkable ones:
a. Glutathione Peroxidases (GPxs)
Fig4. Structure of GPx1 selenoprotein obtained by crystallization
In mammals, there are eight GPx paralogs, from which five (GPx1, GPx2, GPx3, GPx4, and GPx6) contain a Sec residue in their active site. In the other three GPx homologs (GPx5, GPx7, and GPx8), the active-site Sec is replaced by Cys.
GPxs play a wide range of physiological functions in organisms and are involved in hydrogen peroxide (H2O2) signaling, detoxification of hydroperoxides, and maintaining cellular redox homeostasis [1].
The first selenoprotein identified was mammalian glutathione peroxidase 1 (GPx1) [11] (Fig.4).
b. Thyroid Hormone Deodinases (DIOs or DIs)
The iodothyronine deodinase family of selenoproteins consists of three paralogous proteins in mammals (DI1, DI2, and DI3), which are involved in regulation of thyroid hor- mone activity by reductive deodination.
Homologs of mammalian deodinases occur not only in other vertebrates, but are also found in simple eukaryotes and bacteria. The function of deodinase homologs in these organisms is not known [1].
c. Thioredoxin Reductases (TRs or TrxRs,Txnrds)
Thioredoxin reductases (TRs) are oxidoreductases that, together with thioredoxin (Trx), comprise the major disulfide reduction system of the cell. In mammalian cells, there are three TR isozymes, all of which are Sec-containing proteins [1].
Ammotragus lervia
Ammotragus lervia is an African ungulate retaining some primitive and unique characteristics. It has the distinction of being the only wild sheep species in Africa, and the only species in the genus Ammotragus [12].
Common names for the species are: aoudad, aroui, arui, waddan, arruis or Barbary sheep, in English; arruí in Spanish and be de Berberia in Catalan [13].
1. Taxonomy

| Kingdom | Animalia |
| Phylum | Chordata |
| Class | Mammalia |
| Order | Artiodactyla |
| Suborder | Ruminantia |
| Infraorder | Pecora |
| Family | Bovidae |
| Subfamily | Caprinae |
| Genus | Ammotragus Blyth, 1840 |
| Species | A. lervia Pallas, 1777 |
2. Distribution
The Barbary sheep was formerly widespread in rugged and mountainous terrain from deserts and semi-deserts to open forests in North Africa, where it is distributed from Morocco and Western Sahara to Egypt and Sudan [14].
Six subspecies have been described, mainly according to their distribution:
- Ammotragus lervia lervia Pallas, 1777. Distributed in Morocco, northern Algeria and Tunisia. Exemplars of this subspecies were introduced to Spain.
- Ammotragus lervia ornata I. Geoffrey Saint-Hilaire, 1827. Formerly quite widespread throughout the Eastern and Western Desert of Egypt and was actually thought to be extinct. However, evidence of its presence in Western Desert of Egypts was reported.
- Ammotragus lervia sahariensis Rothschild, 1913. Widely distributed in the Western Sahara.
- Ammotragus lervia blainei Rothschild, 1913. Located only in northeast Sudan.
- Ammotragus lervia angusi Rothschild, 1921. Found in Mali, Algeria, Niger and Chad.
- Ammotragus lervia fassini Lepri, 1930. Found only in southern Tunisia and in Libya [12].
Furthermore, Barbary sheep have been introduced to south-eastern Spain, the southwestern United States, Hawaii, Mexico and some parts of Africa. It was introduced to Spain in 1970, more specifically to the Sierra Espuña Regional Park in Murcia and has now spread to Alicante, Almeria and Granada [12].
3. Description
In appearance, it is somewhat of an intermediate between a sheep and a goat. It is a stocky, heavily built animal, with short legs and a long face. The coat, which is generally a sandy-brown colour, is woolly during the winter, but moults to a finer, sleek coat at summer. Both sexes have horns that sweep backwards and outwards in an arch; those of the male are much thicker, longer and more heavily ridged than the slenderer horns of the female. Males also differ from females by their significantly heavier weight, (up to twice that of females), and the notably longer curtain of hair that hangs from the throat, chest and upper part of the forelegs. On males, this mane of long, soft hairs almost touches the ground. The short tail, which is hairless on the underside, has scent glands [14].
The Barbary sheep has a head-body length of 130 – 165 cm and a tail length of 12 – 25 cm. Males weight between 100 to 140 kg, whereas female weight about 40 to 55 kg [14].
4. Reproduction
Fourteen-months-old males and nine-months-old females can be regarded as sexually mature. The mean gestation period is 5.5 months and mating season peak occurs from September to November, so that breeding season tends to be focused in spring. One or two young are born at a time, and lie in a secluded site with the mother for the first few days of life, before joining the rest of the group. Barbary sheeps in captivity have been known to live for 24 years [14].
5. Feeding
Its diet is constituted by lichens, grass, herbs, seeds and leaves from shrubs, bushes and trees such acacias [15].
6. Behavioural ecology

The Ammotragus lervia is a gregarious species [15]. This sheep lives in small groups of three to six individuals, comprising a single adult male, several adult females, and their offspring. Occasionally, such as in the dry season, several of these groups may congregate, forming parties of up to 20 individuals. Adult males must earn their position as head of a group of females through intimidation displays, with males showing their mane of hair on their foreparts, and fights between males [14].
The Barbary sheep feeds primarily at dusk, dawn and during the night. By feeding at night, when plants accumulate moisture from the atmosphere or become covered in dew, the Barbary sheep obtains water, enabling it to survive without drinking water during dry periods in its arid habitat [14].
Another adaptation to this dry and unproductive terrain can be seen in the Barbary sheep’s reaction to threats; with an almost total lack of sufficient vegetation to hide behind, the Barbary sheep will instead remain motionless when threatened, their sandy-brown coat enabling them to blend into their surroundings [14].
7. Habitat
Barbary sheep are found in arid hill and mountain habitats. Within this rocky, rugged terrain, the Barbary sheep selects areas where there is some shade, either caves, rocky overhangs or trees, to which it can retreat during the hottest hours of the day [13].
8. Status and conservation
In Africa this species is endangered due to overhunting, habitat degradation -caused by rapid desertification- and to hard competition for water and food, with domestic cattle.
It is classified as Vulnerable (VU) on the IUCN Red List and listed on Appendix II of CITES [14].
While the Barbary sheep is protected by law throughout most of its range, the lack of enforcement of these laws is a serious problem for the conservation of this species. This relates to the unfortunate fact that most countries in which the Barbary sheep occurs have little funds available to conserve these animals.
On the other hand, it has been introduced in many countries, Spain among them, where populations have multiplied, to the point of becoming a threat to environment and native species. Barbary sheep, as a matter of fact, is listed in the Spanish list and catalogue of invasive alien species [15].

For further information see this Wikipedia entry.
Materials and Methods
The aim of this project was to identify and annotate the selenoproteins and the machinery required for their synthesis encoded in Ammotragus lervia genome. In order to achieve this goal, an homology-based approach has been carried out considering the Homo sapiens the phylogenetically closest species with a very-well-annotated selenoproteome. In some cases, this analysis was performed with the Bos taurus genome.
Queries acquisition
The Homo sapiens and Bos Taurus genomes were chosen to identify the selenoproteins in Ammotragus lervia since they are both mammals and therefore phylogenetically close to our case of study.
The human amino acids sequences of all the queries (selenocysteines proteins, homologous and selenoprotein machinery) were obtained from SelenoDB 1.0 database. Every sequence was copied into an EMACS file named as PROT-human.fa, were “PROT” is the abbreviation of the protein named used on SelenoDB 1.0.
Time after, it was noticed that some proteins were missing, mostly enzymatic machinery of selenoproteins, which were not annotated on SelenoDB 1.0. For this reason, the proteins of Bos Taurus which were not present on the list of human proteins of SelenoDB 1.0, were also copied into an EMACS file and, in this case, were named as PROT-taurus.fa. These proteins were obtained from SelenoDB 2.0 database. In one case, the Selenophosphate synthetase 2 (SEPHS2), the sequence present on the SelenoDB 2.0 database was not correctly annotated so it was extracted from the Uniprot database.
The different software packages needed for this analysis do not recognise the character “U” as an amino acid, despite selenocysteines are represented with it. This is why all the “U” were replaced by an “X”, which represents any possible amino acid. Moreover, all the symbols ( #, @, etc) found at the end of the sequences, which are not recognised by the softwares, were removed.
Exploratory and homology selenoprotein prediction
In order to carry out all the steps necessary for the exploration of potential selenoproteins in Ammotragus lervia in an easier and faster way, a semiautomatic bash program was developed [program].
This program performed all needed steps except for both genome and query acquisition and the SECIS prediction. The different steps were:
- tBLASTn prediction
- Extraction of fetch of interest
- Generation of a subseq in fasta format
- Exonerate prediction
- Translation into protein format
- T-Coffee alingment
- Genewise prediction
The name of the query, the ID of the scaffold, the origin of the scaffold (without substracting 50.000 nucleotides) and the end of the scaffold (without adding 50.000 nucleotides), needed for the calculation of the length, were introduced manually.
We considered a scaffold adequate if one or more of its hits had both an E-value equal or minor to 0,001 and an identity equal or higher than 50%. Being the e-value (expected value) a parameter that describes the number of hits one can expect to see by chance as good as the hit observed; and the % identity the proportion of identical residues between two sequences.
A more detailed explanation of the program steps and commands could be read here.
Some scaffolds generated a multi-protein exonerate prediction, which produced a conflict with the program steps 5 and 6. In order to resolve this issue, it was developed another program which remade the exonerate with the option “-n 1”. This option is used to extract only the best prediction of all [program].
SECIS and Selenoprotein prediction
SECIS are elements associated with selenocysteine proteins and are essential for their synthesis. These elements are located in the 3’ -UTR region. The prediction of the SECIS has been done using two programs: Seblastian and SECISearch3.
SECISearch3 only looks for SECIS elements while Seblastian searchs for SECIS elements as well as tries to find an associated selenoprotein to them. For this reason, these programs were used to confirm the presence of the selenocysteine proteins found with our program.
Both programs were running with default parameters. Output details from both programs and parameters can be found here.
In numerous cases, SECISearch3 predicted more than one possible SECIS. In order to choose the more adequate one some considerations have been followed:
- SECIS must be on the 3’-UTR of the gene.
- SECIS has to be localized on the same strand (+ or -) as the coding gen for the selenocysteine protein.
Once it was chosen, another program was used to extract it from the output generated by SECISearch3, which included other predicted SECIS, in a new file with 'secis.select' extension.
To facilitate the SECISearch3 and the Seblastian step, it was developed a program that removed the fasta head line (‘>...’)and redirected the output in a new file named “subseq.secis”. The previous step was necessary due to the fact that both programs cannot work if the head line is provided. [program]
In order to complement the results of the predictions obtained in the case of selenoprotein families, a phylogenetic tree has been constructed using a tool available in phylogeny.fr. The output provides the correlation between the sequence queries (from Homo sapiens) and the predicted proteins in Ammotragus lervia.
Pipeline output analysis and web development
Once all query proteins were analysed by different programs and SECISearch3 and Seblastian programs were ran, a manual analysis of the results was done in order to choose the best scaffold for each protein.
To carry out this procedure, Seblastian was very useful since this program predicted selenoproteins for each scaffold and it could also provide which type of selenoprotein was, based on other proteins of its database. Every scaffold in which Seblastian found an association with a query protein was chosen directly. Those proteins from which Seblastian could not obtain a prediction, were chosen following the next criteria:
- A same scaffold could not be assigned by Seblastian to more than one protein of the same family and, moreover, its genomic region could not overlap with the region of another assigned scaffold.
- (Non restrictive) T-Coffee of exonerate prediction matched X (selenocisteins) or C.
- Exonerate obtained a good protein which was aligned correctly with the query protein on T-Coffee output (>80%).
- (Non restrictive) The predicted protein started with M.
- The predicted protein could not contain a stop codon bewtween the N- and C-terminus.
Conditions 2-4 were analyzed with the help of a program that showed matches on the entire T-Coffee aligment. However, the output of this program was manually verified.
All the results were annotated in a multi-array that contained all the searched proteins, its selected scaffold, gene location, the selenocistein or homologue (Sec, Cys...) residue and a SECIS ID from SECISearch3 (if it was found) for each query protein.
Once this array of related protein-scaffolds was developed, it was given to a last Python-Bash program [program] which generated the results of the following section (“Results”) in html format. This file was revised and readjusted manually. In addition, a final Bash script [program] was used to generate a v2.0 GFF file with absolute positions of all predicted proteins. These GFF files were sent to our external project evaluators.
Results
The following tables show, for each query, the query protein abreviatured name, its origin, if it contains selenoproteins, its tblastn output, the Exonerate prediction for the protein, the GeneWise prediction for the protein, the Scaffold of best prediction and gene location in scaffold, the T-Coffee alignment for the best prediction and the SECIS element prediction by Seblastian and SECISearch3.



Selenoproteins
| PROTEIN | SPECIES | RESIDUE | BLAST | EXONERATE | GENEWISE | SCAFFOLD | Gene Location (-/+) | Predicted protein/ T-Coffee | SEBLASTIAN | SECIS candidate | SECIS Image |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Iodothyronine deiodinases | |||||||||||
| DI1 |  | Sec (=) |  |  |  | NIVO01071179.1 | 648741 - 642044 (-) |  |  |  |  |
| DI2 |  | Sec (=) |  |  |  | NIVO01066248.1 | 6732164 - 6723152 (-) |  |  |  |  |
| DI3 |  | Sec (=) |  |  |  | NIVO01034600.1 | 4247675 - 4248509 (+) |  |  |  |  |
| Glutathione peroxidases | |||||||||||
| GPx1 |  | Sec (=) |  |  |  | NIVO01040474.1 | 1832678 - 1833524 (+) |  |  |  |  |
| GPx2 |  | Sec (=) |  |  |  | NIVO01055543.1 | 22496 - 19447 (-) |  |  |  |  |
| GPx3 |  | Sec (=) |  |  |  | NIVO01038251.1 | 1168719 - 1161463 (-) |  |  |  |  |
| GPx4 |  | Sec (=) |  |  |  | NIVO01030317.1 | 604591 - 606687 (+) |  |  |  |  |
| GPx5 |  | Cys (=) |  |  |  | NIVO01038789.1 | 484731 - 489829 (+) |  |  |  |  |
| GPx6 |  | Sec (=) |  |  |  | NIVO01038789.1 | 461041 - 456728 (-) |  |  |  |  |
| GPx7 |  | Cys (=) |  |  |  | NIVO01025624.1 | 1405646 - 1412609 (+) |  |  |  |  |
| GPx8 |  | Cys (=) |  |  |  | NIVO01006926.1 | 1291158 - 1286791 (-) |  |  |  |  |
| Methionine sulfoxide reductase A | |||||||||||
| MsrA |  | Cys (=) |  |  |  | NIVO01055729.1 | 63554 - 171582 (+) |  |  |  |  |
| 15-kDa selenoprotein | |||||||||||
| Sel15 |  | Sec (=) |  |  |  | NIVO01070360.1 | 721121 - 756771 (+) |  |  |  |  |
| Selenoprotein H | |||||||||||
| SelH |  | Sec (=) |  |  |  | NIVO01034600.1 | 5029013 - 5028438 (-) |  |  |  |  |
| Selenoprotein I | |||||||||||
| SelI |  | Sec (=) |  |  |  | NIVO01032443.1 | 1395967 - 1433662 (+) |  |  |  |  |
| Selenoprotein K | |||||||||||
| SelK |  | Sec (=) |  |  |  | NIVO01057317.1 | 1822121 - 1788813 (-) |  |  |  |  |
| Selenoprotein M | |||||||||||
| SelM |  | Sec (=) |  |  |  | NIVO01044519.1 | 786172 - 783634 (-) |  |  |  |  |
| Selenoprotein N | |||||||||||
| SelN |  | *Sec (deletion?, =) |  |  |  | NIVO01052487.1 | 965129 - 975153 (+) |  |  |  |  |
| Selenoprotein O | |||||||||||
| SelO |  | Sec (=) |  |  |  | NIVO01039759.1 | 211008 - 230377 (+) |  |  |  |  |
| Selenoprotein P | |||||||||||
| SelP |  | *Sec (8=,-2?) |  |  |  | NIVO01048714.1 | 510433 - 516419 (+) |  |  |  |  |
| Methionine sulfoxide reductase B or Selenoprotein R | |||||||||||
| SelR1 |  | Sec (=) |  |  |  | NIVO01014683.1 | 1290902 - 1287262 (-) |  |  |  |  |
| SelR2 |  | Cys (=) |  |  |  | NIVO01003597.1 | 1801149 - 1822586 (+) |  |  |  |  |
| SelR3 |  | Cys (=) |  |  |  | NIVO01040223.1 | 1554578 - 1676368 (+) |  |  |  |  |
| MSRB1 |  | Sec (=) |  |  |  | NIVO01014683.1 | 1290902 - 1288557 (-) |  |  |  |  |
| MSRB2 |  | Cys (=) |  |  |  | NIVO01003597.1 | 1808743 - 1822586 (+) |  |  |  |  |
| MSRB3 |  | Cys (=) |  |  |  | NIVO01040223.1 | 1497075 - 1676368 (+) |  |  |  |  |
| Selenoprotein S | |||||||||||
| SelS |  | Sec (=) |  |  |  | NIVO01025374.1 | 1333749 - 1333179 (-) |  |  |  |  |
| Selenoprotein T | |||||||||||
| SelT |  | Sec (=) |  |  |  | NIVO01008816.1 | 544233 - 568960 (+) |  |  |  |  |
| Selenoprotein U | |||||||||||
| SelU1 |  | Cys (=) |  |  |  | NIVO01064043.1 | 30496 - 22805 (-) |  |  |  |  |
| SelU2 |  | Cys (=) |  |  |  | NIVO01069066.1 | 35255 - 23477 (-) |  |  |  |  |
| SelU3 |  | Cys (=) |  |  |  | NIVO01017783.1 | 904227 - 901833 (-) |  |  |  |  |
| Selenoprotein V | |||||||||||
| SelV |  | *Ser (Sec) |  |  |  | NIVO01013187.1 | 388309 - 391391 (+) |  |  |  |  |
| Selenoprotein W | |||||||||||
| SelW1 |  | Sec (=) |  |  |  | NIVO01052466.1 | 360962 - 363398 (+) |  |  |  |  |
| SelW2 |  | Cys (=) |  |  |  | NIVO01065774.1 | 5401081 - 5402051 (+) |  |  |  |  |
| Thioredoxin reductases | |||||||||||
| TR1 |  | Sec (=) |  |  |  | NIVO01039896.1 | 945273 - 907892 (-) |  |  |  |  |
| TR2 |  | Sec (=) |  |  |  | NIVO01019577.1 | 234523 - 260407 (+) |  |  |  |  |
| TR3 |  | Sec (=) |  |  |  | NIVO01001699.1 | 4487940 - 4566814 (+) |  |  |  |  |
| TXNRD2 |  | Sec (=) |  |  |  | NIVO01019577.1 | 236203 - 260407 (+) |  |  |  |  |
| TXNRD2-UniProt |  | Sec (=) |  |  |  | NIVO01019577.1 | 234532 - 260407 (+) |  |  |  |  |
| TXNRD3 |  | Sec (=) |  |  |  | NIVO01001699.1 | 4540623 - 4566814 (+) |  |  |  |  |
Selenoprotein machinery
This table shows the presence of the genes encoding the machinery proteins.
| PROTEIN | SPECIES | RESIDUE | BLAST | EXONERATE | GENEWISE | SCAFFOLD | Gene Location (-/+) | Predicted protein/ T-Coffee | SEBLASTIAN | SECIS candidate | SECIS Image |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Eukaryotic elongation factor | |||||||||||
| eEFSec |  | - (=) |  |  |  | NIVO01001699.1 | 3621595 - 3511308 (-) |  |  |  |  |
| SECIS binding protein 2 | |||||||||||
| SBP2 |  | - (=) |  |  |  | NIVO01049792.1 | 1600400 - 1635783 (+) |  |  |  |  |
| Selenophosphate synthetases | |||||||||||
| SPS1 |  | Tyr (=) |  |  |  | NIVO01060204.1 | 73009 - 50691 (-) |  |  |  |  |
| SPS2 |  | Sec (=) |  |  |  | NIVO01016939.1 | 905194 - 903847 (-) |  |  |  |  |
| Phosphoseryl-tRNA kinase | |||||||||||
| PSTK |  | - (=?) |  |  |  | NIVO01034609.1 | 2485067 - 2471748 (-) |  |  |  |  |
| tRNA Sec 1 associated protein 1 | |||||||||||
| SECp43 |  | - (=?) |  |  |  | NIVO01076485.1 | 881203 - 860169 (-) |  |  |  |  |
| Selenocysteine synthase | |||||||||||
| SecS |  | - (=?) |  |  |  | NIVO01007502.1 | 1713228 - 1678486 (-) |  |  |  |  |
Description of selenoproteins
Selenoproteins
Iodothyronine deiodinases
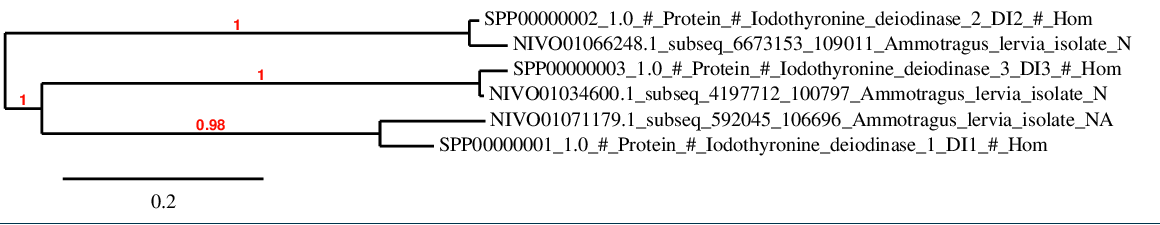
For DI family, there is a strong correlation between predicted proteins and the corresponding human sequences as it is shown in the phylogenetic tree.
DI1
In Ammotragus lervia, DI1 gene is located in the scaffold NIVO01071179.1 between the position 648741 and the position 642044, in the negative strand. This gene has 3 exons (according to Exonerate output) detailed below:
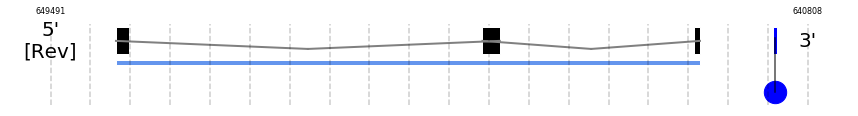
- Exon 1: From position 648741 to 648588
- Exon 2: From position 644539 to 644340
- Exon 3: From position 642110 to 642045
A SECIS candidate was predicted by SECISearch3 [full output] in the 3’-UTR region between the positions 641184 and 641114 in the reverse (-) strand. Seblastian predicts a selenoprotein that matches with 'deiodinase 1, partial [Ovis aries].'
DI2
In Ammotragus lervia, DI2 gene is located in the scaffold NIVO01066248.1 between the position 6732164 and the position 6723152, in the negative strand. This gene has 2 exons (according to Exonerate output) detailed below:
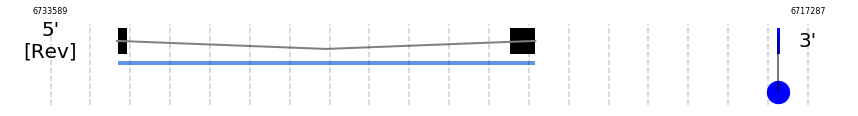
- Exon 1: From position 6732164 to 6731943
- Exon 2: From position 6723725 to 6723153
A SECIS candidate was predicted by SECISearch3 [full output] in the 3’-UTR region between the positions 6717932 and 6717860 in the reverse (-) strand. However no selenoprotein was predicted by Seblastian.
DI3
In Ammotragus lervia, DI3 gene is located in the scaffold NIVO01034600.1 between the position 4247675 and the position 4248509, in the positive strand. This gene has 1 exons (according to Exonerate output) detailed below:
- Exon 1: From position 4247676 to 4248509
A SECIS candidate was selected from 2 predicted by SECISearch3 [full output] in the 3’-UTR region between the positions 4249118 and 4249198 in the forward (+) strand. Seblastian predicts a selenoprotein that matches with 'PREDICTED: thyroxine 5-deiodinase [Bos mutus].'
Glutathione peroxidases
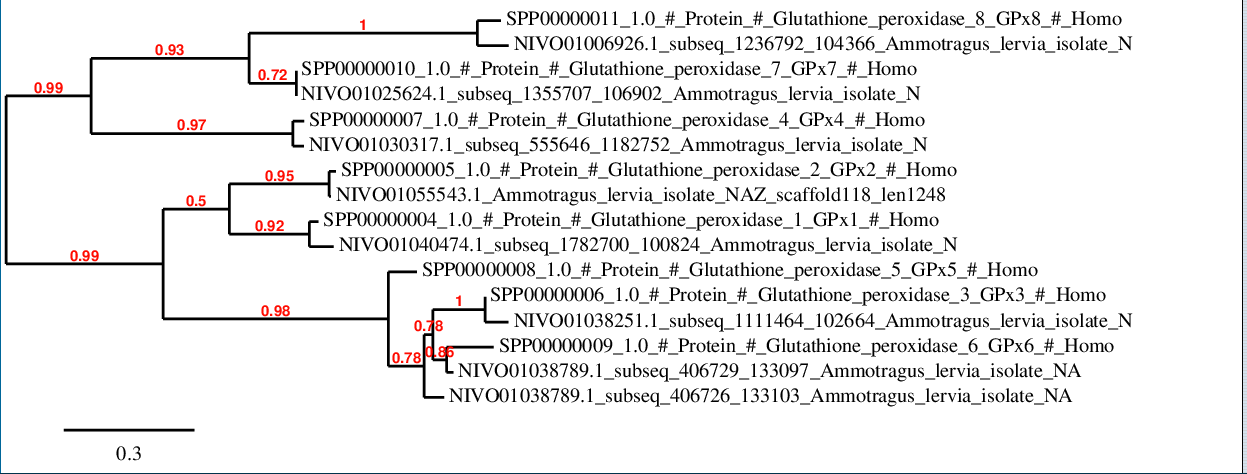
In this case, it is observed a notable grade of conservation between the protein sequences from human and those predicted in Ammotragus lervia. On the one hand, it is observed a closer relation between GPx4, GPx7 and GPx8 respect to the rest of family members. On the other hand, GPx1, GPx2, GPx3, GPx5 and GPx6 maintain a closer relation between them.
GPx1
In Ammotragus lervia, GPx1 gene is located in the scaffold NIVO01040474.1 between the position 1832678 and the position 1833524, in the positive strand. This gene has 2 exons (according to Exonerate output) detailed below:
- Exon 1: From position 1832679 to 1832912
- Exon 2: From position 1833174 to 1833524
A SECIS candidate was selected from 2 predicted by SECISearch3 [full output] in the 3’-UTR region between the positions 1833576 and 1833646 in the forward (+) strand. Seblastian predicts a selenoprotein that matches with 'PREDICTED: glutathione peroxidase 1 [Ovis aries musimon].'
GPx2
In Ammotragus lervia, GPx2 gene is located in the scaffold NIVO01055543.1 between the position 22496 and the position 19447, in the negative strand. This gene has 2 exons (according to Exonerate output) detailed below:
- Exon 1: From position 22496 to 22275
- Exon 2: From position 19795 to 19448
A SECIS candidate was selected from 10 predicted by SECISearch3 [full output] in the 3’-UTR region between the positions 19238 and 19174 in the reverse (-) strand. Seblastian predicts a selenoprotein that matches with 'glutathione peroxidase 2 [Bos taurus].'
GPx3
In Ammotragus lervia, GPx3 gene is located in the scaffold NIVO01038251.1 between the position 1168719 and the position 1161463, in the negative strand. This gene has 5 exons (according to Exonerate output) detailed below:
- Exon 1: From position 1168719 to 1168633
- Exon 2: From position 1164125 to 1163972
- Exon 3: From position 1162762 to 1162645
- Exon 4: From position 1162204 to 1162105
- Exon 5: From position 1161682 to 1161464
A SECIS candidate was selected from 3 predicted by SECISearch3 [full output] in the 3’-UTR region between the positions 1160853 and 1160780 in the reverse (-) strand. Seblastian predicts a selenoprotein that matches with 'PREDICTED: glutathione peroxidase 3 [Ovis aries musimon].'
GPx4
In Ammotragus lervia, GPx4 gene is located in the scaffold NIVO01030317.1 between the position 604591 and the position 606687, in the positive strand. This gene has 7 exons (according to Exonerate output) detailed below:
- Exon 1: From position 604592 to 604675
- Exon 2: From position 605646 to 605740
- Exon 3: From position 605818 to 605962
- Exon 4: From position 606101 to 606252
- Exon 5: From position 606364 to 606388
- Exon 6: From position 606529 to 606588
- Exon 7: From position 606661 to 606687
A SECIS candidate was selected from 5 predicted by SECISearch3 [full output] in the 3’-UTR region between the positions 606745 and 606817 in the forward (+) strand. Seblastian predicts a selenoprotein that matches with 'phospholipid hydroperoxide glutathione peroxidase, mitochondrial precursor [Capra hircus].'
GPx5
In Ammotragus lervia, GPx5 gene is located in the scaffold NIVO01038789.1 between the position 484731 and the position 489829, in the positive strand. This gene has 4 exons (according to Exonerate output) detailed below:
- Exon 1: From position 484732 to 484891
- Exon 2: From position 487812 to 487929
- Exon 3: From position 488411 to 488510
- Exon 4: From position 489626 to 489829
No SECIS candidate was predicted by SECISearch3 [full output] in the 3’-UTR region in the forward (+) strand. Seblastian predicts a selenoprotein that matches with 'PREDICTED: glutathione peroxidase 6 [Capra hircus].'
GPx6
In Ammotragus lervia, GPx6 gene is located in the scaffold NIVO01038789.1 between the position 461041 and the position 456728, in the negative strand. This gene has 4 exons (according to Exonerate output) detailed below:
- Exon 1: From position 461041 to 460885
- Exon 2: From position 459054 to 458937
- Exon 3: From position 457780 to 457681
- Exon 4: From position 456929 to 456729
A SECIS candidate was selected from 2 predicted by SECISearch3 [full output] in the 3’-UTR region between the positions 456240 and 456165 in the reverse (-) strand. Seblastian predicts a selenoprotein that matches with 'PREDICTED: glutathione peroxidase 6 [Capra hircus].'
GPx7
In Ammotragus lervia, GPx7 gene is located in the scaffold NIVO01025624.1 between the position 1405646 and the position 1412609, in the positive strand. This gene has 3 exons (according to Exonerate output) detailed below:
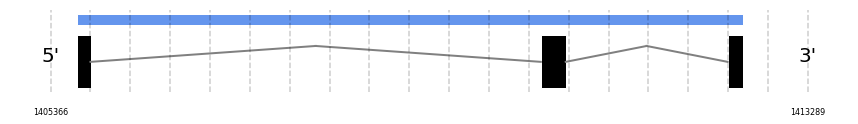
- Exon 1: From position 1405647 to 1405784
- Exon 2: From position 1410495 to 1410756
- Exon 3: From position 1412449 to 1412609
No SECIS candidate was predicted by SECISearch3 [full output] in the 3’-UTR region in the forward (+) strand. However no selenoprotein was predicted by Seblastian.
GPx8
In Ammotragus lervia, GPx8 gene is located in the scaffold NIVO01006926.1 between the position 1291158 and the position 1286791, in the negative strand. This gene has 3 exons (according to Exonerate output) detailed below:
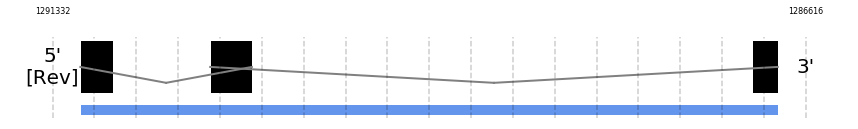
- Exon 1: From position 1291158 to 1290955
- Exon 2: From position 1290345 to 1290084
- Exon 3: From position 1286952 to 1286792
Any SECIS were predicted by SECISearch3 and Seblastian was not available to find any selenoprotein.
Methionine sulfoxide reductase A
MsrA
In Ammotragus lervia, MsrA gene is located in the scaffold NIVO01055729.1 between the position 63554 and the position 171582, in the positive strand. This gene has 4 exons (according to Exonerate output) detailed below:
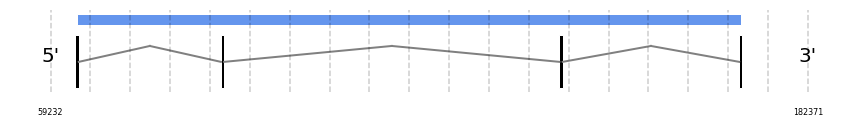
- Exon 1: From position 63555 to 63624
- Exon 2: From position 87198 to 87317
- Exon 3: From position 142240 to 142344
- Exon 4: From position 171473 to 171582
No SECIS candidate was predicted by SECISearch3 [full output] in the 3’-UTR region in the forward (+) strand. However no selenoprotein was predicted by Seblastian.
15-kDa selenoprotein
Sel15
In Ammotragus lervia, Sel15 gene is located in the scaffold NIVO01070360.1 between the position 721121 and the position 756771, in the positive strand. This gene has 4 exons (according to Exonerate output) detailed below:
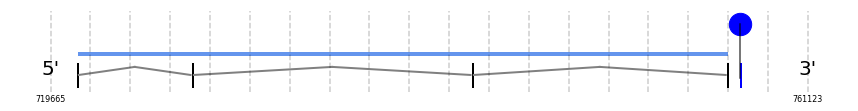
- Exon 1: From position 721122 to 721205
- Exon 2: From position 727420 to 727587
- Exon 3: From position 742748 to 742811
- Exon 4: From position 756716 to 756771
A SECIS candidate was selected from 2 predicted by SECISearch3 [full output] in the 3’-UTR region between the positions 757418 and 757495 in the forward (+) strand. Seblastian predicts a selenoprotein that matches with 'PREDICTED: 15 kDa selenoprotein, partial [Octodon degus].'
Selenoprotein H
SelH
In Ammotragus lervia, SelH gene is located in the scaffold NIVO01034600.1 between the position 5029013 and the position 5028438, in the negative strand. This gene has 3 exons (according to Exonerate output) detailed below:

- Exon 1: From position 5029013 to 5028889
- Exon 2: From position 5028805 to 5028660
- Exon 3: From position 5028536 to 5028439
A SECIS candidate was selected from 2 predicted by SECISearch3 [full output] in the 3’-UTR region between the positions 5027675 and 5027608 in the reverse (-) strand. Seblastian predicts a selenoprotein that matches with 'PREDICTED: selenoprotein H isoform X2 [Ovis aries musimon].'
Selenoprotein I
SelI
In Ammotragus lervia, SelI gene is located in the scaffold NIVO01032443.1 between the position 1395967 and the position 1433662, in the positive strand. This gene has 10 exons (according to Exonerate output) detailed below:
- Exon 1: From position 1395968 to 1396024
- Exon 2: From position 1413449 to 1413517
- Exon 3: From position 1413969 to 1414077
- Exon 4: From position 1416322 to 1416396
- Exon 5: From position 1420443 to 1420705
- Exon 6: From position 1421608 to 1421716
- Exon 7: From position 1427920 to 1427968
- Exon 8: From position 1429968 to 1430148
- Exon 9: From position 1431353 to 1431535
- Exon 10: From position 1433567 to 1433662
No SECIS candidate was predicted by SECISearch3 [full output] in the 3’-UTR region in the forward (+) strand. However no selenoprotein was predicted by Seblastian.
Selenoprotein K
SelK
In Ammotragus lervia, SelK gene is located in the scaffold NIVO01057317.1 between the position 1822121 and the position 1788813, in the negative strand. This gene has 5 exons (according to Exonerate output) detailed below:
- Exon 1: From position 1822121 to 1822103
- Exon 2: From position 1819008 to 1818918
- Exon 3: From position 1817445 to 1817362
- Exon 4: From position 1816793 to 1816704
- Exon 5: From position 1788814 to 1788814
A SECIS candidate was predicted by SECISearch3 [full output] in the 3’-UTR region between the positions 1816331 and 1816242 in the reverse (-) strand. Seblastian predicts a selenoprotein that matches with 'PREDICTED: selenoprotein K [Ceratotherium simum simum].'
Selenoprotein M
SelM
In Ammotragus lervia, SelM gene is located in the scaffold NIVO01044519.1 between the position 786172 and the position 783634, in the negative strand. This gene has 5 exons (according to Exonerate output) detailed below:
- Exon 1: From position 786172 to 786053
- Exon 2: From position 784573 to 784538
- Exon 3: From position 784340 to 784306
- Exon 4: From position 783946 to 783868
- Exon 5: From position 783790 to 783635
A SECIS candidate was selected from 6 predicted by SECISearch3 [full output] in the 3’-UTR region between the positions 783595 and 783523 in the reverse (-) strand. Seblastian predicts a selenoprotein that matches with 'PREDICTED: selenoprotein M, partial [Dasypus novemcinctus].'
Selenoprotein N
SelN
In Ammotragus lervia, SelN gene is located in the scaffold NIVO01052487.1 between the position 965129 and the position 975153, in the positive strand. This gene has 10 exons (according to Exonerate output) detailed below:
- Exon 1: From position 965130 to 965285
- Exon 2: From position 968029 to 968241
- Exon 3: From position 968420 to 968544
- Exon 4: From position 969120 to 969257
- Exon 5: From position 970830 to 970911
- Exon 6: From position 971530 to 971718
- Exon 7: From position 972091 to 972196
- Exon 8: From position 973263 to 973375
- Exon 9: From position 973467 to 973568
- Exon 10: From position 974986 to 975153
A SECIS candidate was selected from 2 predicted by SECISearch3 [full output] in the 3’-UTR region between the positions 976265 and 976338 in the forward (+) strand. However no selenoprotein was predicted by Seblastian.
Selenoprotein O
SelO
In Ammotragus lervia, SelO gene is located in the scaffold NIVO01039759.1 between the position 211008 and the position 230377, in the positive strand. This gene has 9 exons (according to Exonerate output) detailed below:
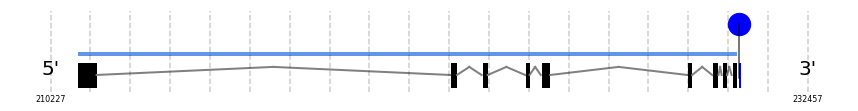
- Exon 1: From position 211009 to 211580
- Exon 2: From position 221955 to 222158
- Exon 3: From position 222879 to 223059
- Exon 4: From position 224157 to 224287
- Exon 5: From position 224616 to 224896
- Exon 6: From position 228912 to 229062
- Exon 7: From position 229649 to 229834
- Exon 8: From position 229927 to 230083
- Exon 9: From position 230219 to 230377
A SECIS candidate was predicted by SECISearch3 [full output] in the 3’-UTR region between the positions 230447 and 230517 in the forward (+) strand. Seblastian predicts a selenoprotein that matches with 'PREDICTED: selenoprotein O isoform X2 [Ovis aries musimon].'
Selenoprotein P
SelP
In Ammotragus lervia, SelP gene is located in the scaffold NIVO01048714.1 between the position 510433 and the position 516419, in the positive strand. This gene has 5 exons (according to Exonerate output) detailed below:
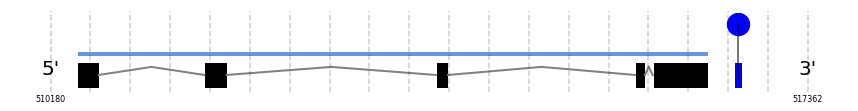
- Exon 1: From position 510434 to 510636
- Exon 2: From position 511638 to 511850
- Exon 3: From position 513833 to 513950
- Exon 4: From position 515724 to 515817
- Exon 5: From position 515890 to 516419
A SECIS candidate was selected from 7 predicted by SECISearch3 [full output] in the 3’-UTR region between the positions 516663 and 516734 in the forward (+) strand. Seblastian predicts a selenoprotein that matches with 'PREDICTED: selenoprotein P isoform X1 [Ovis aries].'
Methionine sulfoxide reductase B or Selenoprotein R

As it is shown, a remarkable correlation is maintained between Ammotragus lervia and Homo sapiens sequences.It seems that SelR2 and SelR3 are more closer between them in respect to SelR1.
SelR1
In Ammotragus lervia, SelR1 gene is located in the scaffold NIVO01014683.1 between the position 1290902 and the position 1287262, in the negative strand. This gene has 4 exons (according to Exonerate output) detailed below:
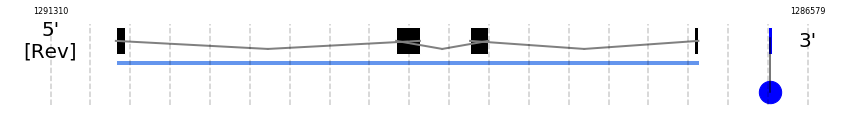
- Exon 1: From position 1290902 to 1290848
- Exon 2: From position 1289151 to 1289003
- Exon 3: From position 1288689 to 1288575
- Exon 4: From position 1287288 to 1287263
A SECIS candidate was predicted by SECISearch3 [full output] in the 3’-UTR region between the positions 1286816 and 1286746 in the reverse (-) strand. Seblastian predicts a selenoprotein that matches with 'PREDICTED: methionine-R-sulfoxide reductase B1 [Ovis aries].'
SelR2
In Ammotragus lervia, SelR2 gene is located in the scaffold NIVO01003597.1 between the position 1801149 and the position 1822586, in the positive strand. This gene has 5 exons (according to Exonerate output) detailed below:
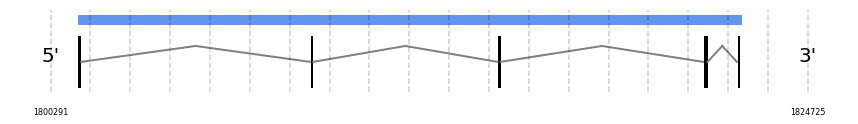
- Exon 1: From position 1801150 to 1801282
- Exon 2: From position 1808715 to 1808815
- Exon 3: From position 1814763 to 1814839
- Exon 4: From position 1821344 to 1821491
- Exon 5: From position 1822488 to 1822586
Any SECIS were predicted by SECISearch3 and Seblastian was not available to find any selenoprotein.
SelR3
In Ammotragus lervia, SelR3 gene is located in the scaffold NIVO01040223.1 between the position 1554578 and the position 1676368, in the positive strand. This gene has 5 exons (according to Exonerate output) detailed below:
- Exon 1: From position 1554579 to 1554712
- Exon 2: From position 1556327 to 1556404
- Exon 3: From position 1601107 to 1601135
- Exon 4: From position 1666298 to 1666395
- Exon 5: From position 1676207 to 1676368
A SECIS candidate was selected from 2 predicted by SECISearch3 [full output] in the 3’-UTR region between the positions 1718523 and 1718598 in the forward (+) strand. However no selenoprotein was predicted by Seblastian.
MSRB1
In Ammotragus lervia, MSRB1 gene is located in the scaffold NIVO01014683.1 between the position 1290902 and the position 1288557, in the negative strand. This gene has 3 exons (according to Exonerate output) detailed below:
- Exon 1: From position 1290902 to 1290848
- Exon 2: From position 1289151 to 1289003
- Exon 3: From position 1288689 to 1288558
A SECIS candidate was predicted by SECISearch3 [full output] in the 3’-UTR region between the positions 1286816 and 1286746 in the reverse (-) strand. Seblastian predicts a selenoprotein that matches with 'PREDICTED: methionine-R-sulfoxide reductase B1 [Ovis aries].'
MSRB2
In Ammotragus lervia, MSRB2 gene is located in the scaffold NIVO01003597.1 between the position 1808743 and the position 1822586, in the positive strand. This gene has 4 exons (according to Exonerate output) detailed below:
- Exon 1: From position 1808744 to 1808815
- Exon 2: From position 1814763 to 1814839
- Exon 3: From position 1821344 to 1821491
- Exon 4: From position 1822488 to 1822586
Any SECIS were predicted by SECISearch3 and Seblastian was not available to find any selenoprotein.
MSRB3
In Ammotragus lervia, MSRB3 gene is located in the scaffold NIVO01040223.1 between the position 1497075 and the position 1676368, in the positive strand. This gene has 6 exons (according to Exonerate output) detailed below:
- Exon 1: From position 1497076 to 1497172
- Exon 2: From position 1554604 to 1554712
- Exon 3: From position 1556327 to 1556404
- Exon 4: From position 1601107 to 1601135
- Exon 5: From position 1666298 to 1666395
- Exon 6: From position 1676207 to 1676368
A SECIS candidate was selected from 2 predicted by SECISearch3 [full output] in the 3’-UTR region between the positions 1718523 and 1718598 in the forward (+) strand. However no selenoprotein was predicted by Seblastian.
Selenoprotein S
SelS
In Ammotragus lervia, SelS gene is located in the scaffold NIVO01025374.1 between the position 1333749 and the position 1333179, in the negative strand. This gene has 1 exons (according to Exonerate output) detailed below:

- Exon 1: From position 1333749 to 1333180
A SECIS candidate was predicted by SECISearch3 [full output] in the 3’-UTR region between the positions 1332833 and 1332754 in the reverse (-) strand. However no selenoprotein was predicted by Seblastian.
Selenoprotein T
SelT
In Ammotragus lervia, SelT gene is located in the scaffold NIVO01008816.1 between the position 544233 and the position 568960, in the positive strand. This gene has 5 exons (according to Exonerate output) detailed below:
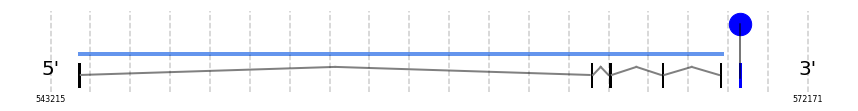
- Exon 1: From position 544234 to 544373
- Exon 2: From position 563892 to 564002
- Exon 3: From position 564610 to 564736
- Exon 4: From position 566621 to 566708
- Exon 5: From position 568839 to 568960
A SECIS candidate was selected from 13 predicted by SECISearch3 [full output] in the 3’-UTR region between the positions 569571 and 569645 in the forward (+) strand. However no selenoprotein was predicted by Seblastian.
Selenoprotein U

The phylogenetic tree shows the grade of correlation between the queries sequences and the predicted proteins is very high. Specifically, SelU1 and SelU3 maintain a higher correlation between them.
SelU1
In Ammotragus lervia, SelU1 gene is located in the scaffold NIVO01064043.1 between the position 30496 and the position 22805, in the negative strand. This gene has 5 exons (according to Exonerate output) detailed below:
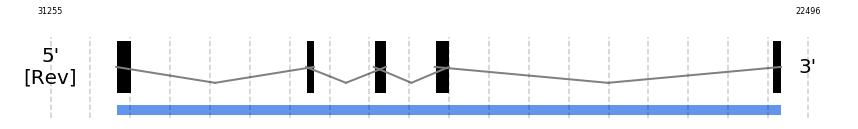
- Exon 1: From position 30496 to 30319
- Exon 2: From position 28300 to 28209
- Exon 3: From position 27516 to 27376
- Exon 4: From position 26811 to 26647
- Exon 5: From position 22916 to 22806
A SECIS candidate was selected from 5 predicted by SECISearch3 [full output] in the 3’-UTR region between the positions 20816 and 20733 in the reverse (-) strand. However no selenoprotein was predicted by Seblastian.
SelU2
In Ammotragus lervia, SelU2 gene is located in the scaffold NIVO01069066.1 between the position 35255 and the position 23477, in the negative strand. This gene has 6 exons (according to Exonerate output) detailed below:
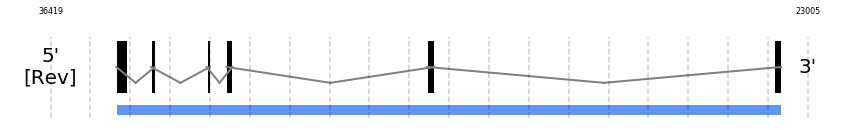
- Exon 1: From position 35255 to 35070
- Exon 2: From position 34640 to 34572
- Exon 3: From position 33678 to 33625
- Exon 4: From position 33303 to 33198
- Exon 5: From position 29754 to 29623
- Exon 6: From position 23602 to 23478
No SECIS candidate was predicted by SECISearch3 [full output] in the 3’-UTR region in the reverse (-) strand. However no selenoprotein was predicted by Seblastian.
SelU3
In Ammotragus lervia, SelU3 gene is located in the scaffold NIVO01017783.1 between the position 904227 and the position 901833, in the negative strand. This gene has 6 exons (according to Exonerate output) detailed below:
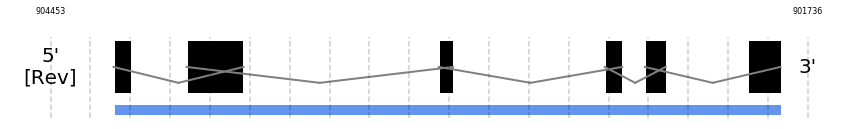
- Exon 1: From position 904227 to 904165
- Exon 2: From position 903965 to 903761
- Exon 3: From position 903060 to 903009
- Exon 4: From position 902465 to 902402
- Exon 5: From position 902321 to 902246
- Exon 6: From position 901952 to 901834
A SECIS candidate was selected from 12 predicted by SECISearch3 [full output] in the 3’-UTR region between the positions 855653 and 855581 in the reverse (-) strand. However no selenoprotein was predicted by Seblastian.
Selenoprotein V
SelV
In Ammotragus lervia, SelV gene is located in the scaffold NIVO01013187.1 between the position 388309 and the position 391391, in the positive strand. This gene has 5 exons (according to Exonerate output) detailed below:
- Exon 1: From position 388310 to 388370
- Exon 2: From position 388623 to 389367
- Exon 3: From position 390960 to 391032
- Exon 4: From position 391143 to 391217
- Exon 5: From position 391317 to 391391
No SECIS candidate was predicted by SECISearch3 [full output] in the 3’-UTR region in the forward (+) strand. However no selenoprotein was predicted by Seblastian.
Selenoprotein W

As it can be seen, the correlation between human and Ammotragus lervia is highly maintained in this selenoprotein family. Furthermore, it is observed a closer relation between SelV and SelW1 in the phylogenetic tree.
SelW1
In Ammotragus lervia, SelW1 gene is located in the scaffold NIVO01052466.1 between the position 360962 and the position 363398, in the positive strand. This gene has 5 exons (according to Exonerate output) detailed below:
- Exon 1: From position 360963 to 360991
- Exon 2: From position 362752 to 362776
- Exon 3: From position 362895 to 362948
- Exon 4: From position 363146 to 363220
- Exon 5: From position 363324 to 363398
A SECIS candidate was predicted by SECISearch3 [full output] in the 3’-UTR region between the positions 364588 and 364666 in the forward (+) strand. Seblastian predicts a selenoprotein that matches with 'PREDICTED: selenoprotein W, partial [Marmota marmota marmota].'
SelW2
In Ammotragus lervia, SelW2 gene is located in the scaffold NIVO01065774.1 between the position 5401081 and the position 5402051, in the positive strand. This gene has 4 exons (according to Exonerate output) detailed below:
- Exon 1: From position 5401082 to 5401170
- Exon 2: From position 5401275 to 5401372
- Exon 3: From position 5401807 to 5401883
- Exon 4: From position 5401971 to 5402051
Any SECIS were predicted by SECISearch3 and Seblastian was not available to find any selenoprotein.
Thioredoxin reductases
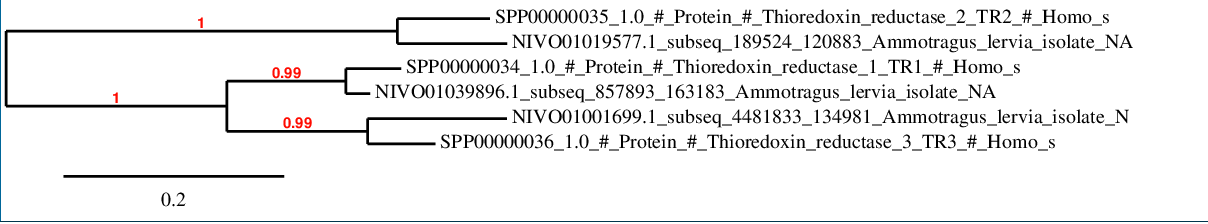
The phylogenetic tree shows as all the predicted proteins in Ammotragus lervia have a high grade of correlation with the corresponding ones in Homo sapiens.
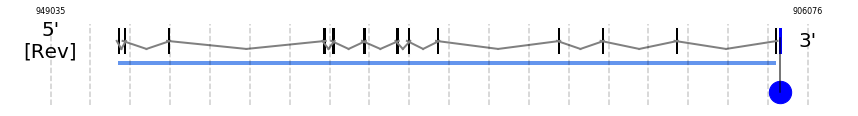
TR1
In Ammotragus lervia, TR1 gene is located in the scaffold NIVO01039896.1 between the position 945273 and the position 907892, in the negative strand. This gene has 13 exons (according to Exonerate output) detailed below:
- Exon 1: From position 945273 to 945187
- Exon 2: From position 944926 to 944854
- Exon 3: From position 942453 to 942334
- Exon 4: From position 933661 to 933519
- Exon 5: From position 933132 to 933017
- Exon 6: From position 931369 to 931144
- Exon 7: From position 929468 to 929376
- Exon 8: From position 928801 to 928725
- Exon 9: From position 927232 to 927076
- Exon 10: From position 920322 to 920215
- Exon 11: From position 917832 to 917737
- Exon 12: From position 913677 to 913543
- Exon 13: From position 907958 to 907893
A SECIS candidate was selected from 5 predicted by SECISearch3 [full output] in the 3’-UTR region between the positions 907662 and 907584 in the reverse (-) strand. Seblastian predicts a selenoprotein that matches with 'thioredoxin reductase 1, cytoplasmic [Bos taurus].'
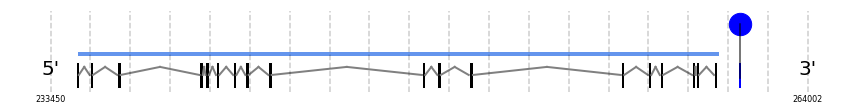
TR2
In Ammotragus lervia, TR2 gene is located in the scaffold NIVO01019577.1 between the position 234523 and the position 260407, in the positive strand. This gene has 18 exons (according to Exonerate output) detailed below:
- Exon 1: From position 234524 to 234602
- Exon 2: From position 235093 to 235116
- Exon 3: From position 236208 to 236273
- Exon 4: From position 239525 to 239581
- Exon 5: From position 239700 to 239841
- Exon 6: From position 240190 to 240264
- Exon 7: From position 240882 to 240960
- Exon 8: From position 241375 to 241437
- Exon 9: From position 242303 to 242373
- Exon 10: From position 248499 to 248518
- Exon 11: From position 249122 to 249213
- Exon 12: From position 250320 to 250494
- Exon 13: From position 256458 to 256594
- Exon 14: From position 257620 to 257715
- Exon 15: From position 258095 to 258187
- Exon 16: From position 259401 to 259472
- Exon 17: From position 259542 to 259639
- Exon 18: From position 260281 to 260407
A SECIS candidate was predicted by SECISearch3 [full output] in the 3’-UTR region between the positions 261256 and 261324 in the forward (+) strand. Seblastian predicts a selenoprotein that matches with 'thioredoxin reductase 2, mitochondrial precursor [Bos taurus].'
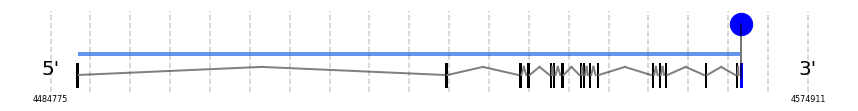
TR3
In Ammotragus lervia, TR3 gene is located in the scaffold NIVO01001699.1 between the position 4487940 and the position 4566814, in the positive strand. This gene has 17 exons (according to Exonerate output) detailed below:
- Exon 1: From position 4487941 to 4487988
- Exon 2: From position 4531836 to 4532048
- Exon 3: From position 4540633 to 4540693
- Exon 4: From position 4541592 to 4541701
- Exon 5: From position 4544355 to 4544459
- Exon 6: From position 4544673 to 4544745
- Exon 7: From position 4545640 to 4545759
- Exon 8: From position 4547904 to 4548046
- Exon 9: From position 4548215 to 4548330
- Exon 10: From position 4548930 to 4549155
- Exon 11: From position 4549844 to 4549936
- Exon 12: From position 4556418 to 4556494
- Exon 13: From position 4557230 to 4557386
- Exon 14: From position 4557999 to 4558106
- Exon 15: From position 4562734 to 4562829
- Exon 16: From position 4566475 to 4566609
- Exon 17: From position 4566749 to 4566814
A SECIS candidate was predicted by SECISearch3 [full output] in the 3’-UTR region between the positions 4566952 and 4567022 in the forward (+) strand. Seblastian predicts a selenoprotein that matches with 'thioredoxin reductase 3 [Oplegnathus fasciatus].'
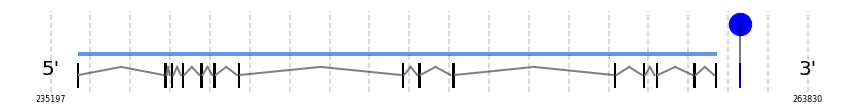
TXNRD2
In Ammotragus lervia, TXNRD2 gene is located in the scaffold NIVO01019577.1 between the position 236203 and the position 260407, in the positive strand. This gene has 15 exons (according to Exonerate output) detailed below:
- Exon 1: From position 236204 to 236273
- Exon 2: From position 239525 to 239581
- Exon 3: From position 239700 to 239841
- Exon 4: From position 240190 to 240264
- Exon 5: From position 240882 to 240960
- Exon 6: From position 241375 to 241437
- Exon 7: From position 242303 to 242373
- Exon 8: From position 248499 to 248518
- Exon 9: From position 249122 to 249213
- Exon 10: From position 250320 to 250494
- Exon 11: From position 256458 to 256594
- Exon 12: From position 257620 to 257715
- Exon 13: From position 258095 to 258206
- Exon 14: From position 259531 to 259639
- Exon 15: From position 260281 to 260407
A SECIS candidate was predicted by SECISearch3 [full output] in the 3’-UTR region between the positions 261256 and 261324 in the forward (+) strand. Seblastian predicts a selenoprotein that matches with 'thioredoxin reductase 2, mitochondrial precursor [Bos taurus].'
TXNRD2-UniProt
In Ammotragus lervia, TXNRD2-UniProt gene is located in the scaffold NIVO01019577.1 between the position 234532 and the position 260407, in the positive strand. This gene has 17 exons (according to Exonerate output) detailed below:
- Exon 1: From position 234533 to 234602
- Exon 2: From position 236208 to 236273
- Exon 3: From position 239525 to 239581
- Exon 4: From position 239700 to 239841
- Exon 5: From position 240190 to 240264
- Exon 6: From position 240882 to 240960
- Exon 7: From position 241375 to 241437
- Exon 8: From position 242303 to 242373
- Exon 9: From position 248499 to 248518
- Exon 10: From position 249122 to 249213
- Exon 11: From position 250320 to 250494
- Exon 12: From position 256458 to 256594
- Exon 13: From position 257620 to 257715
- Exon 14: From position 258095 to 258187
- Exon 15: From position 259401 to 259472
- Exon 16: From position 259542 to 259639
- Exon 17: From position 260281 to 260407
A SECIS candidate was predicted by SECISearch3 [full output] in the 3’-UTR region between the positions 261256 and 261324 in the forward (+) strand. Seblastian predicts a selenoprotein that matches with 'thioredoxin reductase 2, mitochondrial precursor [Bos taurus].'
TXNRD3
In Ammotragus lervia, TXNRD3 gene is located in the scaffold NIVO01001699.1 between the position 4540623 and the position 4566814, in the positive strand. This gene has 15 exons (according to Exonerate output) detailed below:
- Exon 1: From position 4540624 to 4540693
- Exon 2: From position 4541592 to 4541701
- Exon 3: From position 4544355 to 4544459
- Exon 4: From position 4544673 to 4544745
- Exon 5: From position 4545640 to 4545759
- Exon 6: From position 4547904 to 4548046
- Exon 7: From position 4548215 to 4548330
- Exon 8: From position 4548930 to 4549155
- Exon 9: From position 4549844 to 4549936
- Exon 10: From position 4556418 to 4556494
- Exon 11: From position 4557230 to 4557386
- Exon 12: From position 4557999 to 4558106
- Exon 13: From position 4562734 to 4562829
- Exon 14: From position 4566475 to 4566609
- Exon 15: From position 4566749 to 4566814
A SECIS candidate was predicted by SECISearch3 [full output] in the 3’-UTR region between the positions 4566952 and 4567022 in the forward (+) strand. Seblastian predicts a selenoprotein that matches with 'thioredoxin reductase 3 [Oplegnathus fasciatus].'
Selenoprotein machinery
Eukaryotic elongation factor
eEFSec
In Ammotragus lervia, eEFSec gene is located in the scaffold NIVO01001699.1 between the position 3621595 and the position 3511308, in the negative strand. This gene has 7 exons (according to Exonerate output) detailed below:
- Exon 1: From position 3621595 to 3621298
- Exon 2: From position 3576058 to 3575851
- Exon 3: From position 3568634 to 3568538
- Exon 4: From position 3567750 to 3567586
- Exon 5: From position 3529760 to 3529104
- Exon 6: From position 3525023 to 3524867
- Exon 7: From position 3511493 to 3511309
SECISearch3 and Seblastian was not available to find anything as it is not a selenoprotein, it is a protein involved in the selenoproteins machinery.
SECIS binding protein 2
SBP2
In Ammotragus lervia, SBP2 gene is located in the scaffold NIVO01049792.1 between the position 1600400 and the position 1635783, in the positive strand. This gene has 16 exons (according to Exonerate output) detailed below:
- Exon 1: From position 1600401 to 1600552
- Exon 2: From position 1605275 to 1605532
- Exon 3: From position 1605731 to 1605864
- Exon 4: From position 1607063 to 1607292
- Exon 5: From position 1610383 to 1610464
- Exon 6: From position 1612012 to 1612217
- Exon 7: From position 1614874 to 1614996
- Exon 8: From position 1616303 to 1616392
- Exon 9: From position 1617669 to 1617801
- Exon 10: From position 1624650 to 1624816
- Exon 11: From position 1625521 to 1625660
- Exon 12: From position 1626600 to 1626758
- Exon 13: From position 1627399 to 1627619
- Exon 14: From position 1634352 to 1634506
- Exon 15: From position 1634826 to 1635018
- Exon 16: From position 1635683 to 1635783
No SECIS candidate was predicted by SECISearch3 [full output] in the 3’-UTR region in the forward (+) strand. Seblastian was not available to find anything as it is not a selenoprotein, it is a protein involved in the selenoproteins machinery.
Selenophosphate synthetases
SPS1
In Ammotragus lervia, SPS1 gene is located in the scaffold NIVO01060204.1 between the position 73009 and the position 50691, in the negative strand. This gene has 8 exons (according to Exonerate output) detailed below:
- Exon 1: From position 73009 to 72817
- Exon 2: From position 67416 to 67313
- Exon 3: From position 65378 to 65271
- Exon 4: From position 64638 to 64484
- Exon 5: From position 62691 to 62601
- Exon 6: From position 60723 to 60624
- Exon 7: From position 52608 to 52396
- Exon 8: From position 50903 to 50692
No SECIS candidate was predicted by SECISearch3 [full output] in the 3’-UTR region in the reverse (-) strand. Seblastian was not available to find anything as it is not a selenoprotein, it is a protein involved in the selenoproteins machinery.
SPS2
In Ammotragus lervia, SPS2 gene is located in the scaffold NIVO01016939.1 between the position 905194 and the position 903847, in the negative strand. This gene has 1 exons (according to Exonerate output) detailed below:

- Exon 1: From position 905194 to 903848
A SECIS candidate was predicted by SECISearch3 [full output] in the 3’-UTR region between the positions 903275 and 903201 in the reverse (-) strand. Seblastian was not available to find anything as it is not a selenoprotein, it is a protein involved in the selenoproteins machinery.
Phosphoseryl-tRNA kinase
PSTK
In Ammotragus lervia, PSTK gene is located in the scaffold NIVO01034609.1 between the position 2485067 and the position 2471748, in the negative strand. This gene has 6 exons (according to Exonerate output) detailed below:
- Exon 1: From position 2485067 to 2484852
- Exon 2: From position 2482534 to 2482279
- Exon 3: From position 2481826 to 2481628
- Exon 4: From position 2478353 to 2478278
- Exon 5: From position 2475772 to 2475679
- Exon 6: From position 2471945 to 2471749
SECISearch3 and Seblastian was not available to find anything as it is not a selenoprotein, it is a protein involved in the selenoproteins machinery.
tRNA Sec 1 associated protein 1
SECp43
In Ammotragus lervia, SECp43 gene is located in the scaffold NIVO01076485.1 between the position 881203 and the position 860169, in the negative strand. This gene has 8 exons (according to Exonerate output) detailed below:
- Exon 1: From position 881203 to 881085
- Exon 2: From position 875113 to 875014
- Exon 3: From position 874402 to 874350
- Exon 4: From position 872087 to 871956
- Exon 5: From position 870444 to 870325
- Exon 6: From position 867422 to 867260
- Exon 7: From position 867054 to 867021
- Exon 8: From position 860279 to 860170
SECISearch3 and Seblastian was not available to find anything as it is not a selenoprotein, it is a protein involved in the selenoproteins machinery.
Selenocysteine synthase
SecS
In Ammotragus lervia, SecS gene is located in the scaffold NIVO01007502.1 between the position 1713228 and the position 1678486, in the negative strand. This gene has 11 exons (according to Exonerate output) detailed below:
- Exon 1: From position 1713228 to 1713115
- Exon 2: From position 1711744 to 1711590
- Exon 3: From position 1709857 to 1709739
- Exon 4: From position 1709426 to 1709268
- Exon 5: From position 1706796 to 1706643
- Exon 6: From position 1702591 to 1702489
- Exon 7: From position 1694305 to 1694176
- Exon 8: From position 1694050 to 1693959
- Exon 9: From position 1682031 to 1681938
- Exon 10: From position 1680201 to 1680111
- Exon 11: From position 1678784 to 1678487
No SECIS candidate was predicted by SECISearch3 [full output] in the 3’-UTR region in the reverse (-) strand. Seblastian was not available to find anything as it is not a selenoprotein, it is a protein involved in the selenoproteins machinery.
Discussion
During this project, a total of 38 proteins from Homo sapiens (obtained from SelenoDB 1.0) have been compared with the genome of Ammotragus lervia. From these 38 proteins: 25 are described as selenoproteins, 10 as Cystein-containing homologs, 1 as other amino acid-containing homologs and 2 as protein machinery involved in selenoproteins synthesis. The selenoproteome of Homo sapiens was chosen since both Homo sapiens and Ammotragus lervia are placental mammals and, therefore, are phylogenetically close. Moreover, another reason is that the Homo sapiens selenoproteome is the best annotated one.
Nevertheless, 8 proteins from Bos taurus have also been compared against Ammotragus lervia in order to obtain a more accurate prediction of these ones, given the closer phylogenetic relation between these two organisms.
All in all, a total of 46 proteins of the two species mentioned above have been compared to the genome of Ammotragus lervia to infer homology between them and, ultimately, characterize the selenoproteome of the studied species.
The reason why human has been chosen as the main reference species is due to the fact that its selenoproteome is much more accurately annotated in the SelenoDB 1.0 database. Although, a 2.0 version of this database provides a larger number of selenoproteins for human, the 1.0 version has been manually annotated which can provide a more reliable first approach to the selenoproteome of Ammotragus lervia. However, when looking for selenoprotein machinery proteins, in SelenoDB 1.0 some proteins present in the 2.0 version were missing. This is why in those cases where these proteins were missing the 2.0 version was used. Concretely, since Bos Taurus selenoproteome is annotated with a similar accuracy than the annotation of the selenoproteins of Homo sapiens, and there is less phylogenetic distance between Bos taurus and Ammotragus lervia, it was chosen to compare these selenoprotein machinery proteins from Bos taurus with the Ammotragus lervia genome.
Moreover, in some punctual cases, the UniProt sequence of the protein was searched and used since it was better annotated than in the SelenoDB database.
The discussion was carried out taking into account that, in order to be a functional selenoprotein, there must be the presence of a Sec residue, the prediction of SECIS elements in the 3’-UTR region, the presence of a Met residue at the first position of the predicted protein and a stop codon before the conserved Sec residue. Also, it was made sure that the gene locations of the different predicted proteins did not colocalize. Seblastian prediction was strongly taken into account when drawing conclusions.
Next, a brief discussion for each protein is provided in order to analyse and give possible explanations for the results obtained.
Note: Homo sapiens proteins have been used for the analysis except for when it is explicitly indicated the use of the Bos taurus selenoproteome.
Selenoproteins and Cys-homologs
Iodothyronine deiodinases (DIs)
The iodothyronine deiodinases (DI) regulate activation and inactivation of thyroid hormones. DI1 and DI2 convert the inactive form of thyroid hormone, thyroxine (T4), to the active one, 3,3′,5-triiodothyronine (T3). In turn, DI3 can inactivate T3 and T4 leading to formation of inactive T2 and reverse T3 (rT3). There are three Dio enzymes known in mammals, all of which contain Sec: DI1, DI2, DI3. The deiodinases possess a thioredoxin-fold and show significant intrafamily homology. Interestingly, all detected DI3 genes (including Dio3b) are intronless [1, 16].
DI1
DI1 has been found in the Ammotragus lervia genome, since a selenocysteine (Sec) aligns with the Sec residue of the human sequence. Moreover, a SECIS element has been located in the 3’-UTR region and also Seblastian prediction was positive for this protein. Although the conservation grade is quite low, especially in the N-terminal region, reason why the predicted protein does not start by a Met residue, no stop codon was found in the sequence. These findings lead to the conclusion that DI1 is also a selenoprotein in Ammotragus lervia.
DI2
For this protein, it can be reported its existance in Ammotragus lervia. Both a Sec in the protein sequence and SECIS element in the 3’-UTR region have been found. Furthermore, the protein sequence obtained stars with a Met residue and no stop codon was found before the end of the predicted protein. The conservation grade of the alignment between the two sequences was quite optimal. Nevertheless, no Seblastian prediction was obtained, which can be a case of false negative. However, our findings allow the confirmation of the presence of this protein in this species.
DI3
In this case, a properly conserved alignment has been found with the human protein. Both a Sec aligned residue and a SECIS element located in the 3’-UTR region have been detected. A remarkable conservation grade of the alignment between the two sequences was obtained. Moreover, as well as in DI2, the protein sequence contains a Met residue in the first position and no stop codons before the C-terminus. Seblastian prediction was positive. Therefore the presence of the DI3 protein in the Ammotragus lervia genome is hypothesized.
The high homology obtained strengthen the fact that there is a high conservation grade in human and Ammotragus lervia regarding to the member of the iodothyronine deiodinases family.
Glutathione peroxidases (GPx)
Glutathione peroxidases are the largest selenoprotein family in vertebrates. Mammals have 8 GPx homologs, 5 of which are selenoproteins: GPx1-4, GPx6; and three that are GPx homologous for Cys: GPx5, GPx7-8. Moreover, GPx6 homologs in some mammals are not selenoproteins and have a Cys in the active site. All mammalian Sec-containing GPx genes are highly conserved. GPxs play a wide range of physiological functions in organisms and are involved in hydrogen peroxide (H2O2) signaling, detoxification of hydroperoxides, and maintaining cellular redox homeostasis.
GPx1 is the most abundant selenoprotein in mammals and the first mammalian protein whose gene was found to contain Sec-encoding UGA in the open reading frame. GPx2 is primarily found in the epithelium of the gastrointestinal tract. whereas GPx3 is secreted primarily from kidney and is the major GPx form in plasma. GPx4 is expressed in a wide range of cell types and tissues, and GPx6 is only found in olfactory epithelium and during embryonic development [1, 16].
GPx1
The alignment against the human sequence shows a Sec residue conserved in the same position. Moreover, a SECIS element in the 3’-UTR region has been found. Although the predicted protein does not start by a Met residue, no stop codon was found in the sequence. Our finding also adjusts correctly to the Seblastian prediction. Therefore, it is predicted that GPx1 is part of the Ammotragus lervia selenoproteome.
GPx2
For GPx2, an alignment with a conserved Sec in the predicted protein sequence, which in the initial position contains a Met residue, has been obtained. A SECIS element in the 3’-UTR region has also been localized and no stop codon has been located before the C-terminus. Analyzing the T-Coffee output, a high conservation between both sequences was observed. The Seblastian prediction is also positive for this protein. All in all, it is proposed the presence of this protein in terms of a selenoprotein in Ammotragus lervia.
GPx3
In this case, a well conserved alignment has been found due to the conservation of a Sec residue in the same position as in the human sequence as well as the conservation of the Met in the first position. Furthermore, a SECIS element in the 3’-UTR region has also been localized, unlike the presence of a stop codon before the C-terminus, what allows the proposition of the existence of this selenoprotein in Ammotragus lervia. In this case, it has also been obtained the corresponding Seblastian prediction.
GPx4
As for the T-Coffee output for GPx4, it showed a conserved Sec residue and the alignment, where the protein started with Met and no stop codons were found before the C-terminus. The result of the SECISearch provided more than one possible SECIS element in the 3’-UTR region. Nevertheless, it has been chosen the closest to the gene sequence which presents an A grade. It can be concluded that there is a high likelihood for the existence of this protein in the selenoproteome of Ammotragus lervia.
GPx5 and GPx6
As it has been previously described, GPx5 and GPx6 are the most recently evolved GPxs, which appeared from a tandem duplication of GPx3 in placental mammals. Furthermore, no Sec-containing GPx5 has been identified in mammals [16]. Our findings match with these statements since the scaffolds in which GPx5 and GPx6 have been located are the same ones in Ammotragus lervia.
As for GPx5, a Cys residue has been found in the same position than the Sec residue in GPx6, what reinforces the tandem duplication event and the posterior lost of the Sec residue in GPx5. Moreover, no SECIS element in the 3’-UTR region nor Sec residue have been found for GPx5. Although the predicted protein did not start with a Met residue, the third residue is a Met, which could be the real start of the protein in Ammotragus lervia. These findings allow the proposal of this protein to be a Cys-containing homologous. Besides, as no Sec residue was found, no stop codon was found either. On the other hand, the output of the Seblastian prediction corresponds to GPx6 from another organism of the same subfamily (Caprinae) of Ammotragus lervia.
Due to these findings it can not be concluded if there is a GPx5 protein in Ammotragus lervia genome.
In GPx6, both a Sec residue and a SECIS element in the 3’-UTR region have been obtained. There was no stop codon before the C-terminus. However, the protein did not start with a Met residue and there was not any Met residue before the Sec, so it is hypothesised that a Met could be present in a position previous than the predicted start of the protein. Also, Seblastian predicted this protein in Ammotragus lervia. Therefore, all outputs predict the presence of this selenoprotein in this species genome.
GPx7 and GPx8
These two proteins have been also previously classified as Cys-containing homologous evolved from a GPx4-like selenoprotein ancestor. In human, both proteins are considered homologous for Cys [16]. Indeed, in Ammotragus lervia Cys residues are conserved in both T-Coffee alignments what confirms the previous findings in mammals. Moreover, no Seblastian prediction was positive in neither of the two proteins.
Although, a SECIS element was found for GPx7, the fact that it doesn’t have a Sec residue lead us to propose this protein as a Cys-containing homologous in Ammotragus lervia. As well, the same conclusion can be draw regarding to GPx8. Moreover, the predicted GPx7 although did not contain a Met residue in the first position, did contain this residue in the second position, therefore this seconds position is proposed as the start of the GPx7 protein in Ammotragus lervia. In the case of the GPx8, the predicted protein already started with a Met residue. In both cases, as there was not any Sec, nor there was any stop codon in the protein sequence.
Regarding to the conservation, in all eight sequences, a highly conservation grade of the alignment between the two sequence was obtained. This matches the high conservation described in the literature of these proteins.
Methionine sulfoxide reductase A (MsrA)
MsrA, along with MsrBs, is a repA protein that protects cells against oxidative stress and has been implicated in delaying the aging process and protecting against neurodegeneration. It catalyses the reduction of the s-form of MetSO (Met-s-SO) residues in proteins to methionine. It can also reduce free Met-s-SO [17].
This protein is classified as a Cys-containing homologous in human. In the case of Ammotragus lervia, the Cys residue is conserved in the alignment. Moreover, as no Sec residue is found in the predicted protein sequence, no stop codon can be found. However, the protein does not contain a Met residue in the first position, although there is, in fact, a Met a few positions beyond the predicted start of the protein in the alignment, which could be the real start of the protein. All in all, we propose the presence of the MsrA protein in Ammotragus lervia as a Cys-containing homologous.
Nevertheless, the global alignment between both organisms is not completed at the start and at the end of the sequences. Our hypothesis is that this can be due to the fact that in the annotation of genomes, these extreme regions tend to be more difficult to annotate. For this reason, the middle region aligns correctly but not the other ones.
15-kDa selenoprotein (Sel15)
Sep15 was identified experimentally as a 15-kDa selenoprotein of unknown function. Later this protein was proposed to mediate the cancer prevention effect of dietary and regulation of redox homeostasis in the ER. Expression of this selenoprotein was detected in a wide range of mammalian tissues, the highest level of expression were in prostate, liver, kidney, and testis [1].
This selenoprotein has been also found in Ammotragus lervia since a Sec residue is conserved in the alignment with the human sequence. Also, SECISearch found a SECIS element in the 3’-UTR region. The Seblastian software predicted a selenoprotein structure that matches the one obtained with the Exonerate program. Its sequence, although does not start with a Met residue, contains one Met in the fourth position, which could be the actual start of the Sel15 protein. Moreover, there are not any additional UGA codons, apart from the one coding for the Sec, so there are no stop codons inside the protein sequence. These findings allow the prediction of the presence of this selenoprotein in Ammotragus lervia.
Selenoprotein H (SelH)
SelH belongs to the Rdx family of selenoproteins along with SelW, SelT, and SelV. These proteins have a thioredoxin-like fold and are characterized by the presence of a conserved Cys-x-x-Sec motif. SelH has a unique subcellular localization pattern, it localizes specifically in the nucleoli. SelH possesses glutathione peroxidase activity and it is implicated in the regulation of transcription of a group of genes that are involved in de novo glutathione synthesis and phase II detoxification enzymes [1].
The T-Coffee output shows how the predicted SelH presents a Sec residue conserved in the alignment with human sequence, and a SECIS element properly located in the 3’-UTR regions has been found by SECISearch. Furthermore, the Seblastian prediction adjusts notably with the one obtained with the Exonerate program. Moreover, the predicted sequence starts with the Met residue and does not contain any stop codon before the C-terminus. Due to these findings, it can be concluded that SelH is a selenoprotein in Ammotragus lervia.
Selenoprotein I (SelI)
Selenoprotein I (SelI) is a recently evolved selenoprotein, which is found only in vertebrates.It contains a highly conserved CDP-alcohol phosphatidyltransferase domain. There are three aspartic acids, which are critical for function. The entire active region is highly similar between SelI and its homologs. The most prominent difference between SelI and its homologs is a C-terminal extension in SelI, which contains Sec [1].
The alignment against human shows a high conservation grade of the SelI protein, a finding that agrees with what it described in the literature. The Sec residue is found, concretely, in the C-terminal region of the protein, matching previously findings in other studies. Moreover, the predicted protein starts with a Met and does not contain a stop codon before the C-terminus. Interestingly, only one SECIS element has been predicted with the SECISearch software. However, this SECIS element is located between the two first exons, whereas in the Homo sapiens homologous protein the SECIS element is located in the 3’-UTR region. Moreover, Seblastian did not predict any protein. Therefore, it can not be concluded that this protein functions as a selenoprotein in Ammotragus lervia, although the protein sequence is highly conserved.
Selenoprotein K (SelK)
SelK, along with SelS, is localized in the ER membrane and belongs to a type III group of transmembrane proteins that contain a single transmembrane domain, with the COOH-terminal end of the protein facing the cytosol. SelK has been implicated in ER-associated degradation (ERAD) of misfolded proteins. The ERAD machinery consists of multiprotein complexes that are involved in recognition, ubiquitination, and translocation of protein substrates from the ER to the cytosol, and their subsequent degradation by the ubiquitin and proteasome system [1].
In the T-Coffee output, a conserved Sec has been identified in the alignment. Although for this protein, the Exonerate program predicted 5 exons, the last one only contained one residue and was located very far from the previous exon. This is the reason why we have not considered it to be a real exon. Moreover, Seblastian output shows a prediction of 4 exons what matches with our hypothesis. Taking into account these considerations, a SECIS element has been correctly found in the 3’-UTR region. Moreover, the protein starts with the Met residue and does not contain any stop codons before the C-terminus. The conservation grade between the two protein is remarkable high. All in all, and assuming our consideration, it could be proposed the presence of SelK in the Ammotragus lervia selenoproteome, although it cannot be strongly affirmed.
Selenoprotein M (SelM)
SelM, along with Sel15, is a thioredoxin-like fold protein localized in the ER. Unlike the Sel15 tissue expression, SelM is highly expressed on the brain. Due to the high expression of SelM in the brain, several studies investigated the possible role of SelM in neuroprotection. Overexpression of SelM in neuronal cells prevented oxidative damage induced by H2O2 treatment, whereas knockdown of SelM using shRNA caused decreased cell viability and a strong apoptotic cell death [1].
For SelM, both a conserved Sec residue and a 3’-UTR region located SECIS element have been found. Moreover, it contains a Met residue in the first position and no stop codon before the C-terminus. Also, Seblastian prediction matches notably with our findings with Exonerate. Due to all these findings the existence of the SelM as a selenoprotein in Ammotragus lervia is proposed.
Selenoprotein N (SelN)
SelN is a transmembrane glycoprotein expressed in the ER that is highly expressed during embryonic development and has a lower expression in adult tissues. Some studies revealed that it has an important role in skeletal muscle. SelN interacts with RyR which is a calcium channel that mediates the release of Ca2+ from the sarcoplasmic reticulum during muscle contraction. This interaction suggests that this selenoprotein might serve as a cofactor of RyR and might be involved in regulation of intracellular calcium mobilization [1].
In human, SelN presents two residues of Sec. Nevertheless, only one of these seems to be conserved in Ammotragus lervia. On the other hand, a SECIS element in 3’-UTR region has been found for this protein. The two proteins do not align in one region of the protein, probably due to a deletion in Ammotragus lervia. In fact, the Sec residue that is not present in the studied species is located in this region that has been deleted. However, when looking at the Bos taurus SelN sequence in SelenoDB 2.0, it can be seen how only one Sec is present, which matches our findings in Ammotragus lervia. Moreover, the predicted protein does contain a Met residue in the first position. Besides, there is no stop codon before the C-terminus. In conclusion, these findings support the existence of the SelN in the Ammotragus lervia selenoproteome.
Selenoprotein O (SelO)
SelO is the largest mammalian selenoprotein with orthologs found in a wide range of organisms, including bacteria and yeast. SelO contains only a Sec residue located in the antepenultimate position at the COOH-terminal end of the protein. However, the majority of SelO homologs contains a Cys residue instead of Sec. It is localized inside the mitochondria. The finding of the CXXU (Cys and Sec separated by two other residues) motif in the human sequence of SelO suggested that, like other selenoproteins, SelO may have redox activity [1, 18].
When comparing the human sequence of SelO with the Ammotragus lervia genome, a high conservation rate has been observed. The Sec residue is conserved in the C-terminal region and SECIS element in the 3’-UTR region has also been found. Moreover, the predicted protein starts with a Met and does not contain any stop codons before the C-terminus. Seblastian prediction was also positive. This allows the prediction of the presence of this selenoprotein in Ammotragus lervia.
Selenoprotein P (SelP)
SelP is abundantly secreted, accounting almost 50% of the total Se in plasma. The SelP has recently evolved, and SelP homologs are found predominantly in vertebrates. A unique feature is the presence of multiple Sec residues on its sequence. The fact that SelP is secreted into the plasma and the presence of multiple Sec residues suggested that this selenoprotein might function as a Se supplier to peripheral tissues [1].
In this case, the comparison between sequences showed a slightly less conserved protein in comparison to other conservation rates obtained when predicting other selenoproteins. Out of the 10 Sec found in Homo sapiens, in the predicted protein in Ammotragus lervia 8 Sec have been found, and the other two left have been replaced by a Cys residue. Although one of these 8 Sec residues could actually be a stop codon, the fact that all of them align with the corresponding Sec in the Homo sapiens sequence leads us to assume that there are no stop codons in the protein before the C-terminus.
SECISearch was positive since a SECIS element in the 3’-UTR region has been obtained. Moreover, the Seblastian prediction matches with the Exonerate prediction, where the predicted protein starts with a Met. In conclusion, there is a strong likelihood for the presence of SelP in the Ammotragus lervia selenoproteome.
Methionine sulfoxide reductase B or Selenoprotein R (MSRB or SelR)
MSRBs, along with MsrA, are MetSO reductases (Msrs) which reduce back methionine sulfoxide (MetSO) residues to methionine protecting cells from oxidative damage. In contrast withs MsrA, MsrB can only reduce the r-form (Met-r-SO) and neither the s-form and free ree Met-s-SO. In mammals, there are three MsrBs, of which MsrB1 is a selenoprotein and MsrB2 and MsrB3 are Cys-containing isozymes [17].
SelR1/ MSRB1
The Sec-containing MsrB1 protein is the major MsrB in mammals, which is primarily localized in the cytosol and nucleus. It has the highest activity in mammalian liver and kidney among three known MsrB enzymes [17].
The alignment of the predicted protein against human SelR1 shows almost a complete homology, where the Sec residue is conserved and the corresponding SECIS element in 3’-UTR region is predicted. Moreover, the Seblastian prediction matches also with the one made with the Exonerate program. Plus, the predicted protein contains a Met residue in its first position and does not contain any additional UGA codons before the C-terminus apart from the one coding for the Sec residue.
The same results regarding to all the considerations mentioned in the previous paragraph were obtained when performing the alignment against the Bos taurus protein MSRB1. Indeed, the same gene location in the same scaffold was obtained in both comparisons. In conclusion, these findings confirm the presence of SelR1 in Ammotragus lervia.
SelR2 /MSRB2
MsrB2 contains a Cys residue in place of the Sec residue in the enzyme's active site and is localized in mitochondria [17].
This protein is classified as a Cys-containing homologous [16], what matches the finding in the alignments against Homo sapiens and Bos taurus. In both comparisons, Cys residues are conserved in Ammotragus lervia and no Sec residue, and therefore no stop codon, are found in the protein.
When comparing to the human sequence, the grade of conservation in the alignment was not so high and this is the reason for comparing it also against Bos taurus. It can be seen that in Bos taurus, the conservation grade is very high with only a couple of substitutions. Furthermore, as expected, no SECIS elements were predicted when comparing in both species and Seblastian did not predict any selenoprotein.
Interestingly the alignments with human and Bos taurus have been obtained in the same scaffold in Ammotragus lervia. Therefore we can highly predict the presence of this protein in this species.
SelR3/MSRB3
MsrB3 also contains a Cys residue in place of Sec in the enzyme's active site and is targeted to the ER [17].
In the alignment against Homo sapiens, the N-terminal and the C-terminal regions do not align properly. For this reason, we proceeded to perform an alignment against Bos taurus in which the conservation was almost complete. Nevertheless, when comparing against both species, the Cys residues were conserved in Ammotragus lervia, what reaffirms the condition of Cys-containing homologous of this protein in this organism also. Moreover, the predicted protein contains a Met residue in the first position and no stop codon before the C-terminus. Seblastian did not predict any selenoprotein.
As in the case of SelR2, the alignments with human and Bos taurus have been obtained in the same scaffold of the Ammotragus lervia genome.
SECIS elements in this scaffold were found in the 3’-UTR. However, as there is no Sec residue, it can only be concluded the presence of this protein as a Cys-containing homologous.
Selenoprotein S (SelS)
SelS, along with SelK commented before, is localized in the ER membrane and belongs to a type III group of transmembrane proteins that contain a single transmembrane domain, with the COOH-terminal end of the protein facing the cytosol. SelS, same as SelK, has been implicated in ER-associated degradation (ERAD) of misfolded proteins, which has been explained before [1].
When comparing the Homo sapiens sequence of SelS with the Ammotragus lervia predicted SelS sequence, a high conservation rate has been observed just for the first exon, since no more exons have been identified in Ammotragus lervia. Interestingly, the Sec residue found in the human sequence is conserved but, however, in Ammotragus lervia two more possible Sec residues are present in the predicted sequence, which align in the human sequence with an Arg and a Glu residue. These Sec residues, which are coded by a UGA codon, could actually be stop codons. In fact, it cannot be affirmed nor denied that these codons are stop codons. However, based on the fact that in the human sequence there are not Cys residues in these positions, which could meant that in the ancestral organism there were Sec in these positions and that in the human they were replaced by Cys whereas in Ammotragus lervia they continued to be Sec, we propose that it is more likely that these UGA are stop codons rather than code for Sec residues.
Therefore, although, the predicted sequence starts with a Met residue and a SECIS element in the 3’-UTR region has been found, we consider that this selenoprotein is not present in Ammotragus lervia, which matches the fact that Seblastian was not able to predict this protein.
Selenoprotein T (SelT)
SelT, along with SelW, SelH, and SelV, belong to the Rdx family of selenoproteins. As each member of this family, it possess a thioredoxin-like fold and is characterized by the presence of a conserved Cys-x-x-Sec motif. This selenoprotein is mostly localized to the ER and Golgi. Some experiments with SelT knockouts suggested the role of SelT in the regulation of Ca2+ homeostasis and neuroendocrine function. More recently, SelT has been also implicated in the regulation of pancreatic β-cell function and glucose homeostasis [1].
It has been obtained a highly conserved alignment against the human sequence. Both a Sec residue and a SECIS element in 3’-UTR region have been found. Moreover, the predicted protein starts with a Met residue and does not contain a stop codon before the C-terminus. Therefore, these results lead to confirm the conservation of SelT as one more selenoprotein in Ammotragus lervia. However, Seblastian did not predict this selenoprotein but we hypothesise that it can be due to the specificity of the software and the fact that this finding could be a false negative case.
Selenoprotein U (SelU)
The members of this family are designated as Cys-containing homologous at least in Homo sapiens. Mammals contain three Cys-containing SelU proteins (SelU1-3). It is believed that the Cys-containing SelU1 protein in mammals evolved from the Sec-containing SelU sequences in other species like fish [16].
SelU1, SelU2 and SelU3
In Ammotragus lervia, the Cys residue is conserved in the same position as in the human sequence for all these proteins. The alignment presents a highly grade of conservation with few residue substitutions in the three cases. Plus, Seblastian prediction was negative in the three cases. Moreover, none of the three predicted proteins contain a stop codon before the C-terminus and all of them start with a Met residue except for the predicted SelU2. However, we hypothesize that there might be an AUG upstream of the the predicted start of the coding sequence in SelU2. Therefore, and doing this last assumption, these findings suggests the phylogenetic stability of this protein family in mammals and the presence of the three of them as Cys-containing homologous in Ammotragus lervia.
Selenoprotein V (SelV)
SelV is one of the least characterized selenoproteins. It recently evolved, most likely by duplication from SelW, and is found only in placental mammals. SelV is larger than SelW due to the presence of an additional NH2-terminal domain. SelV expression is detected only in testes, this indicates that may be involved in male reproduction, but its specific function is not known [16].
In this case, the Sec residue has been replaced by a Ser one. This finding matches with the fact that SelV has been previously described as the least conserved mammalian selenoprotein, that likely arose from a duplication of SelW in the placental stem [16]. However, the alignment against Homo sapiens presents a notable conservation on N- and C-terminal regions. Neither Seblastian prediction nor SECISearch were positive.
Therefore, it can be concluded that SelV is not a selenoprotein in Ammotragus lervia. However, the question that arises is whether the predicted protein could be a Ser-containing homologous or, instead, it is another type protein or not a protein at all. On the one hand, the fact that the protein does not contain any stop codon before the C-terminus and does start with a Met residue makes it plausible for it to be an homologous. However, the low homology in the alignment makes it hard to assert it. Therefore, further analysis must be carried out.
When comparing the scaffolds found for SelW1 and SelV, there is one present in both scaffold predictions made with tblastn. However, this scaffold was discarded in both cases during the analysis because other scaffold provided a better output for each protein.
Selenoprotein W (SelW)
Selenoprotein W, belongs to the Rdx family as explained before. It is highly conserved among mammalian species, with the Sec residue present in the N-terminal region as part of the -CXXU- redox motif of the Rdx family. SelW expression is regulated by selenium levels and is ubiquitously expressed in different tissues having the highest levels in muscle and brain. Its physiological function remain unknown.
There are two SelW proteins, SelW1 and SelW2 which are considered to form part of the ancestral vertebrate selenoproteome. SelW1 is a highly conserved selenoprotein on vertebrates whereas SelW2 is only a selenoprotein in some vertebrates like fishes. In mammals SelW2 is a Cys-containing homologous [1, 16, 19].
SelW1
In Ammotragus lervia the Sec residue is conserved in respect to the human sequence, and a SECIS element is properly located in the 3’-UTR region. Also, the alignment starts with Met and has no stop codons before the C-terminus. Plus, Seblastian prediction was positive. Therefore, we can highly predict the existence of SelW1 in the selenoproteome of this species.
SelW2
A Cys conserved residue has been found in Ammotragus lervia in the alignment against Homo sapiens. Moreover, the predicted proteins contains a Met residue in the first position and no stop codons before the C-terminus. As expected, neither SECISearch nor Seblastian gave positive outputs. These findings confirm the condition of Cys-containing homologous for SelW2 in this species, as it is in human.
Thioredoxin reductases (TR or TXNRD)
TRs control the redox state of thioredoxins, key proteins involved in redox regulation of cellular processes, comprising the major disulfide reduction system of the cell. Mammals have three TR isozymes: cytosolic TR1, mitochondrial TR3, and TGR all of which are Sec-containing proteins. All of them have a Sec residue in the COOH-terminal penultimate position. Both TR1 and TR3 are present in all vertebrates in contrast TR2 is only present in mammals. TR2 is also considered a glutathione reductase due to the presence of an additional glutaredoxin (Grx) domain located at the NH2-terminal part of the protein [1].
TR1
Thioredoxin reductase 1, TR1, is primarily localized in the cytosol and nucleus [1]. The alignment against Homo sapiens provided the conserved Sec residue in Ammotragus lervia. Moreover, the predicted protein starts with a Met residue and does not contain a stop codon before the C-terminus. Besides, SECISearch predicted a SECIS element in 3’-UTR, what finally confirms the presence of this selenoprotein in Ammotragus lervia.
TR2/TXNRD2
When comparing Ammotragus lervia genome against both Homo sapiens the conservation grade was not considered to be optimal enough. That is why we also compared Ammotragus lervia genome against Bos taurus thus obtaining a better conserved alignment. Indeed, the scaffold that provided the best output was the same in both comparisons.
Moreover, when comparing with Homo sapiens, the predicted protein in Ammotragus lervia did not start with a Met residue. However, when comparing with Bos taurus, the predicted protein and TXNRD2 (Bos taurus) started with the same residue, an Ala.
In order to test the possibility of a non-correct annotation in SelenoDB 2.0, the protein sequence of TXNRD2 (Bos taurus) was obtained from the UniProt database. Interestingly, the alignment provided by T-Coffee with this sequence started with a Met residue and did not have any stop codon before the C-terminus. Considering this, it can be concluded that TR2/TXNRD2 is present in the selenoproteome of Ammotragus lervia.
Independently, in all the performed alignments, a Sec residue is conserved in Ammotragus lervia. Also, a SECIS element in the 3’-UTR region has been found and Seblastian prediction was positive.
TR3/TXNRD3
Thioredoxin reductase 3, TR3 is localized in the mitochondria, where it is involved in reduction of mitochondrial thioredoxin [1].
In this case, a comparison with Homo sapiens and Bos taurus has been also performed due to the same reason as in TR2/TXNRD2. Specifically, in the alignment obtained with Bos taurus, both the predicted protein and the one of Bos taurus started with a Ser residue. However, when comparing with the human sequence, it is seen that the predicted protein starts upstream this Ser residue in the N-terminal region. A possible explanation for this finding, could be a non-correct annotation of TXNRD3 in Bos taurus. In fact, in SelenoDB 2.0 the query provided doesn’t start with a Met residue. This could also explain the findings obtained with TR2/TXNRD2.
Furthermore, when doing the same process than in the TR2 protein, the sequence in UniProt had a low annotation score (2/5) and also had a Ser in the first position. This is why comparison against the sequence present in UniProt was not done, as the same output than the already obtained would have been gotten.
All in all, in both comparisons, a Sec residue is conserved in Ammotragus lervia where no stop codon is present before the C-terminus. Furthermore, a SECIS element located in the 3’-UTR region has been found with SECISearch. Moreover, Seblastian prediction was positive. As in the previous protein (TR2/TXNRD2), the scaffold that provided the best output was the same in both comparisons. It can be concluded that it is likely that the TR3 protein is present in the selenoproteome of Ammotragus lervia, however, as it does not start with a Met residue, it cannot be fully affirmed that it is a functional selenoprotein in this species.
Selenoprotein machinery
Eukaryotic elongation factor (eEFSec)
Sec-specific translation elongation factor (eEFSec) is a machinery Selenoprotein which, recruits Sec-tRNA and facilitates incorporation of Sec to the protein which is being synthesised, along with SBP2. eEFSec contains a unique COOH-terminal extension named domain IV, this IV domain is proposed to be involved in interactions with SBP2 and the long variable arm of tRNA [1].
Regarding to the eEFSec protein, the alignment obtained by the T-Coffee program when comparing with the Homo sapiens eEFSec showed that the predicted protein in Ammotragus lervia is quite conserved. As in the human protein, there is no Sec residue as well as no SECIS element prediction. Moreover, the predicted protein contains a Met residue in the first position and no stop codon before the C-terminus. Hence, all these findings let us predict with a high likelihood the presence of the eEFSec protein as selenoprotein machinery in the Ammotragus lervia genome.
SECIS binding protein 2 (SBP2)
SBP2 is a machinery selenoprotein which together with eEFSec helps on the recruiting of SEC-tRNA. SBP2 contains three distinct domains, an NH2-terminal domain whose function is not known, a Sec incorporation domain (SID) in the middle of the protein, and a COOH-terminal RNA-binding domain (RBD). With the RBD domain it binds to SECIS core, this binding is necessary for Sec insertion into the protein sequences which is being synthesised [1].
As for the SBP2 protein, when comparing against the SBP2 human protein the T-Coffee alignment showed less conservation than most of the other proteins analyzed. In fact, no Met residue in the predicted sequence is aligned with the initial Met of the human protein, although a few residues downstream the predicted start of the protein there is a Met that could be the actual start of the protein. As expected, neither a SECIS element was predicted nor a Sec residue present in the predicted sequence protein. Accordingly, we propose that SBP2 could be present as selenoprotein machinery in the indicated positions in the genome of Ammotragus lervia. However, it cannot be firmly asserted that SBP2 is a functional protein since it is not clear that a Met residue is in the first position.
Selenophosphate synthetases (SPS)
There are two selenophosphate synthetase proteins, SPS1 and SPS2. This selenophosphate is necessary for Sec biosynthesis. Diferents analysis revealed that SPS2 could generate selenophosphate in vitro, whereas SPS1 could not indicating that SPS2 is required for de novo synthesis of selenophosphate, while SPS1 may have a possible role in Sec recycling. While SPS1 is not a selenoprotein, SPS2 is itself a selenoprotein. This finding indicates that SPS2 possibly serves as an autoregulator of selenoprotein synthesis [1].
SPS1
The output of the T-Coffee program showed an extremely high conservation grade, with just one mismatch, when comparing the predicted protein with the one of Homo sapiens. This matches the described high conservation described in the literature for this protein [16]. Moreover, the alignment shows how the Tyr is conserved and is present in the predicted protein. Besides, as expected, no SECIS element or Sec residue were found and the predicted protein contains a Met residue in the first position and no stop codon before the C-terminus. These findings endorse our proposition of the presence of the SPS1 protein in terms of a Tyr-containing homologous.
SPS2
Regarding to the SPS2 protein, when aligning the predicted protein against the human SPS2, the T-Coffee output shows a relatively high conservation, although not as much as in the case of the SPS1 protein. Both the Sec residue is conserved and a SECIS element is predicted in the 3’-UTR region. Furthermore, the predicted proteins starts with a Met residue and there is no stop codon before the C-terminus. This allows us to conclude with a high likelihood that the SPS2 protein is part of the selenoproteome of Ammotragus lervia. However, this does not match with Seblastian prediction, which could not predict a selenoprotein.
Phosphoseryl-tRNA[Ser]Sec kinase (PSTK)
Phosphoseryl-tRNA[Ser]Sec kinase is the enzyme that phosphorylates the precursor Serine of the Sec joined to the tRNA, Ser-tRNA[Ser]Sec, giving as a result O-phosphoseryl-tRNA[Ser]Sec. Therefore, this enzyme enables the formation of the Sec residue [1].
In this case, the alignment has been performed against Bos taurus and as it can be observed, the grade of conservation is almost complete. Plus, the predicted protein starts with a Met residue and since it does not have any Sec residues, there is no stop codon in the protein sequence. Moreover, all Cys residues are conserved in Ammotragus lervia which matches with the fact that PSTK is classified as a Cys-containing homologous in the SelenoDB database. On the other hand, no SECIS element and Seblastian prediction have been obtained. This was expected considering the Cys homologous condition of PSTK.
tRNA Sec 1 associated protein 1 (SECp43)
SECp43 was characterized as RNA-binding protein and it was believed that it formed a complex with Sec tRNA[Ser]Sec. Later it was discovered that SECp43 has a role in regulation of selenoprotein expression and participates on the pathway of selenoprotein biosynthesis. Moreover, the nuclear subcellular localization of SECp43 suggests that it may regulate the translocation of the Sec-tRNA[Ser]Sec complex between the nucleus and cytoplasm [20].
The output obtained by the T-Coffee shows an alignment completely conserved in comparison with Bos taurus. SECp43 is classified as Cys-containing homologous what matches with our results. Furthermore, no SECIS element and Sebastian prediction have been obtained. Plus, there are not any stop codons before the C-terminus.
Nevertheless, it does not start with a Met residue. A possible explanation for this finding could be a non-correct annotation of this protein in Bos taurus in SelenoDB 2.0 database, where the Bos taurus protein does not start with a Met residue. However, a few residues beyond the start of the alignment, there is one Met residue so it can be hypothesized that, in fact, this Met residue is the first position of the predicted protein.
Selenocysteine synthase (SecS)
SecS is a pyridoxal phosphate (PLP)-containing protein which converts the serine of the tRNA into selenocysteyl-tRNA[Ser]Sec by incorporating selenophosphate [1].
When comparing the Bos taurus SecS with the Ammotragus lervia genome, the alignment of the T-Coffee program showed a considerably high conservation grade. Moreover, the predicted protein contains a Met residue at the first position. As expected, no Sec residue nor SECIS in the 3’-UTR region element were found. These findings allow us to propose the presence of the SecS protein, in terms of selenoprotein machinery.
Conclusions
The aim of this project was to identify and annotate the selenoproteins and the machinery required for their synthesis encoded in Ammotragus lervia genome. A total of 38 proteins from Homo sapiens and 8 proteins from Bos taurus have been compared with an homology-based approach with the genome of Ammotragus lervia using bioinformatic programs. All in all, a total of 46 proteins have been used to infer homology and, ultimately, characterize the selenoproteome of the studied species.
From a total of 46 proteins, the following characterization has been obtained:
- Selenoproteins: DI1, DI2, DI3, GPx1, GPx2, GPx3, GPx4, GPx6, Sel15, SelH, SelK, SelM, SelN, SelO, SelP, SelR1/MSRB1, SelR3/MSRB3, SelT, SelW1, TR1, TR2/TXNRD2, TR3/TXNRD3, SPS2
- Cysteine-containing homologous: GPx5, GPx7, GPx8, MsrA, SelR2/MSRB2, SelU1, SelU2, SelU3, SelW2, PSTK, SECp43
- Other amino acid-containing homologous: SPS1
- Selenoprotein machinery*: eEFSec, SBP2, SPS1, SPS2, PSTK, SECp43, SecS
- Non-predicted selenoproteins: SelI, SelS, SelV
*Note: some of the selenoprotein machinery proteins are also classified as selenoproteins or cys-containing homologous.
As it can be observed, 43 out of the 46 proteins have an homologous in Ammotragus lervia. Concretely, 23 selenoproteins, 11 cys-containing homologous, 1 other amino acid-containing homologous and 7 selenoprotein machinery proteins have been identified. On the contrary, it has not been found the homologous of 3 proteins (SelI, SelS, SelV).
Further research is needed in order to correctly confirm the presence of some proteins for which a Met residue could not be located near the start of the predicted protein. These were Gx1, GPx6, SelU2 and SECp43. Moreover, some of the proteins which in this study are considered to be in Ammotragus lervia were not predicted by Seblastian, which was attributed to the high specificity of this software and, therefore, to possible false negatives. These were DI2, SelR3/MSRB3, SelT and SPS2. Furthermore, both SelN and SelP contained less Sec residues in the predicted protein in Ammotragus lervia in comparison to the human sequence.
All in all, we consider that a relatively high number of proteins have been characterised in Ammotragus lervia genome. Additionally, we believe our findings represent a small contribution to the current knowledge
of selenoproteins and to the scientific community.
References
[1] Labunskyy VM, Hatfield DL, Gladyshev VN. Selenoproteins: Molecular Pathways and Physiological Roles. Physiol Rev. 2014;94(3):739–77.
[2] Arner ES. Selenoproteins: what unique properties can arise with selenocysteine in place of cysteine? Exp Cell Res. 2010;316(8):1296–303.
[3] Hatfield DL, Choi IS, Ohama T, Jung JE, Diamond AM. Selenocysteine tRNA isoacceptors as central components in selenoprotein biosynthesis in eukaryotes. In: Selenium in Biology and Human Health, edited by Burk RF. New York: Springer-Verlag, 1994, p. 25–44.
[4] Xu XM, Turanov AA, Carlson BA, Yoo MH, Everley RA, Nandakumar R, Sorokina I, Gygi SP, Gladyshev VN, Hatfield DL. Targeted insertion of cysteine by decoding UGA codons with mammalian selenocysteine machinery. Proc Natl Acad Sci USA. 2010;107(50): 21430–34.
[5] Low SC, Berry MJ. Knowing when not to stop: selenocysteine incorporation in eukaryotes. Trends Biochem Sci. 1996;21(6):203-8.
[6] Böck A, Rother M, Leibundgut M, Ban N. Selenium metabolism in prokaryotes. In: Selenium: Its Molecular Biology and Role in Human Health, edited by Hatfield DL, Berry MJ, and Gladyshev VN. New York: Springer, 2006, p. 9–28.
[7] Budiman ME, Bubenik JL, Miniard AC, Middleton LM, Gerber CA, Cash A, Driscoll DM. Eukaryotic initiation factor 4a3 is a selenium-regulated RNA-binding protein that selectively inhibits selenocysteine incorporation. Mol Cell. 2009;35(4):479-89.
[8] Lobanov AV, Fomenko DE, Zhang Y, Sengupta A, Hatfield DL, Gladyshev VN. Evolutionary dynamics of eukaryotic selenoproteomes: large selenoproteomes may associate with aquatic life and small with terrestrial life. Genome Biol . 2007;8(9):R198.
[9] Lobanov AV, Hatfield DL, Gladyshev VN. Reduced reliance on the trace element selenium during evolution of mammals. Genome Biol . 2008;9(3):R62.
[10] SelenoDB: selenoproteins database [Internet]. Selenodb.org. 2017 [cited 21 November 2017]. Available from: http://www.selenodb.org
[11] Flohe L, Gunzler WA, Schock HH. Glutathione peroxidase: a selenoenzyme. FEBS Lett. 1973;32(1):132-134.
[12] Cassinello J. Ammotragus lervia: a review on systematics, biology, ecology and distribution. Ann Zoo Fenn. 1998;35(3):149-62.
[13] Barbary sheep videos, photos and facts - Ammotragus lervia | Arkive [Internet]. Arkive. 2017 [cited 16 November 2017]. Available from: http://www.arkive.org/barbary-sheep/ammotragus-lervia/
[14] Cassinello J, Cuzin F, Jdeidi T, Masseti M, Nader I, Smet K. Ammotragus lervia: The IUCN Red List of Threatened Species. 2008: e.T1151A3288917.
[15] Barbary sheep | Zoo Barcelona [Internet]. Zoo.keclab.com. 2017 [cited 17 November 2017]. Available from: http://zoo.keclab.com/en/animals/barbary-sheep
[16] Mariotti M, Ridge P, Zhang Y, Lobanov A, Pringle T, Guigo R et al. Composition and Evolution of the Vertebrate and Mammalian Selenoproteomes. PLoS ONE. 2012;7(3):e33066.
[17] Aachmann F, Sal L, Kim H, Marino S, Gladyshev V, Dikiy A. Insights into Function, Catalytic Mechanism, and Fold Evolution of Selenoprotein Methionine Sulfoxide Reductase B1 through Structural Analysis. J. Biol. Chem. 2010;285(43):33315-33323.
[18] Han S, Lee B, Yim S, Gladyshev V, Lee S. Characterization of Mammalian Selenoprotein O: A Redox-Active Mitochondrial Protein. PLoS ONE. 2014;9(4):e95518.
[19] Papp L, Lu J, Holmgren A, Khanna K. From Selenium to Selenoproteins: Synthesis, Identity, and Their Role in Human Health. Antioxidants & Redox Signaling. 2007;9(7):775-806.
[20] Xu X, Mix H, Carlson B, Grabowski P, Gladyshev V, Berry M et al. Evidence for Direct Roles of Two Additional Factors, SECp43 and Soluble Liver Antigen, in the Selenoprotein Synthesis Machinery. J. Biol. Chem. 2005;280(50):41568-41575.
Acknowledgments
We would like to thank everyone whose help made this project possible.
First, we would like to thank our tutor, Fernando Cid, for guiding us throughout the process of doing this project. What's more, we would like to thank the rest of the tutors for also helping us in certains aspects, resolving our doubts and answering our questions.
Next, we would like to thank our lecturers Cedric Notredame and Roderic Guigó for giving us the theoretical bases essential for carring out this project.
Furthermore, we would like to acknowledge the help given by our seminar teacher Robert Castelo who taught us how to program in shell, a tool which allowed us to automate parts of this project, thus, simplifying it. On the other hand, we would like to thank again Cedric Notredame for teaching us how to do BLAST searches. Besides, we are grateful for Toni Gabaldón's teaching in webpage building, as well as genome annotations and genome browsers, and for encouraging us to use other resources at our disposal (eg Python, Jupiter Notebooks ...).
Last but not least, we would like to highlight the help given by the rest of our classmates since we all helped each other regardless of being in different groups.

Contact us
The authors of this project are four students from 4th grade of the Human Biology degree at Pompeu Fabra University. We would be delighted to answer any doubts or provide you with more information regarding to this project. Please, do not hesitate to contact us!
- Florencia Evelin Alonso: florenciaevelin.alonso01@estudiant.upf.edu
- Beatriz Calvo Serra: beatriz.calvo04@estudiant.upf.edu
- César Jessé Enríquez-Rodrigez: cesarjesse.enriquez01@estudiant.upf.edu
- Carolina Paola Frías del Pino: carolinapaola.frias01@estudiant.upf.edu