Description of selenoproteins
Iodothyronine deiodinases
DI1
The protein DI1 is located in the scaffold PKMU01008277.1 between the position 1055125 and the position 1063829 in the reverse strand. This gene contains 4 exons. The structure of the gene, analyzed with the Exonerate file, is described below:

Exon 1:1063520-1063829
Exon 2:1061644-1061787
Exon 3:1056148-1056347
Exon 4:1055125-1055184
Seblastian was able to predict a known selenoprotein using the sequence from Chelonoidis abingdonii (DI1). Regarding the SECIS, one grade A SECIS was found between the locations 1053908-1053976. This secis is located in the same strand of our gene, and after it.
DI2
The protein DI2 is located in the scaffold PKMU01005068.1 between the position 5517365 and 5506389 in the reverse strand. This gene contains 2 exons. The structure of the gene, analyzed with the Exonerate file, is described below:
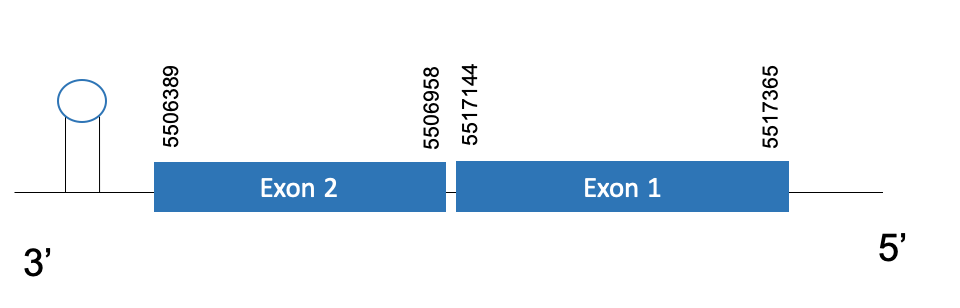
Exon 1:5517144-5517365
Exon 2:5506389-5506958
Seblastian was not able to predict a known selenoprotein using the sequence from Chelonoidis abingdonii. Regarding the SECIS, one grade A SECIS was found between the locations 5501787-5501861. This secis is located in the same strand of our gene, and after it.
DI3
The protein DI2 is located in the scaffold PKMU01005068.1 between the position 5517365 and 5506389 in the reverse strand. This gene contains 2 exons. The structure of the gene, analyzed with the Exonerate file, is described below:

Exon 1:242560-243354
Seblastian was able to predict a known selenoprotein (DI3) using the sequence from Chelonoidis abingdonii. Regarding the SECIs, one grade A SECIS was found between the locations 242096-242179. This secis is located in the same strand of our gene, and after it.
Glutathione peroxidases
GPx1
The protein GPx1 is located in the scaffold PKMU01007833.1. between the position 511801 and 513930 in the forward strand. This gene contains 2 exons. The structure of the gene, analyzed with the Exonerate file, is described below:
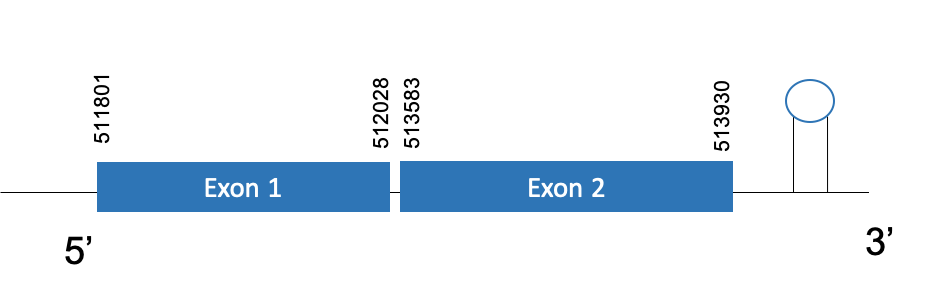
Exon 1:511801-512028
Exon 2:513583-513930
Seblastian was not able to predict a known selenoprotein using the sequence from Chelonoidis abingdonii. Regarding the SECIs, one grade A SECIS was found between the locations 514100-514171. This secis is located in the same strand of our gene, and after it.
GPx2
The protein GPx2 is located in the scaffold PKMU01001079.1. between the position 945493 and 947522 in the forward strand. This gene contains 2 exons. The structure of the gene, analyzed with the Exonerate file, is described below:
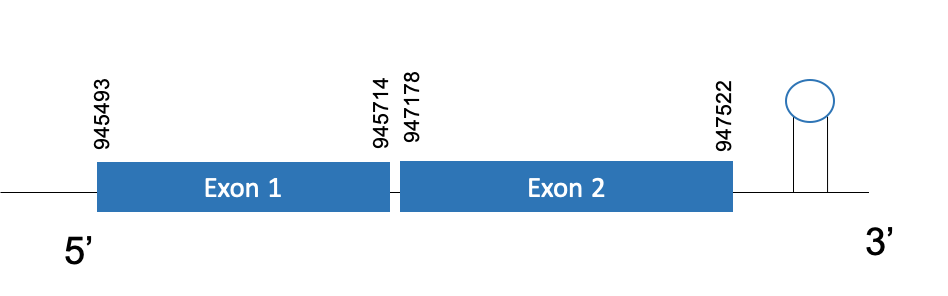
Exon 1:945493-945714
Exon 2:947178-947522
Seblastian was able to predict a known selenoprotein (GPx2) using the sequence from Chelonoidis abingdonii. Regarding the SECIS, one grade A SECIS was found between the locations 947635-947700. This secis is located in the same strand of our gene, and after it.
GPx3
The protein GPx3 is located in the scaffold PKMU01000857.1 between the position 2069251 and 2077792 in the forward strand. This gene contains 5 exons. The structure of the gene, analyzed with the Exonerate file, is described below:

Exon 1:2077706-2077792
Exon 2:2071532-2071685
Exon 3:2070740-2070857
Exon 4:2069570-2069669
Exon 5:2069251-2069445
Seblastian was not able to predict a known selenoprotein using the sequence from Chelonoidis abingdonii. It was also not capable of finding a SECIS.
GPx4
The protein GPx4 is located in the scaffold PKMU01000001.1 between the position 5440036 and 5442834 in the forward strand. This gene contains 5 exons. The structure of the gene, analyzed with the Exonerate file, is described below:

Exon 1:5440036-5440098
Exon 2:5440765-5440859
Exon 3:5441413-5441557
Exon 4:5442285-5442436
Exon 5:5442810-5442834
Seblastian was able to predict a selenoprotein (GPx4) using the sequence from Chelonoidis abingdonii. Regarding the SECIS, one grade A SECIS was found between the locations 5444262-5444334. This SECIS is located in the same strand of our gene, and after it.
GPx7
The protein GPx7 is located in the scaffold PKMU01008789.1 between the position 526415 and 542110 in the reverse strand. This gene contains 3 exons. The structure of the gene, analyzed with the Exonerate file, is described below:
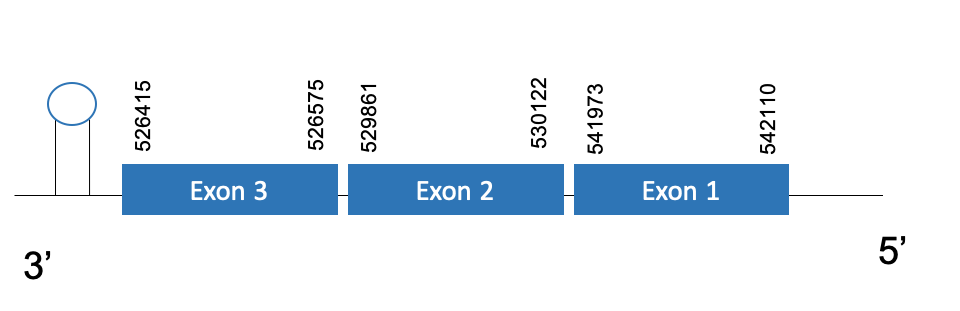
Exon 1:541973-542110
Exon 2:529861-530122
Exon 3:526415-526575
Seblastian was not able to predict a known selenoprotein using the sequence from Chelonoidis abingdonii. Regarding the SECIS, one grade B SECIS was found between the locations 481570-481653. This SECIS is located in the same strand of our gene, and after it.
GPx8
The protein GPx8 is located in the scaffold PKMU01009122.1 between the position 774516 and 779774 in the reverse strand. This gene contains 3 exons. The structure of the gene, analyzed with the Exonerate file, is described below:
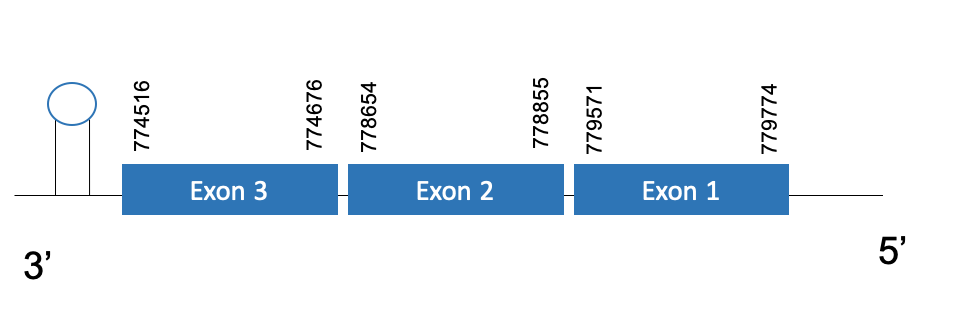
Exon 1:779571-779774
Exon 2:778654-778855
Exon 3:774516-774676
Seblastian was not able to predict a known selenoprotein using the sequence from Chelonoidis abingdonii. The accepted SECIS has a grade B and it is found in the positions 743796-743887. This SECIS is located in the same strand of our gene, and after it.
MsrA
The protein MsrA is located in the scaffold PKMU01001278.1 between the position 963309 and the position 1272944 in the reverse strand. This gene contains 5 exons. The structure of the gene, analyzed with the Exonerate file, is described below:
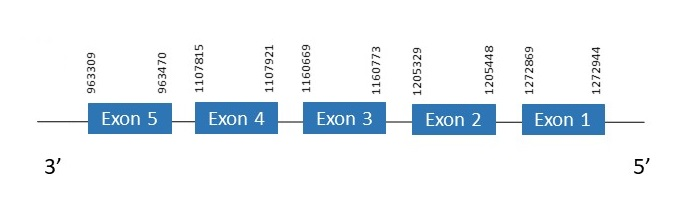
Exon 1:1272869-1272944
Exon 2:1205329-1205448
Exon 3:1160669-1160773
Exon 4:1107815-1107921
Exon 5:963309-963470
Seblastian was not able to predict a known selenoprotein using the sequence from Chelonoidis abingdonii. Regarding the SECIS, one grade B SECIS was found between the locations 1311451-1311522. This SECIS is located in the same strand of our gene, and before it.
Sel15 + SelM
Sel15
The protein Sel15 is located in the scaffold PKMU01001331.1 between the position 129427 and the position 163444 in the forward strand. This gene contains 2 exons. The structure of the gene, analyzed with the Exonerate file, is described below:
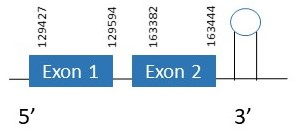
Exon 1:129427-129594
Exon 2:163382-163444
Seblastian was able to predict a known selenoprotein using the sequence from Chelonoidis abingdonii. Regarding the SECIS, one grade A SECIS was found between the locations 176321-176391. This SECIS is located in the same strand of our gene, and after it.
SelM
The protein SelM is located in the scaffold PKMU01008734.1 between the position 2162444 and the position 2176610 in the forward strand. This gene contains 5 exons. The structure of the gene, analyzed with the Exonerate file, is described below:
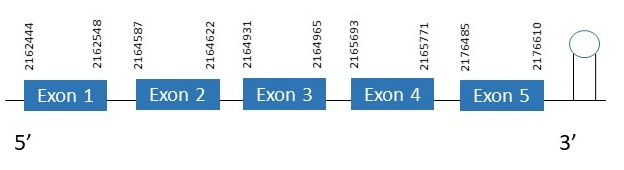
Exon 1:2162444-2162548
Exon 2:2164587-2164622
Exon 3:2164931-2164965
Exon 4:2165693-2165771
Exon 5:2176485-2176610
Seblastian was able to predict a known selenoprotein using the sequence from Chelonoidis abingdonii. Regarding the SECIS, one grade A SECIS was found between the locations 2176985-2177053. This SECIS is located in the same strand of our gene, and after it.
SelH
The protein SelH is located in the scaffold PKMU01001079.1 between the position 270158 and the position 271404 in the forward strand. This gene contains 3 exons. The structure of the gene, analyzed with the Exonerate file, is described below:
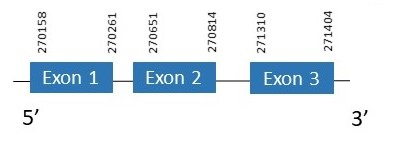
Exon 1:270158-270261
Exon 2:270651-270814
Exon 3:271310-271404
Seblastian was not able to predict a known selenoprotein using the sequence from Chelonoidis abingdonii. Regarding the SECIS, one grade B SECIS was found between the locations 261636-261716. This SECIS is located in the same strand of our gene, and before it.
SelI
The protein SelI is located in the scaffold PKMU01004179.1 between the position 933317 and the position 989453 in thereverse strand. This gene contains 10 exons. The structure of the gene, analyzed with the Exonerate file, is described below:
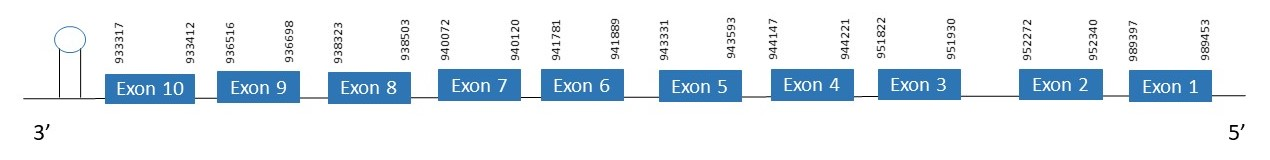
Exon 1:989397-989453
Exon 2:952272-952340
Exon 3:951822-951930
Exon 4:944147-977221
Exon 5:943331-943593
Exon 6:941781-941889
Exon 7:940072-940120
Exon 8:938323-938503
Exon 9:936516-936698
Exon 10:933317-933412
Seblastian was not able to predict a known selenoprotein using the sequence from Chelonoidis abingdonii. Regarding the SECIS, one grade A SECIS was found between the locations 931948-932025. This SECIS is located in the same strand of our gene, and after it.
SelK + SelS
SelK
The protein SelK is located in the scaffold PKMU01003079.1 between the position 139808 and the position 143690 in the reverse strand. This gene contains 4 exons. The structure of the gene, analyzed with the Exonerate file, is described below:
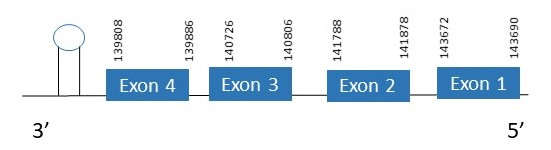
Exon 1:143672-143690
Exon 2:141788-141878
Exon 3:140726-140806
Exon 4:139808-139886
Seblastian was able to predict a known selenoprotein using the sequence from Chelonoidis abingdonii. Regarding the SECIS, one grade A SECIS was found between the locations 138734-138817. This SECIS is located in the same strand of our gene, and after it.
SelS
The protein SelS is located in the scaffold PKMU01007956.1 between the position 982332 and 988379 in the reverse strand. This gene contains 5 exons. The structure of the gene, analyzed with the Exonerate file, is described below:
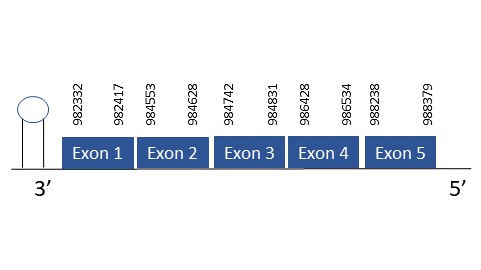
Exon 1:988238-988379
Exon 2:986428-986534
Exon 3:984742-984831
Exon 4:984553-984628
Exon 5:982332-982417
Seblastian was able to predict a known selenoprotein (SelS) using the sequence from Chelonoidis abingdonii. Regarding the SECIS, one grade A SECIS was found between the locations 981336-981416. This SECIS is located in the same strand of our gene, and after it.
SelN
The protein SelN is located in the scaffold PKMU01005290.1 between the position 4718252 and 4730686 in the reverse strand. This gene contains 11 exons. The structure of the gene, analyzed with the Exonerate file, is described below:
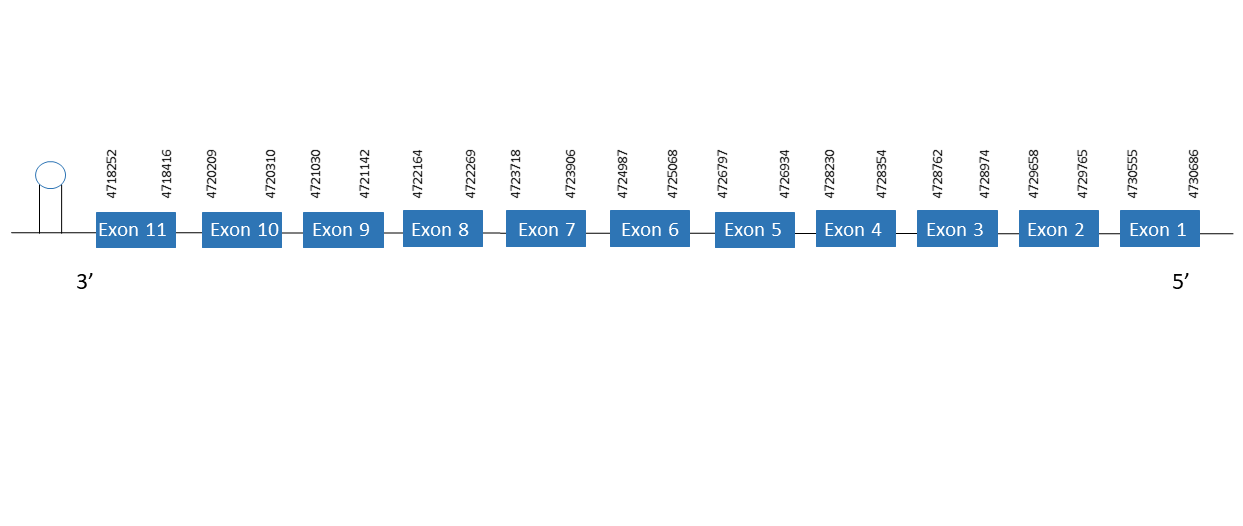
Exon 1:4730555-4730686
Exon 2:4729658-4729765
Exon 3:4728762-4728974
Exon 4:4728230-4728354
Exon 5:4726797-4726934
Exon 6:4724987-4725068
Exon 7:4723718-4723906
Exon 8:4722164-4722269
Exon 9:4721030-4721142
Exon 10:4720209-4720310
Exon 11:4718252-4718416
Seblastian was not able to predict a known selenoprotein using the sequence from Chelonoidis abingdonii. Regarding the SECIS, one grade A SECIS was found between the locations 4713150-4713220. This SECIS is located in the same strand of our gene, and after it.
SelO
The protein SelO is located in the scaffold PKMU01009289.1 between the position 1697628 and 1711296 in the forward strand. This gene contains 7 exons. The structure of the gene, analyzed with the Exonerate file, is described below:
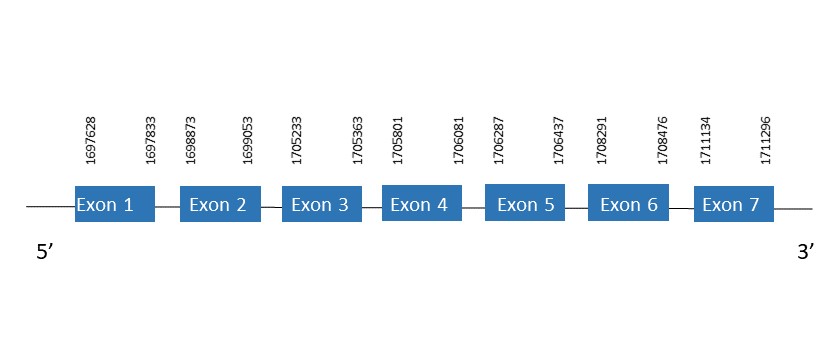
Exon 1:1697628-1697833
Exon 2:1698873-1699053
Exon 3:1705233-1705363
Exon 4:1705801-1706081
Exon 5:1706287-1706437
Exon 6:1708291-1708476
Exon 7:1711134-1711296
Seblastian was not able to predict a known selenoprotein using the sequence from Chelonoidis abingdonii. It was also not capable of finding a SECIS.
SelP
The protein SelP is located in the scaffold PKMU01005800.1 between the position 532753 and the position 536049 in the reverse strand. This gene contains 3 exons. The structure of the gene, analyzed with the Exonerate file, is described below:

Exon 1:535874-536049
Exon 2:535330-535542
Exon 3:532753-532864
Seblastian wasn’t able to predict either a selenoprotein nor a SECIS using the sequence from Chelonoidis abingdonii.
SelPb
The protein SelPb is located in the scaffold PKMU01005800.1 between the position 532717 and 536082 in the reverse strand. This gene contains 3 exons. The structure of the gene, analyzed with the Exonerate file, is described below:
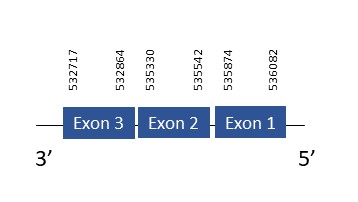
Exon 1:535874-536082
Exon 2:535330-535542
Exon 3:532717-532864
Seblastian was not able to predict a selenoprotein using the sequence from Chelonoidis abingdonii. Regarding the SECIS, one grade A SECIS was found between the locations 747203-747275. This secis is located in the same strand of our gene, and after it.
Methionine-R-sulfoxide reductase 1 (Selenoprotein R)
SelR1
The protein SelR1 is located in the scaffold PKMU01006623.1 between the position 741771 and 743931 in the forward strand. This gene contains 3 exons. The structure of the gene, analyzed with the Exonerate file, is described below:
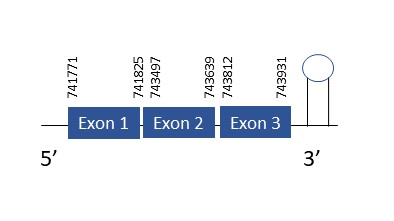
Exon 1:741771-741825
Exon 2:743497-743639
Exon 3:743812-743931
Seblastian was not able to predict a known selenoprotein using the sequence from Chelonoidis abingdonii. The accepted SECIS has a grade B and it is found in the positions 743796-743887. This SECIS is located in the same strand of our gene, and after it.
SelR2
The protein SelR2 is located in the scaffold PKMU01001068.1 between the position 2087949 and 2101199 in the forward strand. This gene contains 4 exons. The structure of the gene, analyzed with the Exonerate file, is described below:
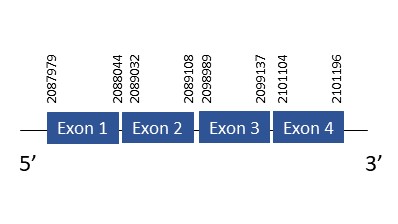
Exon 1:2087949-2088044
Exon 2:2089032-2089108
Exon 3:2098989-2099137
Exon 4:2101104-2101199
Seblastian was not able to predict a selenoprotein using the sequence from Chelonoidis abingdonii. It was also not capable of finding a valid SECIS.
SelR3
The protein SelR3 is located in the scaffold PKMU01002212.1 between the position 5686 and 101324 in the forward strand. This gene contains 5 exons. The structure of the gene, analyzed with the Exonerate file, is described below:
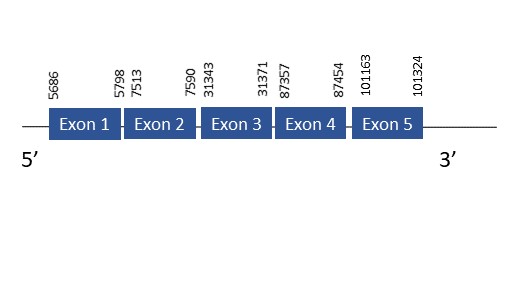
Exon 1:5686-5798
Exon 2:7513-7590
Exon 3:31343-31371
Exon 2:87357-87454
Exon 3:101163-101324
Seblastian was not able to predict a known selenoprotein using the sequence from Chelonoidis abingdonii. It was also not capable of finding a SECIS.
SelT
The protein SelT is located in the scaffold PKMU01006301.1 between the position 1265830 and 1280060 in the forward strand. This gene contains 5 exons. The structure of the gene, analyzed with the Exonerate file, is described below:
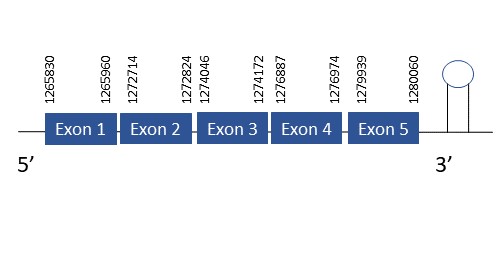
Exon 1:1265830-1265960
Exon 2:1272714-1272824
Exon 3:1274046-1274172
Exon 4:1276887-1276974
Exon 5:1279939-1280060
Seblastian was not able to predict a selenoprotein using the sequence from Chelonoidis abingdonii. Regarding the SECIS, one grade A SECIS was found between the locations 1284080-1284155. This SECIS is located in the same strand of our gene, and after it.
Thioredoxin reductases
TR1
The protein TR1 is located in the scaffold PKMU01001697.1 between the position 81770 and the position 110367 in the forward strand. This gene contains 13 exons. The structure of the gene, analyzed with the Exonerate file, is described below:

Exon 1:81770-81853
Exon 2:83036-83108
Exon 3:83228-83347
Exon 4:84107-84249
Exon 5:84375-84490
Exon 6:86650-86875
Exon 7:90806-90898
Exon 8:93560-93636
Exon 9:96589-96745
Exon 10:98776-98883
Exon 11:101392-101487
Exon 12:104050-104184
Exon 13:110302-110367
Seblastian was able to predict a known selenoprotein using the sequence from Chelonoidis abingdonii. The accepted SECIS has a grade A and it is found in the positions 110620-110696. This SECIS is located in the forward strand, in the 3’UTR region.
TR2
The protein TR2 is located in the scaffold PKMU01010066.1 between the position 69617 and the position 117992 in the reverse strand. This gene contains 15 exons. The structure of the gene, analyzed with the Exonerate file, is described below:
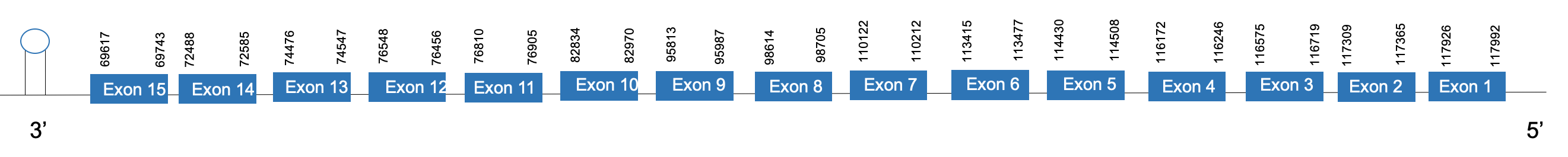
Exon 1:117926-117992
Exon 2:117309-117365
Exon 3:116575-116719
Exon 4:116172-116246
Exon 5:114430-114508
Exon 6:113415-113477
Exon 7:110122-110212
Exon 8:98614-98705
Exon 9:95813-95987
Exon 10:82834-82970
Exon 11:76810-76905
Exon 12:76456-76548
Exon 13:74476-74547
Exon 14:72488-72585
Exon 15:69617-69743
Seblastian was not able to predict a known selenoprotein using the sequence from Chelonoidis abingdonii. The accepted SECIS has a grade A and it is found in the positions 65624-65701. This SECIS is located in the reverse strand, in the 3’UTR region.
TR3
The protein TR3 is located in the scaffold PKMU01001735.1 between the position 7625889 and the position 7643891 in the reverse strand. This gene contains 15 exons. The structure of the gene, analyzed with the Exonerate file, is described below:

Exon 1:7645395-7645458
Exon 2:7643782-7643891
Exon 3:7642249-7642365
Exon 4:7641571-7641643
Exon 5:7640172-7640291
Exon 6:7639507-7639649
Exon 7:7637563-7637678
Exon 8:7637223-7637448
Exon 9:7636864-7636956
Exon 10:7636260-7636336
Exon 11:7634744-7634900
Exon 12:7634370-7634477
Exon 13:7628505-7628600
Exon 14:7626314-7626448
Exon 15:7625889-7625954
Seblastian was not able to predict a known selenoprotein using the sequence from Chelonoidis abingdonii. The accepted SECIs has a grade A and it is found in the positions 7625577-7625661. This secis is located in the reverse strand, in the 3’UTR region.
Selenoprotein U
SelU1
The protein SelU1 is located in the scaffold PKMU01001923.1 between the position 404239 and 417868 in the reverse strand. This gene contains 5 exons. The structure of the gene, analyzed with the Exonerate file, is described below:
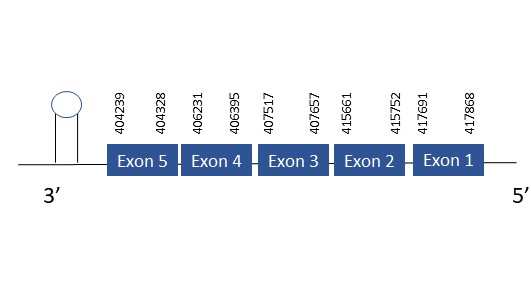
Exon 1:417691-417868
Exon 2:415661-415752
Exon 3:407517-407657
Exon 4:406231-406395
Exon 5:404239-404328
Seblastian was not able to predict a selenoprotein using the sequence from Chelonoidis abingdonii. Regarding the SECIS, one grade A SECIS was found between the locations 403935-404006. This SECIS is located in the same strand of our gene, and after it.
SelU2
The protein SelU2 is located in the scaffold PKMU01002121.1 between the position 38721 and 4335 in the forward strand. This gene contains 6 exons. The structure of the gene, analyzed with the Exonerate file, is described below:
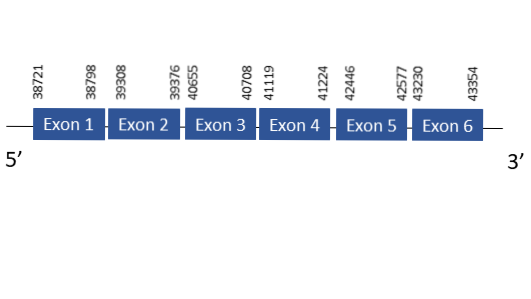
Exon 1:38721-38798
Exon 2:39308-39376
Exon 3:40655-40708
Exon 4:41119-41224
Exon 5:42446-42577
Exon 6:43230-43354
Seblastian was not able to predict a known selenoprotein using the sequence from Chelonoidis abingdonii. It was also not capable of finding a SECIS.
SelU3
The protein SelU3 is located in the scaffold PKMU01007067.1 between the position 4850194 and the position 4862512 in the reverse strand. This gene contains 6 exons. The structure of the gene, analyzed with the Exonerate file, is described below:

Exon 1:4862450-4862512
Exon 2:4860543-4860747
Exon 3:4858582-4858633
Exon 4:4857715-4857778
Exon 5:4857025-4857100
Exon 6:4850194-4850312
Seblastian wasn’t able to predict either a selenoprotein nor a SECIS using the sequence from Chelonoidis abingdonii.
Selenoprotein W
SelW1
The protein SelW1 is located in the scaffold PKMU01003401.1 between the position 1603706 and the position 1606321 in the reverse strand. This gene contains 3 exons. The structure of the gene, analyzed with the Exonerate file, is described below:

Exon 1:1606268-1606321
Exon 2:1605253-1605327
Exon 3:1603706-1603783
Seblastian was not able to predict a known selenoprotein using the sequence from Chelonoidis abingdonii. The accepted SECIS has a grade A and it is found in the positions 1616552-1616462. This SECIS is located in the reverse strand, in the 3’UTR region.
SelW2
The protein SelW2 is located in the scaffold PKMU01002623.1 between the position 172868 and the position 174789 in the forward strand. This gene contains 3 exons. The structure of the gene, analyzed with the Exonerate file, is described below:

Exon 1:172868-172964
Exon 2:174039-174115
Exon 3:174709-174789
Seblastian was not able to predict a known selenoprotein using the sequence from Chelonoidis abingdonii. The accepted SECIS has a grade B and it is found in the positions 175706-175783. This SECIS is located in the forward strand, in the 3’UTR region.
Selenoprotein machinery
eEFSec
The protein eEFSec is located in the scaffold PKMU01005278.1. between the position 625504 and 802532 in the forward strand. This gene contains 7 exons. The structure of the gene, analyzed with the Exonerate file, is described below:

Exon 1:625504-625765
Exon 2:687911-688118
Exon 3:693693-693759
Exon 4:697649-697813
Exon 5:733942-734583
Exon 6:753222-753378
Exon 7:802351-802532
Seblastian was not able to predict a known selenoprotein using the sequence from Chelonoidis abingdonii. Regarding the SECIs, one grade B SECIS was found between the locations 727708-727760. This SECIS is located in the other DNA strand and inside the predicted gene. That’s why we don’t have taken it into account in the figure and in the table of results.
SBP2
The protein SBP2 is located in the scaffold PKMU01000807.1 between the position 1070817 and the position 1100301 in the forward strand. This gene contains 16 exons. The structure of the gene, analyzed with the Exonerate file, is described below:

Exon 1:1070817-1071412
Exon 2:1072609-1072828
Exon 3:1077634-1077788
Exon 4:1078484-1078704
Exon 5:1079336-1079498
Exon 6:1081170-1081302
Exon 7:1082223-1082392
Exon 8:1083522-1083663
Exon 9:1083830-1083997
Exon 10:1085131-1085214
Exon 11:1085721-1085852
Exon 12:1092181-1092296
Exon 13:1093316-1093552
Exon 14:1096701-1096842
Exon 15:1097979-1098303
Exon 16:1100120-1100301
Seblastian wasn’t able to predict either a selenoprotein nor a SECIS using the sequence from Chelonoidis abingdonii.
PSTK
The protein PSTK is located in the scaffold PKMU01003645.1 between the position 1863877 and the position 1870699 in the reverse strand. This gene contains 6 exons. The structure of the gene, analyzed with the Exonerate file, is described below:
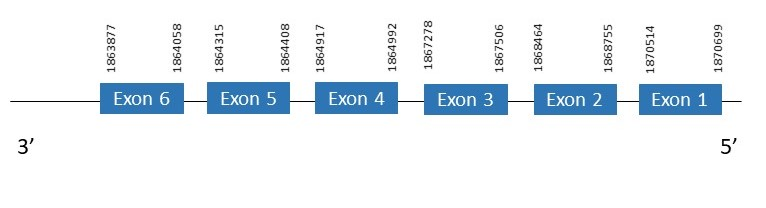
Exon 1:1870514-1870699
Exon 2:1868464-1868755
Exon 3:1867278-1867506
Exon 4:1864917-1864992
Exon 5:1864315-1864408
Exon 6:1863877-1864058
Seblastian wasn’t able to predict a known selenoprotein using the sequence from Chelonoidis abingdonii. Regarding the SECIS, none of them was found.
SECp43
The protein SECp43 is located in the scaffold PKMU01005290.1 between the position 1867920 and the position 1881788 in the forward strand. This gene contains 8 exons. The structure of the gene, analyzed with the Exonerate file, is described below:
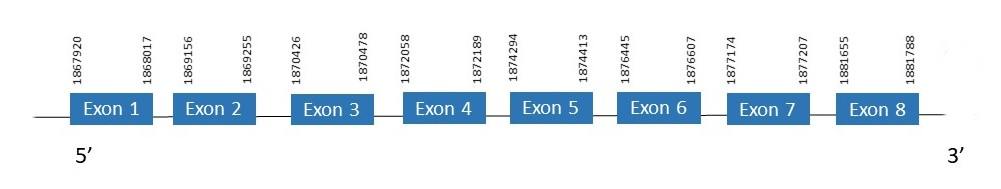
Exon 1:1867920-1868017
Exon 2:1869156-1869255
Exon 3:1870426-1870478
Exon 4:1872058-1872189
Exon 5:1874294-1874413
Exon 6:1876445-1876607
Exon 7:1877174-1877207
Exon 8:1881655-1881788
Seblastian was not able to predict a known selenoprotein using the sequence from Chelonoidis abingdonii. Regarding the SECIs, one grade B SECIS was found between the locations 1930863-1930952. This SECIS is not located in the same strand of our gene.
SecS
The protein SecS is located in the scaffold PKMU01006445.1 between the position 939558 and the position 992103 in the forward strand. This gene contains 11 exons. The structure of the gene, analyzed with the Exonerate file, is described below:
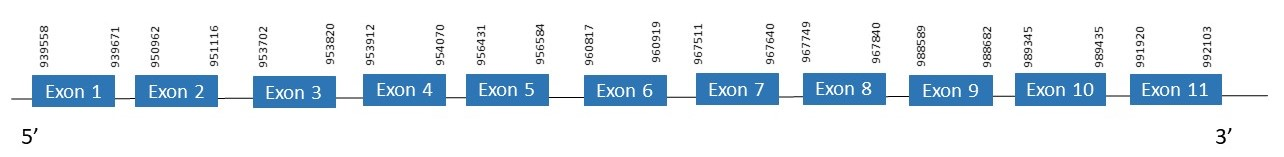
Exon 1:939558-939671
Exon 2:950962-951116
Exon 3:953702-953820
Exon 4:953912-954070
Exon 5:956431-956584
Exon 6:960817-960919
Exon 7:967511-967640
Exon 8:967749-967840
Exon 9:988589-988682
Exon 10:989345-989435
Exon 11:991920-992103
Seblastian was not able to predict a known selenoprotein using the sequence from Chelonoidis abingdonii. Regarding the SECIs, one grade B SECIS was found between the locations 972401-972474. This SECIS is located inside the gene.
SPS1
The protein SPS1 is located in the scaffold PKMU01001170.1 between the position 242377 and the position 263996 in the forward strand. This gene contains 8 exons. The structure of the gene, analyzed with the Exonerate file, is described below:

Exon 1:242377-242569
Exon 2:245419-245522
Exon 3:251707-251814
Exon 4:252834-252988
Exon 5:253914-254004
Exon 6:256919-257018
Exon 7:259573-259785
Exon 8:263785-263996
Seblastian was not able to predict a known selenoprotein using the sequence from Chelonoidis abingdonii. The accepted SECIs has a grade B and it is found in the positions 213852-213924. This SECIS is located in the forward strand, in the 5’UTR region.
SPS2
The protein SPS2 is located in the scaffold PKMU01001233.1 between the position 112584 and the position 118501 in the reverse strand. This gene contains 7 exons. The structure of the gene, analyzed with the Exonerate file, is described below:

Exon 1:118385-118501
Exon 2:117840-117947
Exon 3:114996-115150
Exon 4:114349-114439
Exon 5:113994-114093
Exon 6:113406-113618
Exon 7:112584-112810
Seblastian was not able to predict a known selenoprotein using the sequence from Chelonoidis abingdonii. The accepted SECIs has a grade A and it is found in the positions 112034-111960. This SECIS is located in the reverse strand, in the 3’UTR region.






































