Results
The following tables show, for each query, if it contains Sec, its tblastn output, the exonerate prediction for the protein, the T-Coffee alignment for the best prediction and the SECIS element prediction by Seblastian and SECISearch3. The legend used is showed below:

Selenoproteins
This table shows the presence of the genes encoding selenoproteins.
| Protein ID | Specie | Sec | BLAST | Exonerate | Scaffold | Gene location | T-Coffee | Seblastian | SECIS | SECIS Image |
|---|---|---|---|---|---|---|---|---|---|---|
| Sel15 |  |
 |
 |
 |
JSZB01012017.1 | 41463 - 85564 (-) |  |
 |
 |
 |
| GPx1 |  |
 |
 |
 |
JSZB01017608.1 | 46967 - 47182 (+) |  |
 |
 |
 |
| GPx2 |  |
 |
 |
 |
JSZB01005938.1 | 70515 - 73386 (+) |  |
 |
 |
 |
| GPx3 |  |
 |
 |
 |
JSZB01000211.1 | 246170 - 253336 (-) |  |
 |
 |
 |
| GPx4 |  |
 |
 |
 |
JSZB01061009.1 | 54870 - 56708 (+) |  |
 |
 |
 |
| GPx5 |  |
 |
 |
 |
JSZB01021545.1 | 158717 - 159004 (-) |  |
 |
 |
 |
| GPx6* |  |
 |
 |
 |
JSZB01002610.1 | 385174 - 388654 (+) |  |
 |
 |
 |
| GPx7 |  |
 |
 |
 |
JSZB01029812.1 | 57016 - 64287 (-) |  |
 |
 |
 |
| GPx8 |  |
 |
 |
 |
JSZB01002610.1 | 384416 - 388678 (+) |  |
 |
 |
 |
| DIO1 |  |
 |
 |
 |
JSZB01004892.1 | 152874 - 165770 (+) |  |
 |
 |
 |
| DIO2 |  |
 |
 |
 |
JSZB01004262.1 | 175841 - 184376 (-) |  |
 |
 |
 |
| DIO3 |  |
 |
 |
 |
JSZB01002288.1 | 210800 - 211633 (+) |  |
 |
 |
 |
| SelenoH |  |
 |
 |
 |
JSZB01009980.1 | 76621 - 77201 (-) |  |
 |
 |
 |
| SelenoI |  |
 |
 |
 |
JSZB01006584.1 | 33790 - 72380 (-) |  |
 |
 |
 |
| SelenoK |  |
 |
 |
 |
JSZB01010020.1 | 261356 - 266607 (-) |  |
 |
 |
 |
| SelenoM |  |
 |
 |
 |
JSZB01032570 | 87232 - 89457 (-) |  |
 |
 |
 |
| SelenoN |  |
 |
 |
 |
JSZB01007029.1 | 160268 - 170742 (+) |  |
 |
 |
 |
| SelenoO |  |
 |
 |
 |
JSZB01004690.1 | 99173 - 106964 (+) |  |
 |
 |
 |
| SelenoP |  |
 |
 |
 |
JSZB01008296.1 | 517465 - 523153 (-) |  |
 |
 |
 |
| MSRB1 |  |
 |
 |
 |
JSZB01066426.1 | 221 - 343 (+) |  |
 |
 |
 |
| MSRB2 |  |
 |
 |
 |
JSZB01014487.1 | 180874 - 192373 (-) |  |
 |
 |
 |
| MSRB3 |  |
 |
 |
 |
JSZB01006555.1 | 411790 - 536660 (-) |  |
 |
 |
 |
| SelenoS |  |
 |
 |
 |
JSZB01022157.1 | 328024 - 335603 (-) |  |
 |
 |
 |
| SelenoT |  |
 |
 |
 |
JSZB01003988.1 | 477781 - 495283 (-) |  |
 |
 |
 |
| SelenoV |  |
 |
 |
 |
JSZB01024090.1 | 5431 - 5670 (+) |  |
 |
 |
 |
| SelenoW1 |  |
 |
 |
 |
JSZB01009414.1 | 28441 - 28578 (-) |  |
 |
 |
 |
| SelenoW2 |  |
 |
 |
 |
JSZB01001359.1 | 256553 - 257521 (-) |  |
 |
 |
 |
| TXNRD1 |  |
 |
 |
 |
JSZB01004433.1 | 379580 - 467345 (-) |  |
 |
 |
 |
| TXNRD2 |  |
 |
 |
 |
JSZB01015288.1 | 538284 - 579154 (-) |  |
 |
 |
 |
| TXNRD3 |  |
 |
 |
 |
JSZB01010522.1 | 6722 - 7135 (+) |  |
 |
 |
 |
Selenoprotein machinery
This table shows the presence of the genes encoding the machinery proteins.
| Protein ID | Specie | Sec | BLAST | Exonerate | Scaffold | Gene location | T-Coffee | Seblastian | SECIS | SECIS Image | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| SecS |  |
 |
 |
 |
JSZB01003377.1 | 34855 - 66215 (+) |  |
 |
 |
 |
|
| SBP2 |  |
 |
 |
 |
JSZB01003442.1 | 614932 - 650362 (-) |  |
 |
 |
 |
|
| PSTK |  |
 |
 |
 |
JSZB01001613.1 | 505832 - 512216 (-) |  |
 |
 |
 |
|
| eEFsec |  |
 |
 |
 |
JSZB01017587.1 | 70383 - 184606 (-) |  |
 |
 |
 |
|
| SEPHS1 |  |
 |
 |
 |
JSZB01027477.1 | 25665 - 43796 (+) |  |
 |
 |
 |
|
| SEPHS2 |  |
 |
 |
 |
JSZB01033562.1 | 17437 - 18889 (+) |  |
 |
 |
 |
|
| SECp43 |  |
 |
 |
 |
JSZB01005395.1 | 105973 - 106793 (+) |  |
 |
 |
 |
|
Selenium metabolism
This table shows the presence of the genes encoding proteins involved in selenium metabolism.
| Protein ID | Specie | Sec | BLAST | Exonerate | Scaffold | Gene location | T-Coffee | Seblastian | SECIS | SECIS Image | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ELAVL1 |  |
 |
 |
 |
JSZB01004296.1 | 151221 - 194155 (+) |  |
 |
 |
 |
|
| CELF1 |  |
 |
 |
 |
JSZB01022723.1 | 137324 - 178212 (-) |  |
 |
 |
 |
|
| EIF4A3 |  |
 |
 |
 |
JSZB01021220.1 | 113597 - 123076 (+) |  |
 |
 |
 |
|
| XPO1 |  |
 |
 |
 |
JSZB01010493.1 | 84479 - 119336 (+) |  |
 |
 |
 |
|
| G6PD |  |
 |
 |
 |
JSZB01008575.1 | 98934 - 113564 (-) |  |
 |
 |
 |
|
| MsrA |  |
 |
 |
 |
JSZB01006427.1 | 69768 - 195297 (-) |  |
 |
 |
 |
|
| RPL30 |  |
 |
 |
 |
JSZB01006600.1 | 4115 - 4376 (-) |  |
 |
 |
 |
|
| SELENBP1 |  |
 |
 |
 |
JSZB01014514.1 | 86863 - 91990 (-) |  |
 |
 |
 |
|
| LRP2* |  |
 |
 |
 |
JSZB01003496.1 | 4698 - 96282 (-) |  |
 |
 |
 |
|
| LRP8 |  |
 |
 |
 |
JSZB01009333.1 | 49673 - 87768 (+) |  |
 |
 |
 |
|
| SelenoU1 |  |
 |
 |
 |
JSZB01001214.1 | 239257 - 239995 (-) |  |
 |
 |
 |
|
| SelenoU2 |  |
 |
 |
 |
JSZB01000054.1 | 159 - 6965 (+) |  |
 |
 |
 |
|
| SelenoU3 |  |
 |
 |
 |
JSZB01006448.1 | 43813 - 45307 (+) |  |
 |
 |
 |
|
| SCLY |  |
 |
 |
 |
JSZB01032272.1 | 72474 - 101511 (+) |  |
 |
 |
 |
|
| SEPSECS |  |
 |
 |
 |
JSZB01003377.1 | 34855 - 66215 (+) |  |
 |
 |
 |
|
| SARS2 |  |
 |
 |
 |
JSZB01004564.1 | 65830 - 75168 (-) |  |
 |
 |
 |
|
| TTPA |  |
 |
 |
 |
JSZB01010511.1 | 18975 - 33967 (-) |  |
 |
 |
 |
|
Analysis of Selenoproteins
All the proteins were predicted blasting Manis javanica’s genome against the human proteins found in the SelenoDB database.
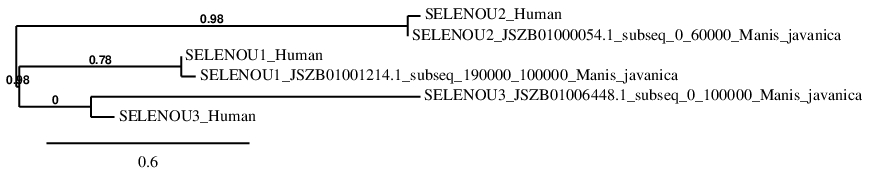
Sel15
In Manis javanica, Sel15 is located in the scaffold JSZB01012017.1 and its gene location is between the position 41463 and the position 85564, in the negative strand. This gene contains 5 exons detailed below, and Sec is located in exon 3, concretely in the positions 55647 (T) and 55645 (A):
Exon 1: From position 85514 to 85564.
Exon 2: From position 77616 to 77783.
Exon 3: From position 55617 to 55680.
Exon 4: From position 44347 to 44396.
Exon 5: From position 41463 to 41591.
A SECIS element of grade A was predicted in the 3’-UTR region between the positions 40878 and 40797 in the negative strand. Though, no Seblastian prediction was found.
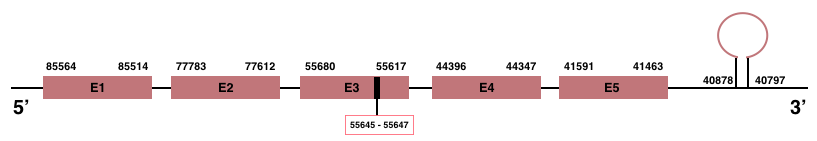
GPx family
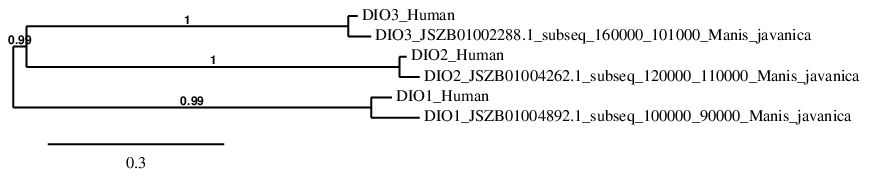
To clearly understand the results of selenoprotein families, a phylogenetic tree was performed. In order to create this tree phylogeny.fr program was used. This program allows to correlate the initial protein queries with the final protein predictions. In the GPx Family tree, the majority of proteins show a correlation in the phylogeny between their sequence in human and in Manis javanica. In the case of GPx5 and GPx6, their sequence appears to be correlated with GPx1 and GPx8, respectively.
GPx1
In Manis javanica, GPx1 is located in the scaffold JSZB01017608.1 and its gene is located between the position 46967 and the position 47182, in the positive strand. This gene has 1 exon detailed below:Exon 1: From position 46967 to 47182.
Even though in human GPx1 is a selenoprotein, in Manis javanica any selenocysteine residue was found.

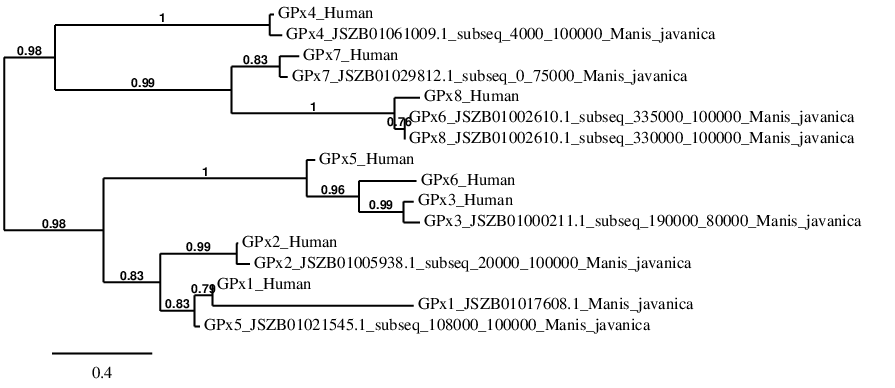
GPx2
In Manis javanica, GPx2 is located in the scaffold JSZB01005938.1 and its gene is located between the position 70515 and the position 73386, in the positive strand. This gene has 2 exons detailed below, and Sec is located in exon 1, concretely in the positions 70632 (T) and 70634 (A):Exon 1: From position 70515 to 70736.
Exon 2: From position 73039 to 73386.
More than one SECIS element was predicted by SECISearch3. A grade A SECIS element was selected in the 3’-UTR region between the positions 73622 and 73686 in the positive strand. Seblastian prediction for GPx2 was also found.
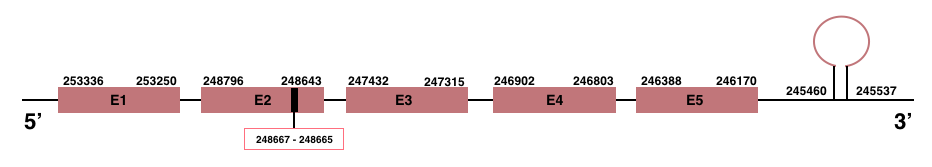
GPx3
In Manis javanica, GPx3 is located in the scaffold JSZB01000211.1 and its gene is located between the position 246170 and the position 253336, in the negative strand. This gene has 5 exons detailed below, and Sec is located in exon 2, concretely in the positions 248667 (T) and 248665 (A):Exon 1: From position 253250 to 253336.
Exon 2: From position 248643 to 248796.
Exon 3: From position 247315 to 247432.
Exon 4: From position 246803 to 246902.
Exon 5: From position 246170 to 246388.
A SECIS element of grade A was predicted in the 3’-UTR region between the positions 245460 and 245537 in the negative strand. Though, no Seblastian prediction was found.
GPx4
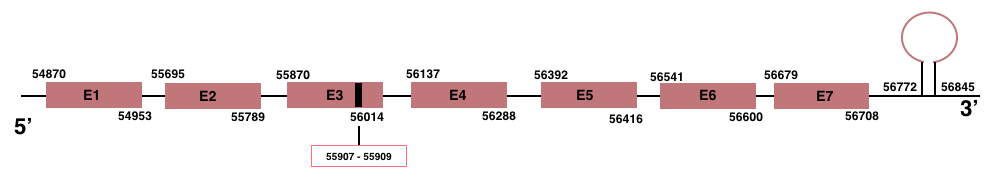
In Manis javanica, GPx4 is located in the scaffold JSZB01061009.1 and its gene is located between the position 54870 and the position 56708, in the positive strand. This gene has 7 exons detailed below, and Sec is located in exon 3, concretely in the positions 55907 (T) and 55909 (A):Exon 1: From position 54870 to 54953.
Exon 2: From position 55695 to 55789.
Exon 3: From position 55870 to 56014.
Exon 4: From position 56137 to 56288.
Exon 5: From position 56392 to 56416.
Exon 6: From position 56541 to 56600.
Exon 7: From position 56679 to 56708.
A SECIS element of grade A was predicted in the 3’-UTR region between the positions 56772 and 56845 in the positive strand. Seblastian prediction for GPx4 was also found.
GPx5
In Manis javanica, GPx5 is located in the scaffold JSZB01021545.1 and its gene is located between the position 158717 and the position 159004, in the negative strand. This gene has 1 exon detailed below:Exon 1: From position 158717 to 159004.
In humans, GPx5 is an homologous for Cys whether in Manis javanica in the same position there is a Leu.

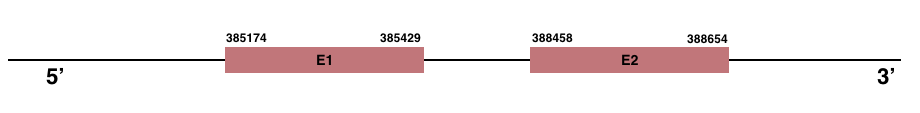
GPx6
In Manis javanica, GPx6 is located in the scaffold JSZB01002610.1 and its gene is located between the position 385174 and the position 388654, in the positive strand. This gene has 2 exons detailed below:Exon 1: From position 385174 to 385429.
Exon 2: From position 388458 to 388654.
Even though in human GPx6 is a selenoprotein, in Manis javanica any selenocysteine residue was found.
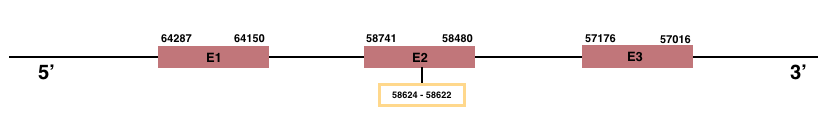
GPx7
In Manis javanica, GPx7 is located in the scaffold JSZB01029812.1 and its gene is located between the position 57016 and the position 64287, in the negative strand. This gene has 3 exons detailed below:Exon 1: From position 64150 to 64287.
Exon 2: From position 58480 to 58741.
Exon 3: From position 57016 to 57176.
In both, human and Manis javanica, GPx7 is an homologous for Cys.
GPx8
In Manis javanica, GPx8 is located in the scaffold JSZB01002610.1 and its gene is located between the position 384416 and the position 388678, in the positive strand. This gene has 3 exons detailed below:Exon 1: From position 384416 to 384619.
Exon 2: From position 385168 to 385429.
Exon 3: From position 388518 to 388678.
In both, human and Manis javanica, GPx8 is an homologous for Cys.

DIO family
To clearly understand the results of selenoprotein families, a phylogenetic tree was performed. In order to create this tree \
phylogeny.fr program was used. This program allows to correlate the initial protein queries with the final protein predictions. In the DIO Family tree, all the selenoproteins show a perfect correlation in the phylogeny between their sequence in human and in Manis javanica.
DIO1
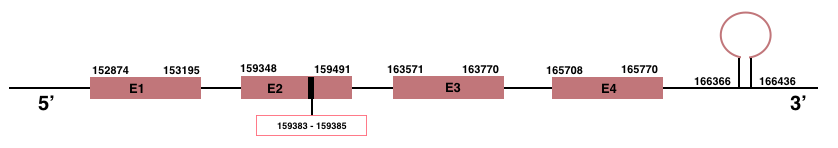
In Manis javanica, DIO1 is located in the scaffold JSZB01004892.1 and its gene is located between the position 152874 and the position 165770, in the positive strand. This gene has 4 exons detailed below and Sec is located in exon 2, concretely in the positions 159383 (T) and 159385 (A):Exon 1: From position 152874 to 153195.
Exon 2: From position 159348 to 159491.
Exon 3: From position 163571 to 163770.
Exon 4: From position 165708 to 165770.
A SECIS element of grade A, predicted in the 3’-UTR region, was found between the positions 166366 and 166436 in the positive strand. Seblastian prediction for DIO1 was also found.
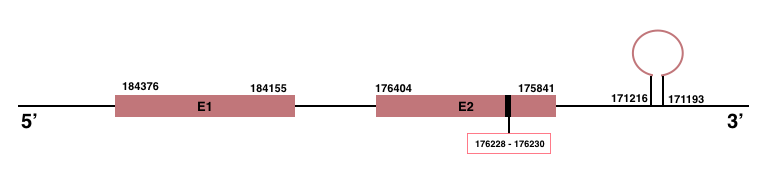
DIO2
In Manis javanica, DIO2 is located in the scaffold JSZB01004262.1 and its gene is located between the position 175841 and the position 184376, in the negative strand. This gene has 2 exons detailed below and Sec is located in exon 2, concretely in the positions 176230 (T) and 176228 (A):Exon 1: From position 184155 to 184376.
Exon 2: From position 175841 to 176404.
A SECIS element of grade A, predicted in the 3’-UTR region, was found between the positions 171216 and 171193 in the negative strand. Though, no Seblastian prediction was found.
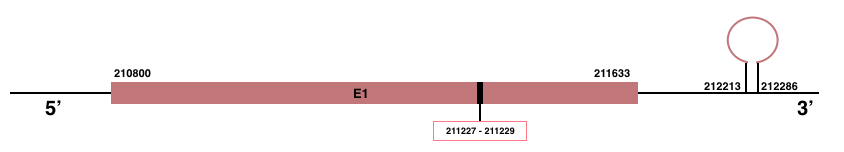
DIO3
In Manis javanica, DIO3 is located in the scaffold JSZB01002288.1 and its gene is located between the position 210800 and the position 211633, in the positive strand. This gene has 1 exon detailed below and Sec is located in this exon, concretely in the positions 211227 (T) and 211229 (A):Exon 1: From position 210800 to 211633.
More than one SECIS element was predicted by SECISearch3. A grade A SECIS element in the 3’-UTR region was selected between the positions 212213 and 212286 in the positive strand. Seblastian prediction was also found.
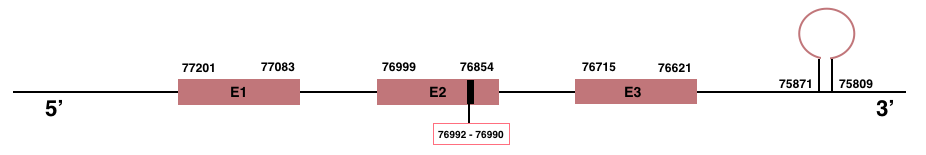
SelenoH
In Manis javanica, SelenoH is located in the scaffold JSZB01009980.1 and its gene is located between the position 76621 and the position 77201, in the negative strand. This gene has 3 exons detailed below and Sec is located in exon 2, concretely in the positions 76990 (T) and 76992 (A):Exon 1: From position 77083 to 77201.
Exon 2: From position 76854 to 76999.
Exon 3: From position 76621 to 76715.
A SECIS element of grade A was predicted in the 3’-UTR region between the positions 75871 and 75809 in the negative strand. Seblastian prediction for SelenoH was also found.
SelenoI
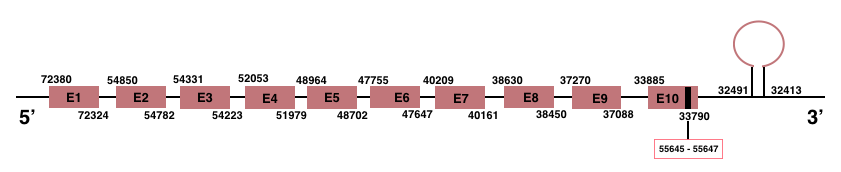
In Manis javanica, SelenoI is located in the scaffold JSZB01006584.1 and its gene is located between the position 33790 and the position 72380, in the negative strand. This gene has 10 exons detailed below and Sec is located in exon 10, concretely in the positions 33819 (T) and 33821 (A) :Exon 1: From position 72324 to 72380.
Exon 2: From position 54782 to 54850.
Exon 3: From position 54223 to 54331.
Exon 4: From position 51979 to 52053.
Exon 5: From position 48702 to 48964.
Exon 6: From position 47647 to 47755.
Exon 7: From position 40161 to 40209.
Exon 8: From position 38450 to 38630.
Exon 9: From position 37088 to 37270.
Exon 10: From position 33790 to 33885.
A SECIS element of grade A was predicted in the 3’-UTR region between the positions 32491 and 32413 in the negative strand. Seblastian prediction was also found.
SelenoK
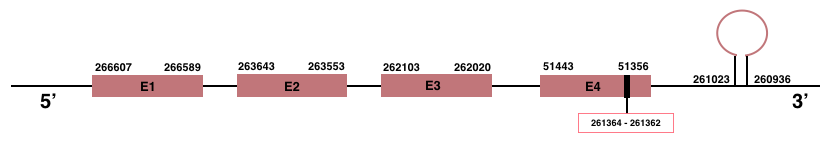
In Manis javanica, SelenoK is located in the scaffold JSZB01010020.1 and its gene is located between the position 261356 and the position 266607, in the negative strand. This gene has 4 exons detailed below and Sec is located in exon 4, concretely in the positions 261364 (T) and 261362 (A):Exon 1: From position 266589 to 266607.
Exon 2: From position 263553 to 263643.
Exon 3: From position 262020 to 262103.
Exon 4: From position 261356 to 261443.
A SECIS element of grade A was predicted in the 3’-UTR region between the positions 261023 and 260936 in the negative strand. Seblastian prediction was also found.
SelenoM
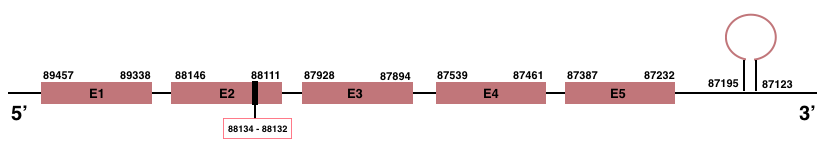
In Manis javanica, SelenoM is located in the scaffold JSZB01032570.1 and its gene is located between the position 87232 and the position 89457, in the negative strand. This gene has 5 exons detailed below and Sec is located in exon 2, concretely in the positions 88134 (T) and 88132 (A):Exon 1: From position 89338 to 89457.
Exon 2: From position 88111 to 88146.
Exon 3: From position 87894 to 87928.
Exon 4: From position 87461 to 87539.
Exon 5: From position 87232 to 87387.
More than one SECIS element was predicted by SECISearch3. A SECIS element of grade A was selected in the 3’-UTR region between the positions 87195 and 87123 in the negative strand. Seblastian prediction was also found.
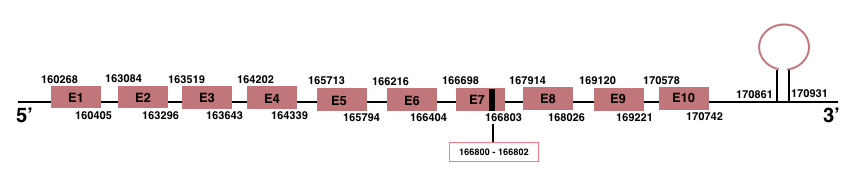
SelenoN
In Manis javanica, SelenoN is located in the scaffold JSZB01007029.1 and its gene is located between the position 160268 and the position 170742, in the positive strand. This gene has 10 exons detailed below and Sec is in exon 7, concretely in the positions 166800 (T) and 166802 (A):Exon 1: From position 160268 to 160405.
Exon 2: From position 163084 to 163296.
Exon 3: From position 163519 to 163643.
Exon 4: From position 164202 to 164339.
Exon 5: From position 165713 to 165794.
Exon 6: From position 166216 to 166404.
Exon 7: From position 166698 to 166803.
Exon 8: From position 167914 to 168026.
Exon 9: From position 169120 to 169221.
Exon 10: From position 170578 to 170742.
A SECIS element of grade A was predicted in the 3’-UTR region between the positions 170861 and 170931 in the positive strand. Though, no Seblastian prediction was found.
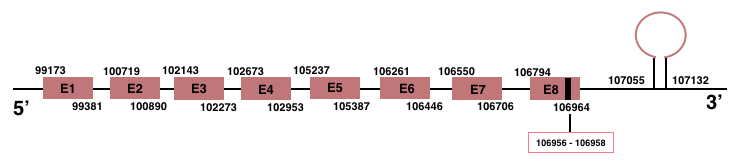
SelenoO
In Manis javanica, SelenoO is located in the scaffold JSZB01004690.1 and its gene is located between the position 99173 and the position 106964, in the positive strand. This gene has 8 exons detailed below and Sec is in exon 8, concretely in the positions 106956 (T) and 106958 (A):Exon 1: From position 99173 to 99381.
Exon 2: From position 100719 to 100890.
Exon 3: From position 102143 to 102273.
Exon 4: From position 102673 to 102953.
Exon 5: From position 105237 to 105387.
Exon 6: From position 106261 to 106446.
Exon 7: From position 106550 to 106706.
Exon 8: From position 106794 to 106964.
More than one SECIS element was predicted by SECISearch3. A grade A SECIS element was selected in the 3’-UTR region between the positions 107055 and 107132 in the positive strand. Seblastian prediction was also found.
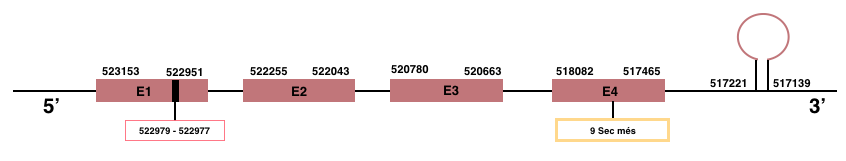
SelenoP
In Manis javanica, SelenoP is located in the scaffold JSZB01008296.1 and its gene is located between the position 517465 and the position 523153, in the negative strand. This gene has 4 exons detailed below and Sec is in exon 1, concretely in the positions 522977 (T) and 522979 (A):Exon 1: From position 522951 to 523153.
Exon 2: From position 522043 to 522255.
Exon 3: From position 520663 to 520780.
Exon 4: From position 517465 to 518082.
More than one SECIS element was predicted by SECISearch3. A grade A SECIS element was selected in the 3’-UTR region between the positions 517139 and 517221 in the negative strand. Seblastian prediction was also found.
MSRB family
To clearly understand the results of selenoprotein families, a phylogenetic tree was performed. In order to create this tree \
\
phylogeny.fr program was used. This program allows to correlate the initial protein queries with the final protein predictions. In the MSRB Family tree, all the selenoproteins show a perfect co\
rrelation in the phylogeny between their sequence in human and in Manis javanica. It also shows that MSRB2 and MSRB3 were separated after the separation from MSRB1.

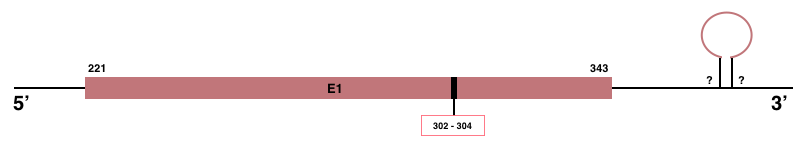
MSRB1
In Manis javanica, MSRB1 is located in the scaffold JSZB01066426.1 and its gene is located between the position 221 and the position 343, in the positive strand. This gene has 1 exon detailed below and Sec is in exon 1, concretely in the positions 302 (T) and 304 (A):Exon 1: From position 221 to 343.
Even though MSRB1 in Manis javanica is a selenoprotein, no SECIS element was predicted.
MSRB2
In Manis javanica, MSRB2 is located in the scaffold JSZB01014487.1 and its gene is located between the position 180874 and the position 192373, in the negative strand. This gene has 4 exons detailed below:Exon 1: From position 192260 to 192373.
Exon 2: From position 187308 to 187384.
Exon 3: From position 181924 to 182071.
Exon 4: From position 180874 to 180975.
In both human and Manis javanica, MSRB2 is an homologous for Cys.
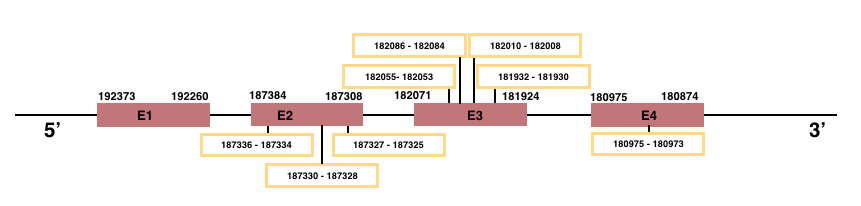
MSRB3
In Manis javanica, MSRB3 is located in the scaffold JSZB01006555.1 and its gene is located between the position 411790 and the position 536660, in the negative strand. This gene has 5 exons detailed below:Exon 1: From position 536551 to 536660.
Exon 2: From position 534885 to 534962.
Exon 3: From position 505862 to 505890.
Exon 4: From position 419984 to 420081.
Exon 5: From position 411790 to 411951.
In both human and Manis javanica, MSRB3 is an homologous for Cys.
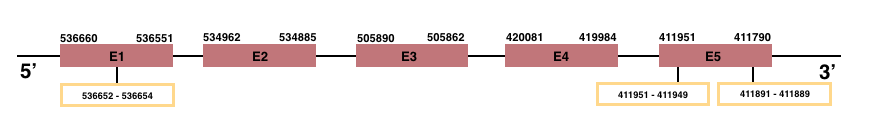
SelenoS
In Manis javanica, SelenoS is located in the scaffold JSZB01022157.1 and its gene is located between the position 328024 and the position 335603, in the negative strand. This gene has 6 exons detailed below and Sec is in exon 6, concretely in the positions 328029 (T) and 328027 (A):Exon 1: From position 335528 to 335603.
Exon 2: From position 334714 to 334848.
Exon 3: From position 332631 to 332737.
Exon 4: From position 330045 to 330134.
Exon 5: From position 329865 to 329943.
Exon 6: From position 328024 to 328106.
A SECIS element of grade A was predicted in the 3’-UTR region between the positions 327680 and 327601 in the negative strand. Seblastian prediction was also found.
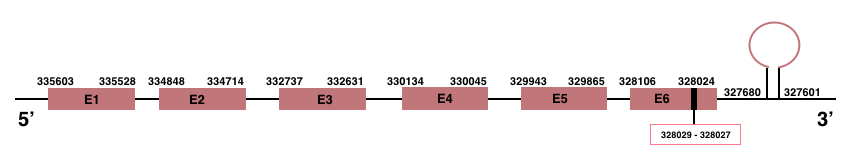
SelenoT
In Manis javanica, SelenoT is located in the scaffold JSZB01003988.1 and its gene is located between the position 477781 and the position 495283, in the negative strand. This gene has 5 exons detailed below and Sec is in exon 2, concretely in the positions 482244 (T) and 482242 (A):Exon 1: From position 495147 to 495283.
Exon 2: From position 482139 to 482249.
Exon 3: From position 481451 to 481577.
Exon 4: From position 479391 to 479478.
Exon 5: From position 477781 to 477902.
A SECIS element of grade A was predicted in the 3’-UTR region between the positions 477221 and 477147 in the negative strand. Seblastian prediction was also found.
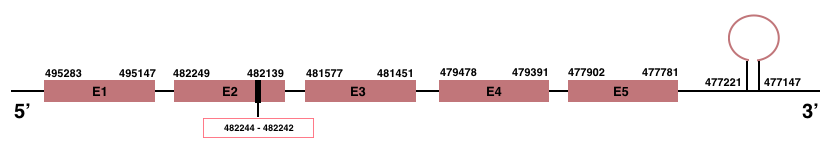
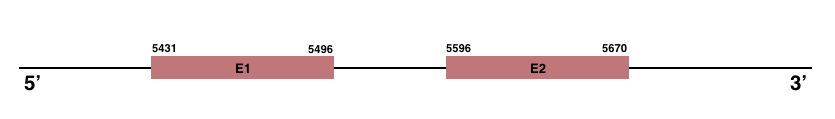
SelenoV
In Manis javanica, SelenoV is located in the scaffold JSZB01024090.1 and its gene is located between the position 5431 and the position 5670, in the positive strand. This gene has 2 exons detailed below:Exon 1: From position 5431 to 5496.
Exon 2: From position 5596 to 5670.
Even though in humans SelenoV is a selenoprotein, in Manis javanica any selenocysteine residue was found.
SelenoW family
To clearly understand the results of selenoprotein families, a phylogenetic tree was performed. In order to create this tree \
\
phylogeny.fr program was used. This program allows to correlate the initial protein queries with the final protein predictions. In the SelenoW Family tree, all the selenoproteins show a perfect co\
rrelation in the phylogeny between their sequence in human and in Manis javanica.

SelenoW1
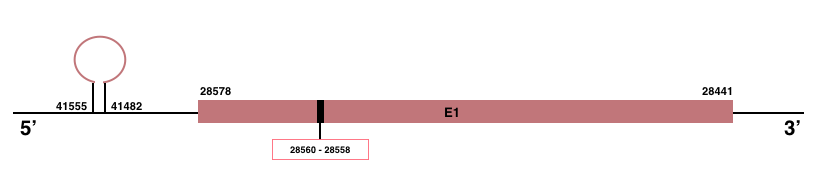
In Manis javanica, SelenoW1 is located in the scaffold JSZB01009414.1 and its gene is located between the position 28441 and the position 28578, in the negative strand. This gene has 1 exon detailed below and Sec is in exon 1, concretely in the positions 28558 (T) and 28560 (A):Exon 1: From position 28441 to 28578.
Although SelenoW1 has a Sec residue, only a SECIS element of grade C was predicted in the 5’-UTR region between the positions 41555 and 41482 in the negative strand. No Seblastian prediction was found.
SelenoW2
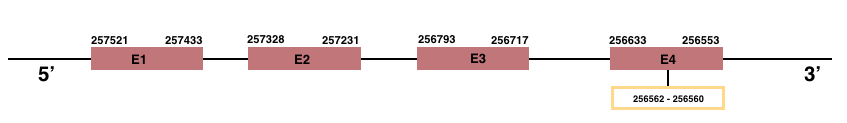
In Manis javanica, SelenoW2 is located in the scaffold JSZB01001359.1 and its gene is located between the position 256553 and the position 257521, in the negative strand. This gene has 4 exons detailed below:Exon 1: From position 257433 to 257521.
Exon 2: From position 257231 to 257328.
Exon 3: From position 256717 to 256793.
Exon 4: From position 256553 to 256633.
In both human and Manis javanica, MSRB2 is an homologous for Cys.
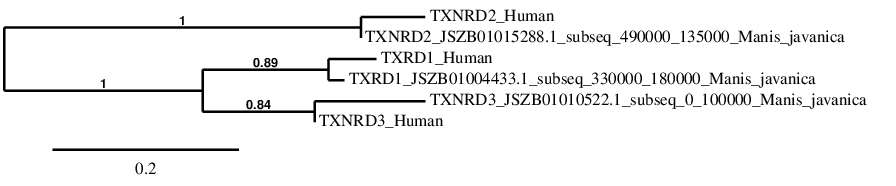
TXNRD family
To clearly understand the results of selenoprotein families, a phylogenetic tree was performed. In order to create this tree \
\
phylogeny.fr program was used. This program allows to correlate the initial protein queries with the final protein predictions. In the TXNRD Family tree, all the selenoproteins show a perfect co\
rrelation in the phylogeny between their sequence in human and in Manis javanica. It also shows that TXNRD1 and TXNRD3 were separated after the separation from TXNRD2.
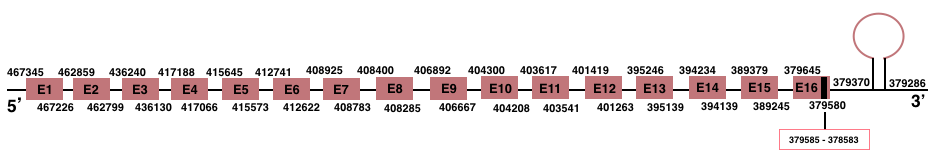
TXNRD1
In Manis javanica, TXNRD1 is located in the scaffold JSZB01004433.1 and its gene is located between the position 379580 and the position 467345, in the negative strand. In this gene has 16 exons detailed below and Sec is in exon 16, concretely in the positions 379585 (T) and 379583 (A):Exon 1: From position 467226 to 467345.
Exon 2: From position 462799 to 462859.
Exon 3: From position 436131 to 436240.
Exon 4: From position 417066 to 417188.
Exon 5: From position 415573 to 415645.
Exon 6: From position 412622 to 412741.
Exon 7: From position 408783 to 408925.
Exon 8: From position 408285 to 408400.
Exon 9: From position 406667 to 406892.
Exon 10: From position 404208 to 404300.
Exon 11: From position 403541 to 403617.
Exon 12: From position 401263 to 401419.
Exon 13: From position 395139 to 395246.
Exon 14: From position 394139 to 394234.
Exon 15: From position 389245 to 389379.
Exon 16: From position 379580 to 379645.
A SECIS element of grade A was predicted in the 3’-UTR region between the positions 379370 and 379286 in the negative strand. Seblastian prediction was also found.
TXNRD2
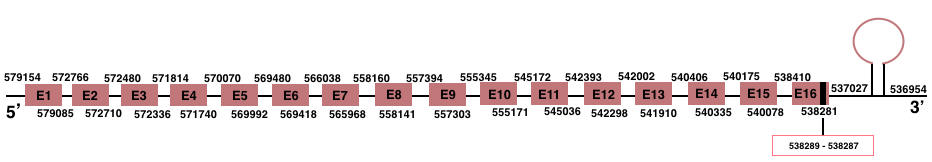
In Manis javanica, TXNRD2 is located in the scaffold JSZB01015288.1 and its gene is located between the position 538284 and the position 579154, in the negative strand. This gene has 16 exons detailed below and Sec is in the exon 16, concretely in the positions 538289 (T) and 538287 (A):Exon 1: From position 579085 to 579154.
Exon 2: From position 572710 to 572766.
Exon 3: From position 572336 to 572480.
Exon 4: From position 571740 to 571814.
Exon 5: From position 569992 to 570070.
Exon 6: From position 569418 to 569480.
Exon 7: From position 565968 to 566038.
Exon 8: From position 558141 to 558160.
Exon 9: From position 557303 to 557394.
Exon 10: From position 555171 to 555345.
Exon 11: From position 545036 to 545172.
Exon 12: From position 542298 to 542393.
Exon 13: From position 541910 to 542002.
Exon 14: From position 540335 to 540406.
Exon 15: From position 540078 to 540175.
Exon 16: From position 538284 to 538410.
More than one SECIS element was predicted by SECISearch3. A grade A SECIS element was selected in the 3’-UTR region between the positions 537027 and 536954 in the negative strand. Seblastian prediction was also found.
TXNRD3
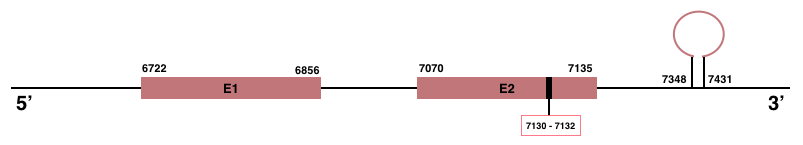
In Manis javanica, TXNRD3 is located in the scaffold JSZB01010522.1 and its gene is located between the position 6722 and the position 7135, in the positive strand. This gene has 2 exons detailed below and Sec is in the exon 2, concretely in the positions 7130 (T) and 7132 (A):Exon 1: From position 6722 to 6856.
Exon 2: From position 7070 to 7135.
A SECIS element of grade A was predicted in the 3’-UTR region between the positions 7348 and 7431 in the positive strand. Seblastian prediction was also found.
Analysis of Selenoprotein machinery
All the proteins were predicted blasting Manis javanica’s genome against the human proteins found in the SelenoDB database.
SecS
In Manis javanica, SecS is located in the scaffold JSZB01003377.1 and its gene is located between the position 34855 and the position 66215, in the positive strand. This gene has 11 exons detailed below:
Exon 1: From position 34855 to 34968.
Exon 2: From position 35765 to 35919.
Exon 3: From position 37964 to 38082.
Exon 4: From position 38440 to 38598.
Exon 5: From position 41065 to 41218.
Exon 6: From position 44194 to 44296.
Exon 7: From position 53383 to 53512.
Exon 8: From position 53645 to 53736.
Exon 9: From position 62683 to 62776.
Exon 10: From position 64450 to 64540.
Exon 11: From position 65912 to 66215.
Since it is a protein involved in the selenoproteins machinery, it does not contain any selenocysteine. For this reason, neither Seblastian nor SECISSearch3 were used to predict selenoprotein or SECIS elements.
SBP2
In Manis javanica, SBP2 is located in the scaffold JSZB01003442.1 and its gene is located between the position 614932 and position 650362, in the negative strand. This gene has 11 exons detailed below:
Exon 1: From position 650270 to 650362.
Exon 2: From position 646998 to 647120.
Exon 3: From position 645699 to 645788.
Exon 4: From position 644340 to 644472.
Exon 5: From position 624558 to 624724.
Exon 6: From position 623777 to 623915.
Exon 7: From position 622418 to 622574.
Exon 8: From position 621572 to 621792.
Exon 9: From position 615838 to 615992.
Exon 10: From position 615354 to 615546.
Exon 11: From position 614932 to 615032.
Since it is a protein involved in the selenoproteins machinery, it does not contain any selenocysteine. For this reason, neither Seblastian nor SECISSearch3 were used to predict selenoprotein or SECIS elements.
PSTK
In Manis javanica, PSTK is located in the scaffold JSZB01001613.1 and its gene is located between the position 505832 and the position 512216, in the negative strand. This gene has 6 exons detailed below.
Exon 1: From position 512001 to 512216.
Exon 2: From position 509719 to 510010.
Exon 3: From position 509282 to 509480.
Exon 4: From position 506852 to 506927.
Exon 5: From position 506233 to 506326.
Exon 6: From position 505832 to 505872.
Since it is a protein involved in the selenoproteins machinery, it does not contain any selenocysteine. For this reason, neither Seblastian nor SECISSearch3 were used to predict selenoprotein or SECIS elements.
eEFsec
In Manis Javanica, eEFsec is located in the scaffold JSZB01017587.1 and its gene is located between the position 70383 and the position 184606, in the negative strand. This gene has 4 exons detailed below:
Exon 1: From position 184297 to 184606.
Exon 2: From position 94584 to 94791.
Exon 3: From position 72888 to 72984.
Exon 4: From position 70383 to 70550.
Since it is a protein involved in the selenoproteins machinery, it does not contain any selenocysteine. For this reason, neither Seblastian nor SECISSearch3 were used to predict selenoprotein or SECIS elements.
SEPHS1
In Manis javanica, SEPHS1 is located in the scaffold JSZB01027477.1 and its gene is located between the position 25665 and the position 43796, in the positive strand. This gene has 7 exons detailed below:
Exon 1: From position 25665 to 25857.
Exon 2: From position 30506 to 30609.
Exon 3: From position 32387 to 32494.
Exon 4: From position 33394 to 33548.
Exon 5: From position 36765 to 36855.
Exon 6: From position 38748 to 38847.
Exon 7: From position 43585 to 43796.
Since it is a protein involved in the selenoproteins machinery, it does not contain any selenocysteine. For this reason, neither Seblastian nor SECISSearch3 were used to predict selenoprotein or SECIS elements.
SEPHS2
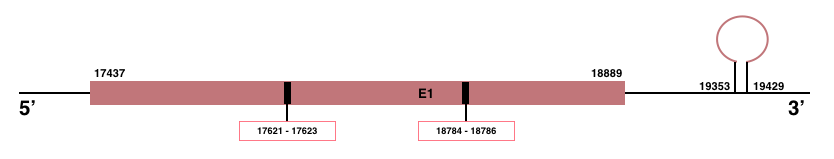
In Manis javanica, SEPHS2 is located in the scaffold JSZB01033562.1 and its gene is located between the position 17437 and the position 18885, in the positive strand. This gene has 1 exons detailed below and Sec are located in the exon 1, concretely in the positions 17623 (T) and 17625 (A) and in the positions 18784 (T) and 18786 (A):
Exon 1: From position 17437 to 18885.
Even though it is a protein involved in the selenoproteins machinery, SEPSHS2 contains a selenocysteine. Then, Seblastian and SECISearch3 were used to predict selenoprotein and SECIS element. A SECIS element of grade A was predicted in the 3’-UTR region between the positions 19353 and 19429 in the positive strand. Seblastian prediction was also found.
SECp43
In Manis javanica, SECp43 is located in the scaffold JSZB01005395.1 and its gene is located between the position 105973 and the position 106793, in the positive strand. This gene has 3 exons detailed below:Exon 1: From position 105973 to 106161.
Exon 2: From position 106296 to 106427.
Exon 3: From position 106692 to 106793.
Since it is a protein involved in the selenoproteins machinery, it does not contain any selenocysteine. For this reason, neither Seblastian nor SECISSearch3 were used to predict selenoprotein or SECIS elements. SECp43 is also an homologous for Cys in humans whether in Manis javanica instead of Cys there is an STOP codon.