Be aware of
The importance of selenoproteines
Selenium and Selenocysteine
Selenium (Se) is a non-metal chemical element with atomic number 34. Although it is toxic in large doses, selenium is an essential micronutrient for animals and with functions in human health. Selenium deficiency has been recognized as a contributing factor to pathophysiological conditions, such as heart disease, neuromuscular disorders, cancer, male infertility and inflammation [Labunskyy et al., 2014].
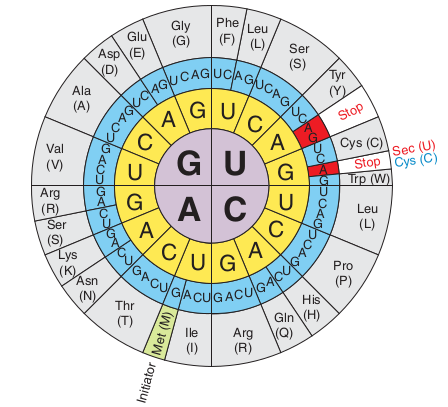
Selenoproteins contain at least one selenocysteine (Sec or U), which is a Se-containing aminoacid [Labunskyy et al., 2014]. These proteins are present in the three domains of life (eukarya, archaea, and bacteria). Sec is considered the 21st aminoacid of the genetic code. It is not found in a standard way in the genetic code, as it is encoded by the codon UGA(Figure 1: Genetic code including Sec codon), which normally acts as a stop codon. For this reason, Sec synthesis requires specific translation machinery.
Selenoproteins
Selenoproteins constitute a family characterized by proteins containing one or more selenocysteines. In eukarya, selenoproteins are largely present in animals, but are absent in higher plants and yeasts.
The function of approximately half of mammalian selenoproteins is not known. Among the functionally characterized selenoproteins, many have a role in redox regulation. Selenoproteins can be broadly classified into two types: housekeeping and stress-related. Housekeeping selenoproteins participate in functions critical to cell survival, whereas stress-related selenoproteins are not essential for survival and often show decreased expression in Se-deficient conditions [Mariotti, 2012]. A set of selenoproteins in an organism is known as the selenoproteome. The human selenoproteome is encoded in 25 selenoprotein genes [Mariotti, 2012].
Comparative analyses of organisms containing large selenoproteomes with those lacking selenoproteins revealed that several groups of terrestrial organisms reduced their dependence on selenium by replacing selenoproteins with Cys homologs or loosing selenoproteins. Cys mutants of selenoproteins can partially preserve their activities. Interestingly, aquatic organisms have large selenoproteomes, indicating that the environment is a key factor in the evolution of the selenoproteome.
In mammals, the replacements were always from Sec to Cys, not the other way round. Although some selenoprotein genes appeared in mammals (GPx6, SelV, GPx5, the latter immediately converted to a Cys version), they evolved by gene duplication.
Sec biosynthesis
Sec is the only known amino acid in eukaryotes whose biosynthesis occurs once the precursor amino acid (serine) is already linked to the tRNA, known as Sec tRNA[Ser]Sec. This tRNA is very different from other tRNAs because it has some variations in its sequence and it also suffers some post/transcriptional modifications [Labunskyy et al., 2014]. The gene for tRNA[Ser]Sec is known as Trsp. It is a gene that is in a single copy in every organism but fishes.
Aminoacylation of tRNA[Ser]Sec by seryl-tRNA synthetase (SerS) is the first step in the byosinthesis of Sec. Serine recognizes the tRNA thanks to the modifications on its structure. Then, this seryl-tRNA[Ser]Sec has to be converted to selenocysteyl-tRNA[Ser]Sec by Sec synthase (SecS), which incorporates selenophosphate, the active form of selenium, into the aminoacid backbone and forms Sec-tRNA. Previously to this step, a phosphoseryl-tRNA has to be formed by phosphoseryl-tRNA kinase (PTSK).
SPS2 enzyme is also implicated in cysteine (Cys) biosynthesis. Cys is an essential aminoacid in mammals, but the synthesis of Cys on tRNA[Ser]Sec is a new de novo discovered pathway for its synthesis. SPS2 is a selenoprotein, and it possibly serves as an autoregulator of selenoprotein synthesis.
Sec is decomposed by Sec lyase (SCL), which catalyses a degradation of Sec to L-Ala and selenium. It is thought that SCL recycles the selenium from Sec during degradation of selenoproteins.
Sec incorporation into proteins
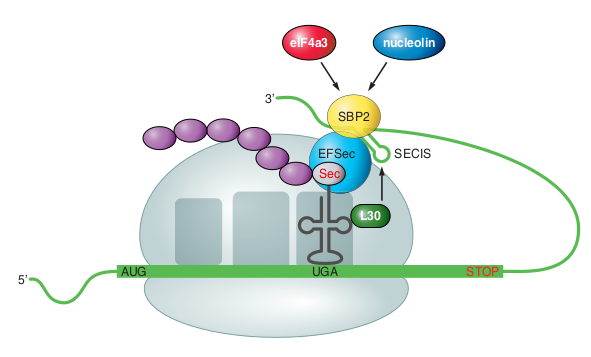
Incorporation of Sec into proteins is dictated by UGA codons present in selenoprotein mRNAs. The special recognition of this mRNA is dependent on the SECIS element [Labunskyy et al., 2014]. When a ribosome encounters the UGA codon, Sec machinery interacts with the translation machinery to increase the coding potential of UGA codons. In response to the SECIS element, Sec-tRNA[Ser]Sec translates UGA as Sec. In this process, the Sec-specific translation elongation factor (eEFSec) is also needed, as well as the SECIS binding protein 2 (SBP2). SBP2 is associated with ribosomes and contains a RNA-binding domain that binds to SECIS elements and interacts with eEFSEc, which recruits Sec-tRNA[Ser]Sec. Additional SECIS-binding proteins are ribosomal protein L30 (which constitutes a part of the basal Sec insertion machinery) nucleolin and eIF4a3 (Figure 2: Representation of the translation machinery necessary for Sec). These proteins serve as regulatory proteins that modulate synthesis of selenoproteins.
SECIS elements are cis-acting stem-loop RNA structures in the 3’-UTR of selenoprotein mRNAs. In bacteria, SECIS elements are located just after the coding region, whereas eukarya and archaea can have them much further. Eukaryotic SECIS elements are formed by two helixes separated by an internal loop, a GA Quartet (SECIS core) structure, and an apical loop or bulge. The GA Quartet is the part that interacts with SBP2. Eukaryotic SECIS can be classified in two classes regarding the presence of the apical bulge.
Selenoproteins and bioinformatics
Since selenoproteins genes contain UGA codons, which are normally used as STOP signals during protein translation, selenoproteins are often missannotated in sequence databases. Bioinformatics is a useful tool for the identification of selenoproteins by two different approaches:
- Every selenoprotein gene has the UGA codon encoding Sec and a SECIS element. However, SECIS have different sequences, so the secondary RNA structure has to be predicted to improve its sensivity. By finding these two elements, a selenoprotein can be predicted.
- Identification of selenoproteins comparing homologs in different species lts us find them in a new genome.
We are going to take advantage of these approaches to identificate the selenoproteins and their machinery in the newly-sequenced genome of Chelonoidis abingdonii.