Annex: Methodology detailed description
This annex describes in detail all the steps carried out by the bash program for the exploration of potential selenoproteins in Ammotragus lervia and all the steps followed for the acquisition of SECIS.
Bash program
Module loading
In order to obtain all the programs (Blast, Exonerate, GeneWise, etc) and the paths required for this analysis their modules need to be loaded using the following commands:
module load modulepath/goolf-1.7.20
module load BLAST+/2.2.30-goolf-1.7.20
module load Exonerate/2.2.0-goolf-1.7.20
module load T-Coffee/11.00.8cbe486-goolf-1.7.20
export PATH=/cursos/20428/BI/bin:$PATH
export PATH=/cursos/20428/BI/soft/genewise/x86_64/bin:$PATH
export WISECONFIGDIR=/cursos/20428/BI/soft/genewise/x86_64/wise2.2.0/wisecfg/
1. tBLASTn predicition (BLAST)
tBLASTn is a BLAST (Basic Local Alignment Search Tool) program which converts the query sequence into a nucleotide one and uses an heuristic algorithm to align this sequence to another one from a database, in our case the Ammotargus lervia genome. Giving as a result the regions with high-homology between both sequences. The command required is:
export WISECONFIGDIR=/cursos/20428/BI/soft/genewise/x86_64/wise2.2.0/wisecfg/
Where:
- PROT-human.fa, is the query sequence.
- bd_ammotragus.fa, is the genome of Ammotragus lervia.
- blast_PROT.fa, is the output of tBLASTn, being PROT the abbreviation of the protein name.
For an easier understanding of the results it is useful to tabulate them by using the -outfmt 6 command and was saved in table.fa.
tblastn –query PROT-human.fa –db bd_ammotragus.fa –out blast_PROT.fa -outfmt 6 -out table.fa
The output of the tBLASTn is a list of scaffolds, regions of high-homology between the genome and the query, some of them with more than one hit. Each hit has its origin and end, a percentage of identity, an e-value, as well as other parameters.
2. Extraction of fetch of interest (Fastafetch)
For this step it is needed an index of the Ammotargus lervia genome, which was provided to us by our bioinformatics teachers following the next command:
/cursos/20428/BI/genomes/2017/Genus_species/genome.index
Fastafetch is done in order to extract the genomic region that may contain the studied gen. Each region of an adequate scaffold will be introduced on a fasta file. We considered a scaffold adequate if one or more of its hits had both an e-value equal or minor to 0,001 and an identity equal or higher than 50%. Being the e-value (expect value) a parameter that describes the number of hits one can expect to see by chance as good as the hit observed; and the % identity the proportion of identical residues between two sequences.:
The command required is:
fastafetch /cursos/20428/BI/genomes/2017/Ammotragus_lervia/genome.fa bd_ammotragus.index contig > contig.fetch
Where:
- /cursos/20428/BI/genomes/2017/Ammotragus_lervia/genome.fa is the Ammotragus lervia genome.
- bd_ammotragus.index, is the genome index.
- contig, is the ID of the scaffold.
- contig.fetch, is the output name, being “contig” the ID of the scaffold.
3. Generation of a subseq in fasta format (Fastasubseq)
Is executed with the purpose to delimit the region of the selected sacaffold. The required command is:
fastasubseq -f contig.fetch -s origin -l length > contig.subseq
Where:
- contig.fetch, is the output of Fastafetch
- origin, corresponds to the lowest nucleotide of all the scaffolds hits, minus 50.000 nucleotides if possible.
- length, corresponds to the difference between the lowest nucleotide and the highest of all the scaffolds hits, plus 50.000 nucleotides if possible
- contig.subseq, is the output being contig the ID of the selected scaffold
4. Exonerate prediction (Exonerate)
This program predicts the gene by comparing the region obtained with fastasubseq with the human protein sequence.
exonerate --exhaustive yes -m p2g --showtargetgff -q PROT-human.fa -t contig.subseq >PROT-contig.exonerate
Where:
- --exhaustive yes, forces the programm to do its function exhaustively.
- -m p2g, makes possible the comparison between the protein and the genome.
- --showtargetgff, allows us to get the output information in a GFF file format.
- -q, indicates that the file used is in a FASTA format.
- PROT-human.fa, is the query sequence.
- -t, indicates the target, the sequences obtained from the Fastasubseq.
- contig.subseq, is the output form the Fastasubseq.
- contig.exonerate, is the output of the exonerate, being “contig” de ID of the scaffold.
Due to the big amount of time that this step took, the Exonerate of some protein were made by removing the command --exhaustive. This proteins were SBP2, SelP, SPS1, SPS2, SelU1, SelU3 and MsrA.
The final step of the Exonerate is to extract the exons, from the first outcome, on a GFF format. The next command is required:
egrep -w exon PROT-contig.exonerate > PROT-contig.exonerate.gff
Where:
- egrep -w exon, extracts the exons regions
- PROT-contig.exonerate, is the outcome from the first step
- PROT-contig.exonerate.gff, is the final outcome, being PROT the abbreviation of the protein name and contig the ID of the selected scaffold
5. Translation into protein format (FastafromGFF + Fastatranslate)
FastafromGFF
Extracts the cDNA sequence encoded on the exonerate outcome and introduces it in a file. The command required is:
fastaseqfromGFF.pl PROT-contig.subseq PROT-contig.exonerate.gff > PROT-contig.nt.fa
Where:
- PROT-contig.subseq, is the output of Fastasubseq.
- PROT-contig.exonerate.gff, is the second output of Exonerate.
- PROT-contig.nt.fa, is the outcome of FastafromGFF, being “PROT” the abbreviation of the protein name and “contig” the ID of the scaffold.
Fastatranslate
This program translates the cDNA sequence into an amino acids one. The command required is:
fastatranslate -f PROT-contig.nt.fa -F 1 > PROT-contig.prot
Where:
- -f is followed by the exon sequences.
- PROT-conting.nt.fa, is the outcome of FastafromGFF.
- -F indicates the reading frame, which is 1 in this case.
- PROT-contig.prot, is the output being “PROT” the abbreviation of the protein name, and “contig” the ID of the scaffold.
6. Prediction alignment (T-coffee)
It is a program that compares the predicted protein, of Ammotragus lervia, obtained with Fasta translate with the human protein (query). The output represents the most probable alignment and the homology between both sequences.
Inside the sequence of the obtained protein with Fasta translate the selenocysteines are represented with an “*”, it is required to replace this symbol with an “X” to be able to use it on T-coffee.
The command required is:
t_coffee PROT-contig.prot PROT-human.fa > PROT-contig.tcoffee_exo 2>../tcoffe_trash
Where:
- PROT-contig.prot, is the output of Fasta translate.
- PROT-human.fa, is the human query sequence.¡
- PROT-contig.tcoffee_exo is the output being PROT the abbreviation of the protein name and contig the ID of the scaffold.
7. Genewise prediction (Genewise)
Is another program that predicts the protein sequences encoded on the Ammotragus lervia genome. It was used to validate the results from Exonerate.
The command required is:
genewise -pep -pretty -cdna -gff -both PROT-human.fa /cursos/20428/BI/genomes/2017/Ammotragus_lervia/genome.fa >PROT-contig.gen
Where:
- -pep, is used to show the predicted peptide sequence.
- -pretty
- -cdna, is used to show the predicted cDNA sequence.
- -gff, indicates that the output is on a GFF format.
- PROT-human.fa, is the query sequence.
- /cursos/20428/BI/genomes/2017/Ammotragus_lervia/genome.fa, is the genome of Ammotragus lervia.
- PROT-contig.gen, is the output of Genewise, being PROT the human protein abbreviation and contig the ID of the scaffold.
SECIS acquisition
SECIS are elements associated with selenocysteine proteins which are essential for their synthesis. This elements are located in the 3’ -UTR region. The prediction of the SECIS has been done by using two programs: Seblastian and SECISearch3.
SECISearch3 looks for SECIS elements and Seblastian use it for search SECIS elements and try to find a selenoprotein. For this reason them were used to confirm the selenocysteine proteins found with the previous programs. Both programs use as input the fastasubseq sequences without header line (‘>’). Next image shows the paremeters used.
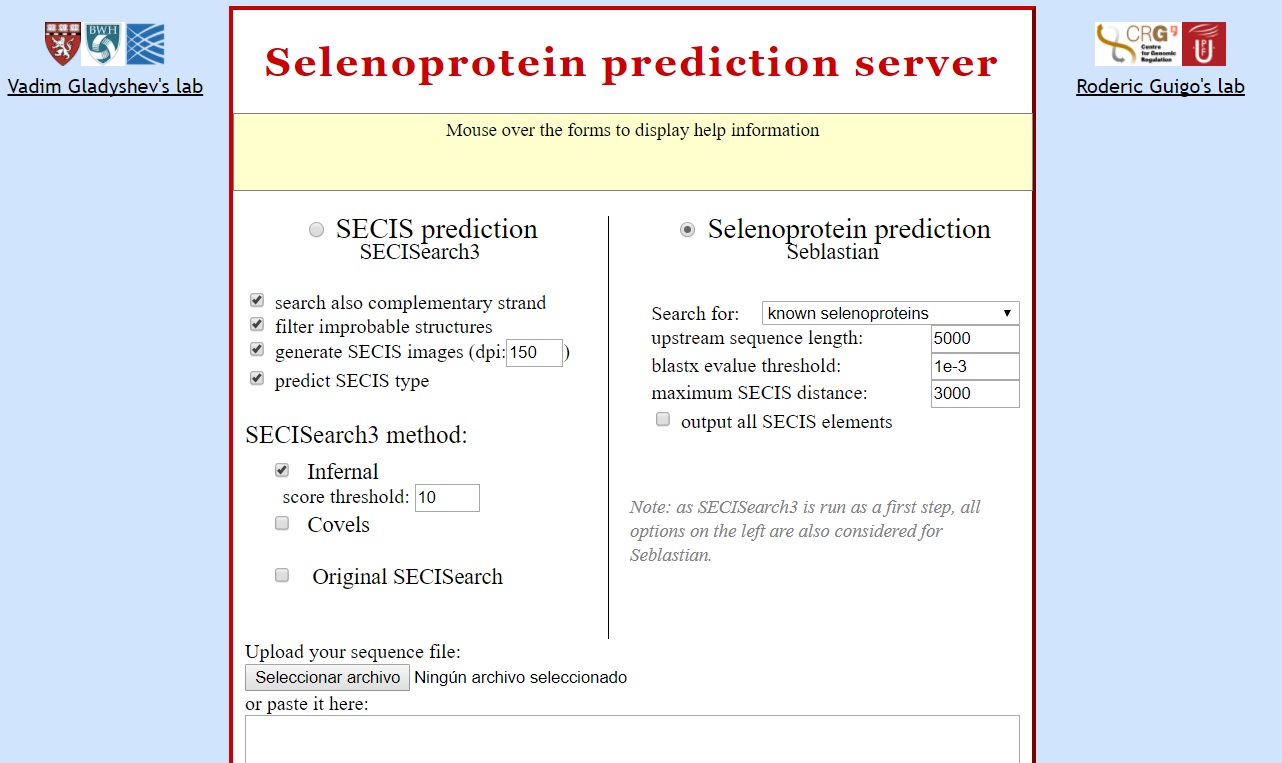
Some times these programs predict more than one possible SECIS, in order to choose the more adequate one some considerations must be followed:
- The first one is that the SECIS must be on the 3’-UTR of the gen.
- Secondly the SECIS has to be localized on the same strand (+ or -) as the coding gen for the selenocysteine protein.
The output provided by SECISearch3 we saved were:
- File “Sequence and structure of all SECIS elements” saved as PROT-contig.secis
- SECIS Images were named as PROT-conting.secis.*#.png,
where:
- PROT is the abbreviation of the protein name
- Conting is the Scaffold ID
- * is the direction of the strand (+ or -)
- # is the number of SECI.
The output provided by Seblastian we use was:
- “summary of predictions” file. It was named PROT-conting.seblast
where:
- PROT is the abbreviation of the protein name
- conting is the Scaffold ID.