Discussion
Selenium
Selenium (Se) is an essential dietary micronutrient for animals including humans, although it was known as a toxic element in the past. In order to avoid its high reactivity, this micronutrient is stored as H2Se in the organism. Selenium must be present in an appropriate quantity since both deficit and accumulation may induce health problems such as cardiovascular and metabolism diseases, that appear since selenium is related with metabolic pathways and it has also an antioxidant response. In particular, deficits in selenium produce Keshan disease, characterised by heart failure and pulmonary edema. Contrarily, an accumulation of this nutrient may cause an intoxication.[4,26] Selenium is also regarded as a protective agent against cancer. Several mechanisms have been proposed, such as antioxidant protection by selenoenzymes, specific inhibition of tumor cell growth by metabolites, modulation of cell cycle and apoptosis and effect on DNA repair.[5,32]
When selenium binds with a cysteine, this amino acid is transformed into a selenocysteine (Sec, U), the 21st amino acid. Although both proteins have the same function, they are structurally different; selenocysteine loses the sulfur group, which is replaced by a selenium molecule. Proteins that contain selenocysteine are named as selenoproteins.[26]
Selenoproteins
The biological effects of selenium are largely mediated by selenium-containing proteins, selenoproteins, that are present in all three domains of life. In eukaryotes, selenoproteins show a mosaic occurrence; for example, vertebrates and algae have dozens of these proteins, while other organisms, such as higher plants and fungi, have lost all selenoproteins during evolution.[18]
Particularly, there are around 45 selenoproteins described in vertebrates but only 25 are found in humans [Figure 1].[20] Even though, selenoproteins are not found in some animals neither plants. Although selenoproteins represent diverse molecular pathways and biological functions, all these proteins contain at least one selenocysteine and most serve oxidoreductase functions.[17]
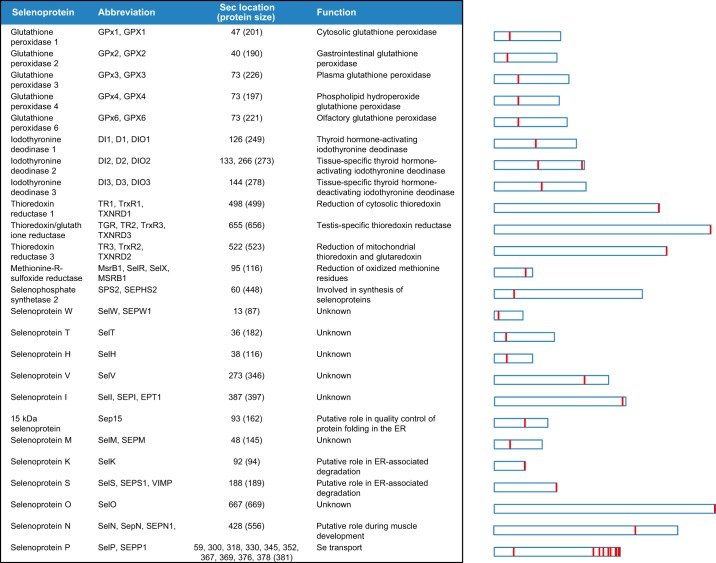
(Labunskyy V et al.,2014)
Selenocysteines are encoded by a UGA codon in the selenoprotein mRNA [Figure 2]. The decoding of UGA as Sec requires the reprogramming of translation since UGA is normally read as a stop codon, whose normal function is to terminate translation. In order to decode UGA codon as Sec, organisms evolved the Sec insertion machinery that allows the incorporation of this amino acid at specific UGA codons in a process requiring a cis-acting Sec insertion sequence (SECIS) element present on the mRNA sequence.[11,12,17] By comparing different organisms, it has been found that in some selenoproteins, selenocysteines have been replaced for cysteines (Cys) during evolution.[20]
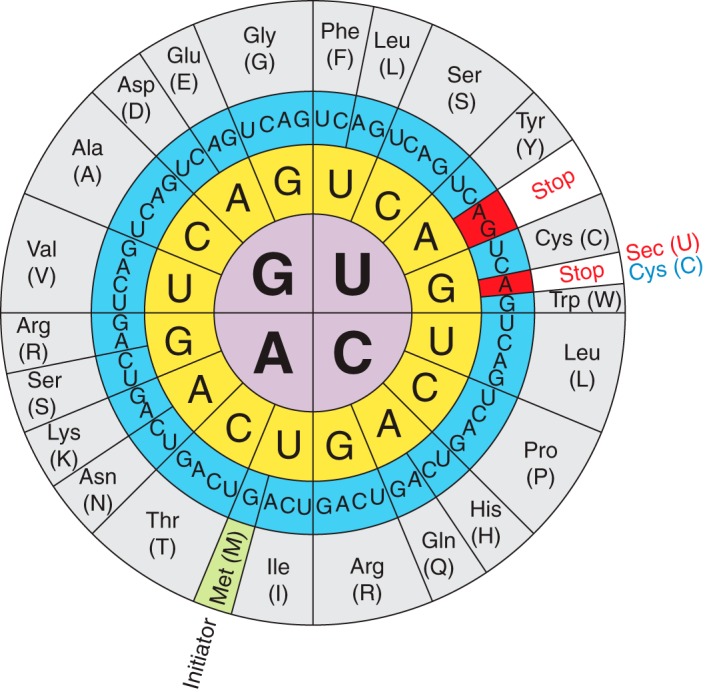
(Labunskyy V et al.,2014)
Biosynthesis of Selenoproteins
The introduction of Sec into proteins requires several genes and a complex machinery that is described below. In eukaryotes, the control of selenoprotein expression is possible due to a set of dedicated cis-acting factors (SECIS elements and the in-frame UGA codon) and trans-acting factors (SPS1, SPS2, SecS, PSTK, eEFsec, SBP2 and the Sec-tRNA[Ser]Sec), as well as a variety of regulatory mechanisms, that enable a high grade of translational control.[34]
In order to synthesize selenoproteins, cell machinery must recognise UGA codon from the selenoprotein mRNA as a new amino acid, and not as a stop codon. Then, a selenocysteine is incorporated by the specific tRNA and the translation continues. In eukaryotes, this mechanism is guided by the 3’-UTR extreme that contains the SECIS element, which allows the binding between the tRNA and the selenocysteine. Sec is unique among other amino acids in the fact that it is the only known amino acid in eukaryotes whose biosynthesis occurs on its own tRNA, the Sec-tRNA[Ser]Sec, and it is produced from serine (Ser) instead of Cys.
Seryl-tRNA synthetase (SerRS), Phosphoseryl-tRNA Kinase (PSTK) and Sec Synthase (SecS) are enzymes needed in the first step of biosynthesis of selenocysteines. The tRNA is initially aminoacylated with serine in a reaction catalysed by SerRS to form Ser-tRNA[Ser]Sec, which is the basis for Sec biosynthesis. Then, the serine found in the seryl-tRNA[Ser]Sec is phosphorylated by the enzyme PSTK to form the phosphorylated intermediate PSer-tRNA[Ser]Sec and, finally, SecS incorporates selenophosphate, the active form of Se, into the amino acid backbone to form Sec-tRNA[Ser]Sec. Selenophosphate (H2SePO3) generated by SPS2 is used as a donor of active Se for SecS. In contrast, de novo synthesis of Cys using the Sec biosynthetic machinery is provided by H2SPO3 [Figure 3]. [2,17,34]
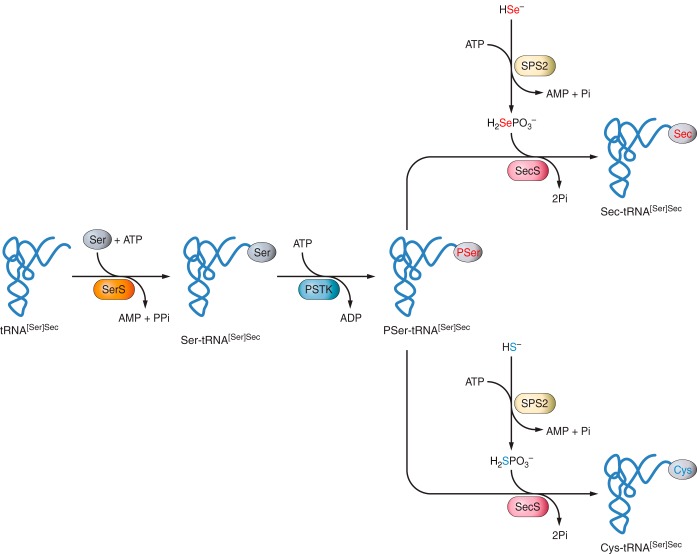
(Labunskyy V et al.,2014)
Once Sec is synthetized, it must be incorporated into proteins in order to form a selenoprotein. Cotranslational incorporation of Sec into proteins is dictated by in-frame UGA codons present in selenoprotein mRNAs. Sec is introduced into selenoproteins by a complex mechanism that requires special trans-acting protein factors, Sec-tRNA[Ser]Sec and a cis-acting Sec insertion sequence (SECIS) element.[12] When a ribosome finds the UGA codon, which normally signals translation termination, Sec machinery interacts with the translation machinery to prevent premature termination. SECIS elements are used as the factors that dictate recoding of UGA as Sec; Sec-tRNA[Ser]Sec, in response to the SECIS element, translates UGA as Sec. In this step, at least two factors are required: SECIS binding protein 2 (SBP2) and Sec-specific translation elongation factor (eEFSec), since this tRNA is not recognized by usual elongation factors. SECIS binds to SBP2 and this union promotes the binding of eEFsec, which recruits Sec-tRNA[Ser]Sec and facilitates the incorporation of Sec into the polypeptide, so the translation will continue [Figure 4].[7,13,14,15]
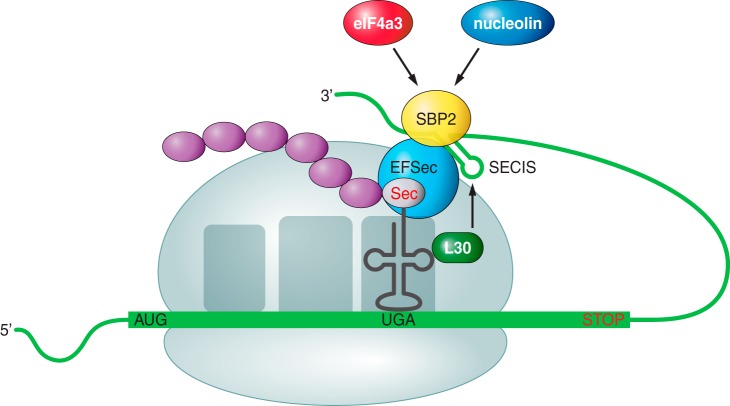
(Labunskyy V et al.,2014)
Phylogeny of Selenoproteins in vertebrates
The following figure shows the phylogeny of selenoproteins in vertebrates. It is useful to predict selenoproteins and to identify possible duplications or deletions. Duplications are represented in green, switches of Sec into Cys are represented in red and, finally, deletions are represented in grey. Moreover, in the left part of the image the ancestral vertebrate selenoproteome is represented [Figure 5].[20]
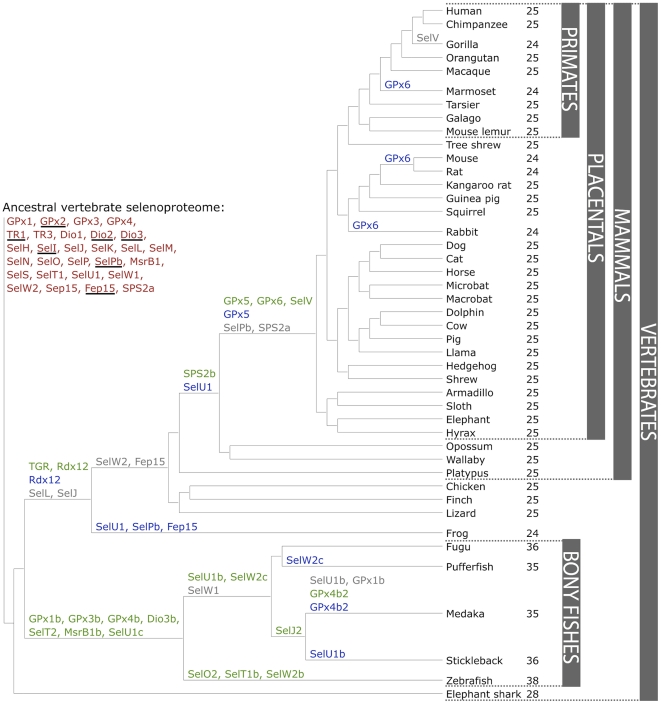
(Mariotti M et al.,2012)
Known Selenoproteins
Selenoproteins are characterised for the presence of Sec residues in their sequence. With a few exceptions, this amino acid is located in the enzyme active sites and performs catalytic redox reactions. The human selenoproteome is encoded by 25 genes [Figure 1].[17,18,20]
15-kDa Selenoprotein and Selenoprotein M
These two proteins are thioredoxin-like fold ER-resident proteins and contain an NH2-terminal signal peptide, consistent with their ER localisation, that form a distinct selenoprotein family with 31% sequence identity.15-kDa Selenoprotein (Sel15): This protein is proposed to mediate cancer prevention effect of dietary Se and regulation of redox homeostasis in the ER. Moreover, it might have a role in disulphide bond formation and quality control of a restricted group of glycoproteins. It is expressed in prostate, liver, kidney and testis.
Selenoprotein M (SelenoM): It is a distant homolog of Sel15 which was identified by bioinformatics approaches. It is highly expressed in brain so it has a possible role in neuroprotection.
Selenoproteins W, V, T, and H
All the following proteins belong to the Rdx family of selenoproteins and all of them possess a thioredoxin-like fold and are characterized by the presence of a conserved Cys-x-x-Sec motif. Based on its motifs, it is proposed that they are thiol-based oxidoreductases, but their exact function remains unknown.Selenoprotein W (SelenoW): Is one of the first identified Sec-containing proteins and is one of the most abundant selenoproteins in mammals. It is localised in the cytosol and is expressed at high levels in muscles and brain. It belongs to the stress-related group of selenoproteins, and its expression is highly regulated by the availability of Se in the diet. SelenoW1 and SelenoW2 are both members of the ancestral proteasome.
Selenoprotein V (SelenoV): Is one of the least characterized selenoproteins. It recently evolved, most likely by duplication from Seleno W and it is found only in placental mammals. Although it is closely related to Seleno W it is a larger protein due to the presence of an additional NH2-terminal domain. It is only expressed in testes.
Selenoprotein T (SelenoT): Is one of the Sec-containing proteins identified using bioinformatics tools. It is predominantly localised to the ER and Golgi and is ubiquitously expressed both during embryonic development and adult tissues. It might be involved in cell structure organisation and cell adhesion properties. It has also been suggested to have a role in the regulation of Ca2+ homeostasis and neuroendocrine function.
Selenoprotein H (SelenoH): This selenoprotein has also a conserved nuclear targeting RKRK motif in the N-terminal sequence. It has a unique subcellular localization pattern and it is localised specifically to the nucleoli. Similar to SelenoW, SelenoH is sensitive to dietary Se intake. It specifically binds to sequences containing heat shock and stress response elements, and it possesses glutathione peroxidases activity implicated in the regulation of transcription.
Selenoprotein I (SelenoI)
Selenoprotein I is one of the least studied selenoproteins since it evolved recently and is only found in vertebrates. It has seven predicted transmembrane domains and it contains a highly conserved CDP-alcohol phosphatidyltransferase domain. Between the first and second transmembrane domains, there are three aspartic acids which are critical for its catalytic activity.Selenoproteins K and S
Although selenoprotein K and selenoprotein S have no significant sequence similarity, they can be assigned as a family of related selenoproteins based on their topology, including a single transmembrane domain in the NH2-terminal sequence, the presence of a glycine-rich segment and a characteristic location of Sec residues in the COOH-terminal end of the protein.It is the most widespread eukaryotic selenoprotein family, whose members are present in nearly all known Se-utilizing organisms from unicellular eukaryotes to humans. Both proteins are located to the ER membrane, with the COOH-terminal and of the protein facing the cytosol. They have been recently implicated in ER-associated degradation of misfolded proteins, so they must have analogous function.
Selenoprotein K (SelenoK): It might be involved in binding some misfolded proteins from the ER and targeting them to the proteasome-dependent degradation.
Selenoprotein S (SelenoS): It has an additional coiled-coil domain in the cytosolic portion of the protein, which has been proposed to mediate the interaction with other proteins or oligomerization of SelenoS.
Selenoprotein O (SelenoO)
Selenoprotein O is one of the least characterised human selenoproteins. It contains a single Sec residue located in the antepenultimate position at the COOH-terminal end of the protein. However, the majority of SelenoO homologous contains a Cys residue instead of Sec (Sec-containing SelenoO sequences are present only in vertebrates). It has a mitochondrial targeting peptide and a putative protein kinase domain.Selenoprotein N (SelenoN)
Selenoprotein N was one of the first selenoproteins identified through bioinformatics approaches. It is an ER-resident transmembrane glycoprotein that is highly expressed during embryonic development and to a lesser extent in adult tissues including skeletal muscle. It might play an important role in satellite cells maintenance and is required for skeletal muscle tissue regeneration after stress or injury.Selenoprotein P (SelenoP)
Selenoprotein P is an abundantly expressed and secreted selenoprotein that accounts for almost 50% of the total Se in plasma. The SelenoP family has recently evolved, and SelenoP homologues are found predominantly in vertebrates. Its unique feature is the presence of multiple Sec residues. For example, human SelenoP gene has 10 in-frame Sec encoding TGA codons and two SECIS elements within its 3’-UTR. Since it is secreted into the plasma and it has multiple Sec residues in its sequence, this selenoprotein might function as a Se supplier to peripheral tissues, particularly brain and testis.Gluthatione Peroxidases (GPx) family
GPx was the first family of selenoproteins discovered. In mammals there are eight GPx paralogues, from which five of them are selenoproteins (GPx1, GPx2, GPx3, GPx4 and GPx6) and the other three are GPx homologues (GPx5, GPx7 and GPx8), in which the active site Sec is replaced by Cys. Moreover, GPx6 homologs in some mammals are not selenoproteins and have a Cys in the active site.It has been found an accurate relation between their activity and the concentration of Se in the enterocytes. They are found in the cytosol, where they play a wide range of physiological functions in organisms; hydrogen peroxide (H2O2) signalling, detoxification of hydroperoxides and cellular redox homeostasis maintaining. They are also a vehicle to collect Se.
GPx1, GPx2, GPx3 and GPx6 are tetrameric vertebrate GPx, and they have specificity for hydrogenperoxide and other soluble, low-molecular-weight hydroperoxides. In contrast, monomeric GPx4 is involved in the reduction of complex phospholipid hydroperoxides that are associated with membranes.
GPx1: It is the most abundant selenoprotein in mammals. It catalyses glutathione (GSH)-dependent reduction of hydrogen peroxide to water. It is expressed in all cell types, with highest expression levels in liver and kidney, and it is highly regulated by Se availability.
GPx2: It is found in the epithelium of the gastrointestinal tract, where it plays a preventive role in the cancer development when its expression is increased in epithelium-derived tumours.
GPx3: It is secreted primarily from kidney and is the major GPx form in plasma.
GPx4: It is expressed in a wide range of cell types and tissues, and plays an important role in preventing oxidative stress-induced apoptosis.
GPx6: It is only found in the olfactory epithelium during embryonic development.
Iodothyronine Deodinases (DIO) family
The DIO family is the second most important family of selenoproteins. It consists of three paralogous proteins in mammals (DIO1, DIO2 and DIO3) which are involved in the regulation of the thyroid hormone activity by reductive deodination. Concretely, they transform the inactive forms T4 and rT3 into the active form T3. These proteins have distinct subcellular localizations and tissue expression; DIO1 and DIO3 are located on the plasma membrane, but DIO2 is localised to the endoplasmic reticulum (ER).They are integral membrane selenoproteins characterized by a thioredoxin fold. All three deiodinase selenoenzymes contain a single transmembrane domain and form a homodimer structure. The active-site Sec residue is located in the NH2-terminal part of the protein. However, in DIO2, an additional Sec whose function is unknown, is present in the COOH-terminal region close to the end of the protein. DIO1 and DIO2 catalyse the reaction to convert T4 into T3, while DIO3 inactivates T3 and T4. They play an important role in the maintaining of thyroid hormone levels and its activity by both activating its prohormone and degrading the biologically active T3. The circulating levels of thyroid hormone are primarily regulated by DIO1 activity, However, DIO2 and DIO3 have been implicated in fine-tuning local intracellular T3 concentrations in a tissue-specific manner, without changing overall serum levels of T3. This local regulation of thyroid hormone activity is important for a number of physiological processes, for example tissue regeneration after injury in specific tissues during development.
Thioredoxin reductase (TXNRD) family
These selenoproteins are oxidoreductase that, together with thioredoxin (Trx), comprise the major disulphide reduction system of the cell. They catalyse the reduction of thioredoxin dependent of NADPH, play a regulatory role in metabolic activity and are associated with the prevention of some cancers. In mammals, there are three TXNRD isoenzymes (TXNRD1, TXNRD2 and TXNRD3), all of them Sec-containing proteins. These proteins contain a Sec residue in the COOH-terminal penultimate position.TXNRD1: It is primarily localised in the cytosol and the nucleus. Although cytosolic thioredoxin (Trx1) acts as the measure substrate for TXNRD1, it can also reduce a variety of low-molecular-weight compounds.
TXNRD2: It is localised in the mitochondria, where it is involved in the reduction of mitochondrial thioredoxin (Trx2) and glutaredoxin 2 (Grx2).
TXNRD3: It differs from the others thioredoxin reductases in that it contains and additional glutaredoxin (Grx) domain located in the NH2-terminal. Due to the presence of this domain, this protein displays Grx activity, suggesting that it is involved in both Trx and GSH systems. It is expressed at high levels in testis after puberty, so it is proposed that TXNRD3 might be involved in the formation of disulphide bonds during sperm maturation.
Selenoprotein R or Methionine-R-Sulfoxide Reductase 1 (MSRB1)
MSRB1 is a zinc-containing enzyme that was initially identified as selenoprotein R and selenoprotein X by searching for putative SECIS element structure using bioinformatics tools. Later, this protein was found to function as a Methionine-R-sulfoxide reductase, which catalyses repair of the R enantiomer of oxidized methionine residues in proteins. Based on its functional similarity to MsrA, it was renamed MSRB1.The Sec-containing MSRB1 protein is the major MSRB in mammals, which is primarily localized in the cytosol and nucleus. It has the highest activity in mammalian liver and kidney. Two additional MSRB homologs (MSRB2 and MSRB3) contain Cys residue in place of Sec in the enzyme’s active site and have different subcellular distributions; MSRB2 is localised in mitochondria, whereas MSRB3 is targeted to the ER.
Known Selenoprotein machinery
The factors that contribute in the machinery of selenoproteins are:[17]
SEPHS1 – Selenophosphate synthetase 1: It is an enzyme that catalyses the synthesis of selenophosphate from selenide and ATP, which is the donor used to synthesize Sec. It acts together with SEPHS2 and SECp43 binding the selenophosphate group to the Cys residue.
SEPHS2 – Selenophosphate synthetase 2: It is an enzyme that catalyses the synthesis of selenophosphate from selenide and ATP, generating the Se donor component for the synthesis of Sec. It is unique among the components of Sec synthesis machinery in that is a selenoprotein by itself.
PSTK – Phosphoseryl-tRNA[Ser]Sec kinase: This enzyme that phosphorilates the precursor Serine of the Sec joined to the tRNA.
SecS – Sec synthase: It is the enzyme that converts the Ser on tRNA to selenocysteyl-tRNA[Ser]Sec by incorporating a selenophosphate, the active form of selenium, into the amino acid backbone to form Sec-tRNA[Ser]Sec.
SBP2 – SECIS-binding protein 2: This enzyme promotes Sec incorporation by associating with SECIS elements and recruiting the eEFsec-selenocysteyl-tRNA[Ser]Sec complex to the ribosome. It is stably associated with ribosomes and contains a distinct L7Ae RNA-binding domain that is known to bind SECIS elements with high affinity and specificity.
eEFsec - Eukaryotic elongation factor: It is the elongation factor that recruits the tRNA with the Sec and binds to SBP2 in order to insert the Sec into nascent polypeptides in response to UGA.
Manis javanica
Malayan pangolin or Sunda pangolin (Manis javanica) is a pangolin specie found in the Southeast of Asia including Thailand, the Philippines, Vietnam, Laos, Cambodia, Malaysia and Singapore. They live in forested areas and plantations, spending most of the time above the trees. It is an exploited specie for traditional medicine and its skin. However, it is a protected specie since it is considered to be under an extreme risk of extinction.[8]
The skin of the pangolin’s feet is granular, although on its front feet some pads can be found. It has thick and powerful claws to dig into the soils. They have poor eyesight but a highly developed sense of smell. Despite their lacking teeth, its long, sticky tongue serves to collect ants and termites. Their body is covered by rows of scales and fibrous hair. Their head-body length can measure from 40 to 65cm, and their tail measures about 35-65cm. Males are larger than females, and its weight is up to 10kg.[27,28]

Pangolins tend to live alone, since they behave shyly. They are mostly visible during the winter season and are nocturnal animals. When they detect any kind of threatening situation, they protect their soft underparts by rolling into balls. Pangolins are great diggers and make big dens full of vegetation in order to insulate near termite mound and ant nests. Their main predators are humans, tigers and clouded leopards.[8]

Click here for more information.