Let's talk about what we have found
Exploring the selenoproteome of Chelonoidis abingdonii
In this study we want to characterize the selenoproteome and the machinery in charge of synthesizing them in the animal species Chelonoidis abingdonii, an extinct tortoise that lived in the Pinta Island (Galapagos, Ecuador).
The selenoproteome consists of all the selenoproteins present in a genome. Selenoproteins contain at least one selenocysteine (Sec). Sec residue is codified by the UGA codon, which normally acts as a stopping translation signal, and thus requires a specific machinery to be produced.
To correctly predict and assess the Chelonoidis abingdonii selenoproteome and the proteins related to its synthesis, we have studied the homology between the already described selenoproteins of Homo sapiens and the proteins encoded by the genes of the tortoise. To do this prediction, our team designed an automatic program that, using the data obtained through tBLASTn, applied Exonerate, GeneWise and T-Coffee to obtain the alignment between the predicted sequences of the selenoproteins in these species and the human (or lizzard) selenoproteins. The predicted genes were analysed through Seblastian to see if they could produce a selenoprotein. SECISearch3 was used to predict if the mRNA produced by our gene could contain any SECIS.
Iodothyronine deiodinases
The iodothyronine deodinase (DI) family of selenoproteins consists of three paralogous proteins in mammals (DI1, DI2 and DI3), which are involved in regulation of thyroid hormone activity by reductive deodination [Labunskyy et al., 2014]. The majority of thyroid hormone that is produced by thyroid is secreted in its inactive form, thyroxine (T4). These proteins metabolize it to inactivate or deactivate it. Surprisingly, homologs of mammalian deodinases occur not only in other vertebrates, but are also found in simple eukaryotes and bacteria. The function of deodinase homologs in these organisms is not known. These proteins have distinct subcellular localizations and tissue expression. All of them are transmembrane proteins which have a thioredoxin fold and form homodimers. The circulating levels of thyroid hormone are primarily regulated by DI1 activity. However, DI2 and DI3 have been implicated in fine-tuning local intracellular T3 concentrations in a tissue-specific manner, without changing overall serum levels of T3. All of the iodothyronine deiodinases contain Sec. The active-site Sec residue is located in the N-terminal part of the protein. However, in DI2, an additional Sec, whose function is unknown, is present in the C-terminal region. Other authors state that it appears that the primary function of the second UGA is to serve as stop codon. This second Sec does not participate in the catalytic mechanism and is dispensable for the D2 functional activity. The 3 DIs have a low sequence identity (50%), but they have similar overall topology and structural organization [Mariotti et al., 2012] The three Iodothyronine deiodinases (DI1, DI2 and DI3) are found in all the vertebrates. Bonny fishes have duplicatd DI3 (producing DI3b).
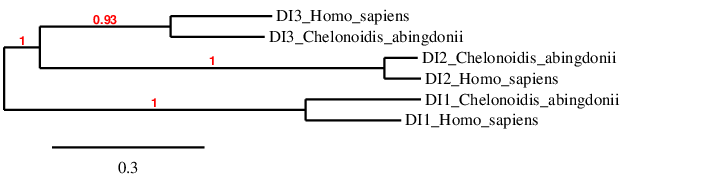
In the phylogeny we see that we have probably predicted the selenoproteins correctly, because each type of DI clusters directly with its human homolog, and afterwards it is related to the other DIs.
DI1
DI1 converts the form of thyroid hormone T4 to the active form T3. It is located in the plasma membrane. [Labunskyy et al., 2014]. The gene in the scaffold PKMU01008277.1 is the only one that was selected from the Tblastn prediction based on our criteria. The gene is located in the reverse strand. This gene contains 4 exons. It contains a Sec in the same position as human DI1 protein does. T_Coffee shows a good identity and an even higher similarity, and the predicted protein has almost the same length that the human one. However, the protein we have predicted lacks the first 9 amino acids. This can be because exonerate has not correctly predicted the structure of the gene, and thus the first 9 amino acids are lacking (including the Met in the beginning of every protein). In the sequence studied, one grade A SECIS was found. This SECIS is predicted after the end of our gene, and in the same strand, so we can accept it. Moreover, Seblastian also predicted DI1 inside the Chelonoidis abingdonii sequence we have studied. For all these reasons we can accept our prediction, confirming that Chelonoidis abingdonii has DI1.
DI2
DI2 converts the form of thyroid hormone T4 to the active form T3. It is located in the endoplasmic reticulum. One of the major effects of DI2 deficiency is observed in skeletal muscle tissue. During normal development, DI2 expression reaches maximum levels in skeletal muscles just after birth and then decreases in the postnatal period. Moreover, activity of DI2 is increased in muscle following injury and is associated with enhanced transcription of T3-dependent genes that are required for muscle differentiation and regeneration implicating DI2 in muscle regeneration process. Another function of DI2 happens in response to cold exposure. In this situation, expression of DI2 in brown adipose tissue increases up to 50-fold leading to activation of thyroid hormone in this tissue to produce a thermogenic effect. [Labunskyy et al., 2014] The gene in the scaffold PKMU01005068.1 is the only one that was selected from the Tblastn prediction based on our criteria. The gene is located in the reverse strand. This gene contains 2 exons. It contains a Sec in the same position as human DI1 protein does. T_Coffee shows a very good identity, with just 6 gaps inside the sequence. The two proteins have almost the same length. The predicted protein has Met at its beginning, therefore it probably corresponds to a real protein. In the sequence studied, two SECIS were predicted. One grade B SECIS was rejected because it was not in the same strand of the predicted gene. Thus, we accepted the grade A SECIS that was found. This SECIS is predicted after the end of our gene, and in the same strand. However, Seblastian did not predict any selenoprotein inside the Chelonoidis abingdonii sequence we have studied. For all these reasons we can accept our prediction, confirming that Chelonoidis abingdonii has DI2.
DI3
DI3, in contrast to DI1 and DI2, can inactivate T3 and T4 leading the formation of T2 and T3 reverse. It is located in the plasma membrane. Regarding its tissue-specific functions, as thyroid hormone signaling affects the balance between proliferation and differentiation, it appears that decreased T3 levels caused by enhanced DI3 expression contribute to high proliferation rates of basal cell carcinoma. Interestingly, all detected DI3 genes (including DI3b) are intronless. [Mariotti et al., 2012]. We decided to study two Tblastn predictions for DI3 gene based on our criteria. We finally accepted the gene in the scaffold PKMU01001135.1. The gene is located in the reverse strand. The rejected Tblastn prediction was in the same positions than the gene that we said to be for DI1. When studying this prediction, both T_Coffee (which showed a better alignment with DI1 than with DI3) and Seblastian (which predicted DI1 in the input sequence) supported the hypothesis that this prediction corresponded to DI1 and not DI3. The accepted gene contains 2 exons. It contains a Sec in the same position as human DI3 protein does. T_Coffee shows a good identity and an even higher similarity, and the predicted protein’s length is a bit shorter that the human one The last part of the predicted gene does not have a good alignment. However, the predicted protein starts with a Met, so it is possible that we have predicted a real gene. In the sequence studied, one viable grade A SECIS was found. This SECIS is predicted after the end of our gene, and in the same strand, therefore we can accept it. Moreover, Seblastian also predicted DI3 inside the Chelonoidis abingdonii sequence we have studied. For all these reasons we can accept our prediction, confirming that Chelonoidis abingdonii has DI3.
Glutathione Peroxidases
Glutathione peroxidases are the largest selenoprotein family in vertebrates. They are found in vertebrates, as well as single-cell eukaryotes and prokaryotes. GPxs play a wide range of physiological functions in organisms and are involved in hydrogen peroxide (H2O2) signaling, detoxification of hydroperoxides and maintaining cellular redox homeostasis. Selenoproteins of the glutathione peroxidase (GPx) family are widespread in all three domains of life. In mammals, there are eight GPx paralogs, from which five (GPx1, GPx2, GPx3, GPx4, and GPx6) contain a Sec residue in their active site. In the other three GPx homologs (GPx5, GPx7, and GPx8), the active-site Sec is replaced by Cys. Moreover, GPx6 homologs in some mammals are not selenoproteins and have a Cys in the active site [Labunskyy et al., 2014]. GPx are highly conserved, and this is even more evident in Sec-containing GPx (in the case of mammalian GPx, they share approximately 80% of identity) [Mariotti et al., 2012]. GPx1, GPx2, GPx3 and possibly GPx6 work as tetramers, whereas GPx4 is a monomer. This protein family includes the two types of selenoproteins mentioned in the introduction of this webpage. GPx1 is a stress-related selenoprotein which is highly regulated by Se availability (its expression decreases drastically when there is Se deficiency), whereas GPx4 is a housekeeping selenoprotein (it less affected by dietary selenium status and often serve functions critical to cell survival) [Labunskyy et al., 2014; Mariotti et al., 2012].
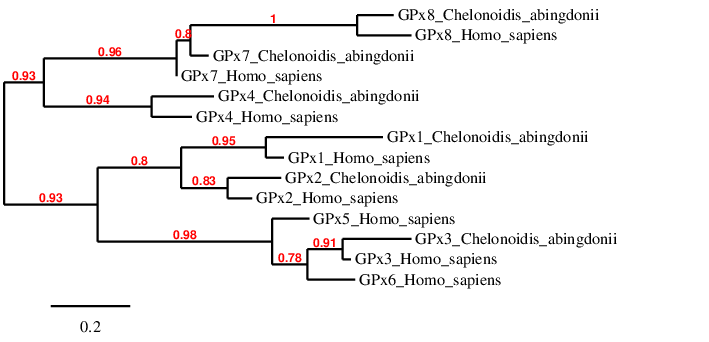
Regarding the phylogenetic tree, we can see that each human protein pairs specifically with their homolog in Chelonoidis abingdonii. This happens with GPx1, GPx2, GPx3, GPx4 and GPx8. Although Chelonoidis abingdonii’s GPx7 is not directly linked to the human GPx7, it is in a very short distance from it, whereas it is very far from far from GPx8. We can hypothesize that this is due to the fact that the program we have used is heuristic and that GPx7 and GPx8 have evolved in a similar way in Chelonoidis abingdonii. It is interesting to see that GPx7 and GPx8 are related to GPx4. This is because GPx7 and GPx8 evolved form GPx4. We can also see that GPx5 and GPx6 are not found in the tortoise, however they are quite similar to GPx3 (because they come from a GPx3 duplication in mammals, and they have not changed significantly).
GPx1
It is the most abundant selenoprotein in mammals and it was the first selenoprotein found in them. Its expression is abundant in the liver and the kidney. It is located in the cytosol and catalyses GSH-dependent reduction of hydrogen peroxide to water (in fact, the Sec residue of the active site is necessary for this function). For this reason it has a protective role under oxidative stress and signalling cascades (because H2O2 functions as an important signaling molecule). However, in some situations, it can also act as a pro-oxidative agent. Interestingly, some authors have found that overexpression of GPx1 in mice may lead to a higher risk of developing type 2 diabetes and obesity [Labunskyy et al., 2014]. We decided to study two Tblastn predictions for GPx1 gene based on our criteria. We finally accepted the gene in the scaffold PKMU01007833.1. The gene is located in the forward strand. The rejected Tblastn prediction was in the same positions than the predicted DI1 gene. When studying this prediction, both T_Coffee (which showed a better alignment with GPx2 than with GPx1) and Seblastian (which predicted GPx2 in the input sequence) supported the hypothesis that this prediction corresponded to GPx2 and not GPx1. This gene contains 2 exons. It contains a Sec in the same position as human GPx1 protein does. T_Coffee shows a good identity and an even higher similarity, and the predicted protein has almost the same length as the human one. However, the gene we have predicted lacks the first 7 and the last 3 residues. This could be because exonerate has not correctly predicted the structure of the gene and, therefore, these residues are lacking (including the Met at the beginning of every protein). In the sequence studied, one grade A SECIS was found. This SECIS is predicted after the end of our gene, and in the same strand, therefore we can accept it. Seblastian did not predict any selenoprotein inside the studied Chelonoidis abingdonii sequence. For all these reasons we can accept our prediction, confirming that Chelonoidis abingdonii has GPx1.
GPx2
It is only found in vertebrates. GPx2 is primarily found in the epithelium of the gastrointestinal tract It is also implicated in the GSH-dependent detoxification of hydrogen peroxide. It is thought to have a role in the development of cancer, however it is not known whether it prevents it (it is controlled by the antioxidant response transcription factor Nrf2) or promotes it (is it regulated by the Wnt pathway, which is involved in cellular proliferation) [Labunskyy et al., 2014].
We decided to study two Tblastn predictions for GPx2 gene based on our criteria. The rejected Tblastn prediction was in the same positions as the predicted GPx1 gene. When studying this prediction, T_Coffee showed a better alignment with GPx1 than with GPx2, supporting the hypothesis that this prediction corresponded to GPx1 and not GPx2. Furthermore, the accepted prediction (in the scaffold PKMU01001079.1.) shows GPx2 in Seblastian. We finally accepted the gene in the scaffold PKMU01001079.1. This gene is located in the forward strand. The accepted gene contains 5 exons. It contains a Sec in the same position as human GPx2 protein does. T_Coffee shows a very good identity and the predicted protein has the same length that the human one (it just lacks the last amino acid). Moreover, the predicted protein has Met at the beginning of it, so it probably corresponds to a real protein. In the sequence studied, one grade A SECIS was found. This SECIS is predicted after the end of our gene, and in the same strand, therefore we can accept it. For all these reasons we can accept our prediction, confirming that Chelonoidis abingdonii has GPx2.
GPx3
GPx3 is secreted primarily from kidney and is the major GPx form in plasma in mammals. It is also implicated in the GSH-dependent detoxification of hydrogen peroxide [Labunskyy et al., 2014].
The gene in the scaffold PKMU01000857.1 is the only one that was selected from the Tblastn prediction based on our criteria. The gene is located in the reverse strand. This gene contains 5 exons. It contains a Sec in the same position as the human GPx3 protein does. T_Coffee shows a good identity and an even higher similarity, and the predicted protein has almost the same length that the human one (except for the last 8 amino acids of our prediction). Furthermore, the predicted protein has Met at the beginning of it, so it probably corresponds to a real protein. In the sequence studied, no SECIS has been found. As we have predicted one Sec, the SECIS should be present. It is possible that this part of the genome was not correctly annotated, therefore the SECIS cannot be detected. Seblastian, has also not predicted any selenoprotein. We can accept our prediction, confirming that Chelonoidis abingdonii has GPx3.
GPx4
GPx4 is involved in the reduction of complex phospholipid hydroperoxides that are associated with membranes. Instead using GSH, it uses protein thiols as donors of electrons. GPx4 has been postulated to play an important role in preventing oxidative stress-induced apoptosis and it is essential during embryo development. GPx4 deficiency leads to increased lipid peroxidation and cell death in knockout cells, which may be a potential mechanism of neurodegeneration. It has also a role in gametogenesis [Labunskyy et al., 2014]. We decided to study two Tblastn predictions for GPx4 gene based on our criteria. The rejected Tblastn prediction was in the same positions than the gene that we said to be for GPx3. When studying this prediction, T_Coffee showed a better alignment with GPx3 than with GPx4, supporting the hypothesis that this prediction corresponded to GPx3 and not GPx4. Furthermore, the accepted prediction shows GPx4 in Seblastian. We finally accepted the gene in the scaffold PKMU01000001. This gene is located in the forward strand. The accepted gene contains 5 exons. It contains a Sec in the same position as the human GPx4 protein does. T_Coffee shows a very good identity in the regions where there are matches. However, the beginning and the end of the tortoise protein are shorter than the human one and the Lizard GPx4 protein is even shorter. This might indicate that this protein is shorter in reptiles than in humans. The predicted protein does not show the first Met residue, however we can assume its existence. In the sequence studied, one grade A SECIS was found. This SECIS is predicted after the end of our gene, and in the same strand, therefore we can accept it. Moreover, Seblastian also predicted GPx4 inside the studied Chelonoidis abingdonii sequence. For all these reasons we can accept our prediction, confirming that Chelonoidis abingdonii has GPx4.
GPx5
It was created by GPx3 duplication in placentals. It immediately suffered a replacement of Sec with Cys. [Mariotti et al., 2012]. The Tblastn prediction only showed one possible gene, in the scaffold PKMU01000857.1. This scaffold and the positions where it is placed the gene correspond to the same ones than GPx3. A Sec is predicted in this sequence, however the human version has Cys instead. Furthermore, the T_coffe alignment is not good as it has many gaps. For these reasons, we can say that GPx5 is not in the genome of Chelonoidis abingdonii, as expected by phylogenetics.
GPx6
GPx6 is only found in olfactory epithelium and during embryonic development. It was created by duplication in placentals. It has independly suffered replacement of Sec with Cys in many species, such as the marmoset, the rat, the mouse and the rabbit. [Mariotti et al., 2012]. It is also implicated in the GSH-dependent detoxification of hydrogen peroxide [Labunskyy et al., 2014]. The Tblastn prediction only showed one possible gene, in the scaffold PKMU01000857.1. This scaffold and the positions where the gene is predicted correspond to the same ones as GPx3’s prediction. A Sec is predicted in this sequence, which exists in the human protein. However, the T_coffe alignment is not good. For these reasons, we can say that GPx6 is not in the genome of Chelonoidis abingdonii, as expected by phylogenetics.
GPx7
Both GPx7 and GPx8 evolved from a GPx4 ancestor, however they lost the selenocysteine residue [Mariotti et al., 2012]. We decided to study two Tblastn predictions for GPx7 gene based on our criteria. The rejected Tblastn prediction was in the same positions than the gene that we said to be for GPx8. When studying this prediction, T_Coffee showed a better alignment with GPx8 than with GPx7, supporting the hypothesis that this prediction corresponded to GPx8 and not GPx7. We finally accepted the gene in the scaffold PKMU01008789.1. The gene is located in the reverse strand. This gene contains 3 exons. It contains a Cys in the same position as human GPx7 protein does (it is a Cys-containing homolog). T_Coffee shows a good identity and an even higher similarity, and the predicted protein has almost the same length as the human one. However, the first amino acid is a Leu instead of a Met. Probably, the first position of this codon was not correctly annotated and the triplet AUG (which stands for Met) was sequenced as a CUG (which codes for Leu). In the sequence studied, one grade B SECIS was found. This SECIS is predicted after the end of our gene, and in the same strand, therefore we can accept it. Seblastian did not find any protein in the analysed sequence. For all these reasons we can accept our prediction, confirming that Chelonoidis abingdonii has GPx8.
GPx8
Both GPx7 and GPx8 evolved from a selenoprotein, a GPx4 ancestor, however they lost the selenocysteine residue [Mariotti et al., 2012]. The gene in the scaffold PKMU01009122.1 is the only one that was selected from the Tblastn prediction based on our criteria. The gene is located in the reverse strand. This gene contains 3 exons. It contains a Cys in the same position as human GPx8 protein does (it is a Cys-containing homolog). T_Coffee shows a very good identity, and the predicted protein has the same length that the human one. Furthermore, the predicted protein starts with a Met, so it is very likely to be a real protein. In the sequence studied, two SECIS were predicted. One grade B SECIS was rejected because it was not in the same strand of the predicted gene. Thus, we accepted the other grade B SECIS that was found. This SECIS is predicted after the end of our gene (perhaps a little bit too far) and in the same strand. However, Seblastian did not predict any selenoprotein inside the studied Chelonoidis abingdonii sequence. For all these reasons we can accept our prediction, confirming that Chelonoidis abingdonii has GPx8.
Thioredoxin reductases
Thioredoxin reductases (TRs) are oxidoreductases that comprise one of the major disulfide reduction system of the cell. In mammalian cells, there are three TR isozymes, all of which are Sec-containing proteins [Labunskyy et al., 2014]. TR1 and TR3 are among the 21 selenoproteins present in all vertebrates and are classified as housekeeping selenoproteins in mammals (less affected by dietary selenium status and often serve functions critical to cell survival) [Mariotti et al., 2012].
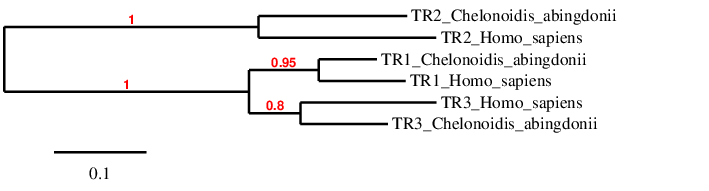
In the phylogeny we see that we have probably predicted the selenoproteins correctly, because each type of TR clusters directly with its human homolog, and afterwards they are related to each other.
TR1
Thioredoxin reductase 1 is primarily localized in the cytosol and nucleus. TR1 reduces a variety of low-molecular-weight compounds, mainly cytosolic thioredoxin (Trx1). Among other physiological roles, TR1 has been implicated in DNA repair, maintaining redox homeostasis and regulation of cell signaling, as well as activating the p53 tumor suppressor [Arner and Holmgren, 2000]. However, TR1 might have an alternative role in promoting tumor progression, as it is required for tumor growth due to high susceptibility of cancer cells to oxidative stress [Fath et al., 2011].
Three tblastn predictions were chosen in the beginning, however two of them were discarded due to bad t_coffee alignments and scaffolds overlapping with other proteins’. The gene in the scaffold PKMU01001697.1 is the only that was finally selected from the Tblastn prediction based on our criteria. The gene is located in the forward strand and contains 13 exons. TR1 is a selenoprotein in humans and the selenocysteine residue has been conserved in C. abingdonii. T_coffee shows a good identity between the human and the tortoise protein, however, there is a 1 residue lacking at the beginning, which could be due to an incorrect prediction of the structure of the gene in exonerate. Furthermore, the length of the predicted protein is almost the same as the human one (499 amino acids). In the sequence studied, one grade A SECIS were predicted. This SECIS is predicted after the end of our gene, in the 3’UTR region, and in the same strand, therefore it can be accepted. Moreover, Seblastian also predicted TR1 selenoprotein inside the Chelonoidis abingdonii sequence we have studied. For all these reasons we can accept our prediction, confirming that Chelonoidis abingdonii has TR1.
TR2
TR2 (also known as TGR) differs from TR1 and TR3 in that TGR contains an additional glutaredoxin (Grx) domain, which suggests that this protein is involved in both Trx and GSH systems. However, its physiological function is still unknown [Labunskyy et al., 2014]. It appeared from an ancestral TR protein, where also TR1 comes from [Mariotti et al., 2012].
Three tblastn predictions were chosen in the beginning, however two of them were discarded due to bad t_coffee alignments and scaffolds overlapping with other proteins’. The gene in the scaffold PKMU01010066.1 is the only that finally was selected from the Tblastn prediction based on our criteria. The gene is located in the reverse strand and contains 15 exons. TR2 is a selenoprotein in humans and the selenocysteine residue has been conserved in C. abingdonii. There is an additional selenocysteine residue in the tortoise’s protein that it is not found in humans. T_coffee shows a few mismatches when aligning the human and the tortoise protein, and also a 30-residue gap at the beginning, which could be due to an incorrect prediction of the structure of the gene in exonerate. In the sequence studied, three grade B SECIS and one A SECIS were predicted. One of them was found in the forward strand and before the start of the gene, in the 5’UTR region, therefore was rejected; another one was found after the end of the gene, in the 3’UTR, but in the forward strand, and was also rejected. The last two were located after the end of the gene, and the one closer to it was chosen. However, Seblastian did not predict any selenoprotein in the Chelonoidis abingdonii sequence we have studied. For all these reasons we can accept our prediction, confirming that Chelonoidis abingdonii has TR2.
TR3
TR3 is localized in the mitochondria, where it is involved in reduction of mitochondrial thioredoxin (Trx2) and glutaredoxin 2 (Grx2) [Labunskyy et al., 2014].
Three tblastn predictions were chosen in the beginning, however two of them were discarded due to bad t_coffee alignments and scaffolds overlapping with other proteins’. The gene in the scaffold PKMU01001735.1 is the only that was finally selected from the Tblastn prediction based on our criteria. The gene is located in the reverse strand and contains 15 exons. TR3 is a selenoprotein in humans and the selenocysteine residue has been conserved in C. abingdonii. T_coffee does not show a good identity between the human and the tortoise protein, there is a 191-residue gap at the beginning, which could be due to an incorrect prediction of the structure of the gene in exonerate. Therefore, the lizard TR3 protein was used and aligned to the predicted amino acid sequence. There is still a gap in the beginning and this time only 41 residues are lacking. This could be seen as a better alignment, however, the number of lacking amino acids is smaller because the lizard protein is smaller as well. The short length of the lizard’s protein could be due to a shortening of the protein after the split of reptiles. In the sequence studied, two SECIS were predicted. One grade B SECIS was rejected because it was found before the start of the gene, in the 5’UTR region, and in the forward strand. Thus, we accepted the A grade SECIS that was found after the gene, in the 3’UTR region, and in the same strand. However, Seblastian did not predict any selenoprotein in the Chelonoidis abingdonii sequence we have studied. For all these reasons we can accept our prediction, confirming that Chelonoidis abingdonii has TR3.
MsrA
MsrA is a sulfoxide reductase, present in humans and some eukaryotic species. As the Methionine amino acid is highly susceptible to oxidation, it ends up being a mixture of methionine-S-sulfoxide and methionine-R-sulfoxide. The function of MsrA, complementary to MsrB1, is the reduction of the methionine-S-sulfoxide levels [Lee et al., 2008]. However, MsrA and MsrB have different structures and belong to different selenoprotein families.
We decided to study two Tblastn predictions for MsrA gene based on our criteria. We accepted the gene in the scaffold PKMU01001278.1, as in the other hit no SECIS were found. The gene is located in the reverse strand and it contains 5 exons. In some organisms, such as the unicellular eukaryotes, Chlamydomonas reinhardtii and In Aureococcus anophagefferens, and an anaerobic bacterium, Clostridium sp., MsrA is a selenoprotein and contains a Sec residue in the active site [Labunskyy et al., 2014]. However, neither in Chelonoidis abingdonii nor in the human query the Sec was found. T_Coffee shows a good identity, but there is a gap in the predicted protein at the beginning of the alignment. Genewise confirmed that we could not predict the first exons of the protein. This can be because exonerate has not correctly predicted the structure of the gene, and thus the first amino acids are lacking (including the Met in the beginning of every protein, being a Leu in our predicted protein). In the sequence studied, five SECIS were predicted. However, none of them was considered valid, as four of them were found inside the exons’ region and the other one was found before the gene. Moreover, Seblastian did not predict any selenoprotein in the sequence. For all these reasons we can accept our prediction on the presence of MsrA in Chelonoidis abingdonii. However, we cannot confirm MrsA as a selenoprotein.
Methionine-R-sulfoxide reductase
MsrB Is a zinc-containing selenoprotein that was initially identified as selenoprotein R (SelR) or selenoprotein X (SelX). This protein was found to function as a stereospecific methionine-R-sulfoxide reductase, which catalyzes repair of the R-enantiomer of oxidized methionine residues in proteins. Based on its functional similarity to methionine-S-sulfoxide reductase A (MsrA), which catalyzes reduction of the other isomer, this selenoprotein was also named MsrB. Although MsrB and MsrA are structurally different and have no sequence similarity, they have complementary functions, each protein acts on only one of the two stereoisomers [Mariotti et al., 2012].
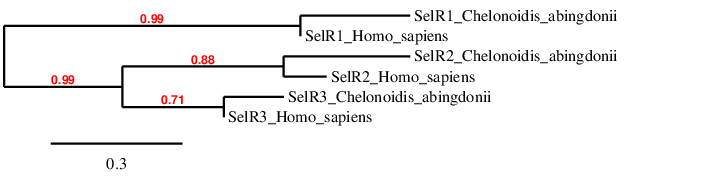
In the phylogeny we see that we have probably predicted the selenoproteins correctly, because each type of R clusters directly with its human homolog, and afterwards it is related to the other Rs.
SelR1
The gene in the scaffold PKMU01006623.1 is the only one that was selected from the Tblastn prediction based on our criteria. The gene is located in the forward strand.
This gene contains 3 exons. It contains a Sec in the same position as human SelR1 protein does.T_Coffee shows a very good identity, and the predicted protein has almost the same length that the human one. However, the protein we have predicted lacks the last 8 amino acids. This can be because exonerate has not correctly predicted the structure of the gene, and these are lacking. The predicted protein starts with a Met, so it is very likely to be a real protein.
In the sequence studied, two SECIS were predicted. One grade B SECIS was rejected because it was too far of our gene. Thus, we accepted the other SECIS a grade A one. This SECIS is predicted after the end of our gene, and in the same strand. However, Seblastian did not predict any selenoprotein inside the Chelonoidis abingdonii sequence we have studied.
For all these reasons we can accept our prediction, confirming that Chelonoidis abingdonii has SelR1.
SelR2
Sel R2 is a cys-containing homolog in human and lizard species. We decided to study two Tblastn predictions for SelR2 gene based on our criteria. After obtaining the t_coffee files, few similarity and low identity could be observed, therefore we decided to find another prediction using the lizard protein as a query.
We finally accepted a gene in the scaffold PKMU01001068.1. The gene is located in the forward strand.This gene contains 4 exons and it contains 5 Sec it could be due to an error in our prediction, as there was not any Sec in neither human or lizard sequences. T_Coffee shows an acceptable identity and similarity, the predicted protein has almost the same length that the human one. However, there are a few gaps in the middle and in the end of the alignment. The first amino acid is a Ser instead of a Met but it matches with the lizard sequence (as it is probably misanotated).
In the sequence studied, one grade A SECIS was found, but this SECIS is predicted before our gene so we decided to discard it. Moreover, Seblastian did not find any protein in the analysed sequence. We can accept our prediction, confirming that Chelonoidis abingdonii has SelR2.
SelR3
We decided to study three Tblastn predictions for SelR3 gene based on our criteria. But After obtaining the t_coffee files, few similarities and low identity could be observed in two of them so we decided to discard them. We finally accepted a gene in the scaffold PKMU01002212.1. This gene is located in the forward strand.
This gene contains 5 exons and also contains 7 Cys in the same position as human SelR3 protein does (it is a Cys-containing homolog). T_Coffee shows a good identity and similarity, and the predicted protein has a length similar to the human one; except for the first 31 amino acids of our prediction. This can be because exonerate has not correctly predicted the structure of the gene, and thus the first amino acids are lacking (including the Met in the beginning of every protein).
In the sequence studied, not any SECIS has been found. As we have not predicted any Sec, it is normal that there is not any SECIS present. Seblastian, has not also predicted any selenoprotein. We can accept our prediction, confirming that Chelonoidis abingdonii has SelR3.
Sel15 and SelM
These two proteins share an approximate sequence identity of a 31% and demonstrate somewhat similar distribution, with homologs present from green algae to humans. They were found by different approaches, however they may be considered a family as both proteins share a common thioredoxin-like domain and contain an NTD signal peptide, consistent with their ER localization. Their shared function is the reduction or rearrangement of disulfide bonds in the ER-localized or secretory proteins. This function was suggested by the presence of redox-active motifs and structural similarities to other thioredoxin-fold oxidoreductases [Labunskyy et al., 2014].
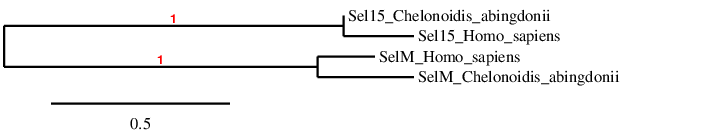
In the phylogeny we see that we have probably predicted the selenoproteins correctly, because Sel15 and SelM clusters directly with its human homolog, and afterwards they are related to each other.
Sel15
Sel15, in addition to the NTD signal peptide, has a Cys-rich domain, which is required for the interaction of Sep15 with its binding partner UDP-glucose:glycoprotein glucosyltransferase (UGGT). It is mainly localized in prostate, liver, kidney and testis, having been implicated in preventing liver, prostate, breast and lung cancers [Labunskyy et al., 2014].
The gene in the scaffold PKMU01008277.1 is the only one that was selected from the Tblastn prediction based on our criteria. The gene is located in the forward strand and it contains 2 exons. As expected, as Sel15 is a selenoprotein, it contains a Sec. T_Coffee shows a good identity. However, the protein we have predicted lacks the firsts 28 amino acids and some of the last ones. This can be because exonerate has not correctly predicted the structure of the gene, and thus the first amino acids are lacking (including the Met in the beginning of every protein). In the sequence studied, as expected, one valid SECIS was confirmed, as it was found after the gene in the same strand. Moreover, Seblastian predicted this selenoprotein in the sequence. For all these reasons we can accept our prediction, confirming that Chelonoidis abingdonii has Sel15.
SelM
This protein has the same N-terminal domain as Sel15, but has a different C-terminal domain: a highly flexible region that has been associated with substrate binding or interaction with other protein factors. It is highly expressed in the brain, and has a role in neuroprotection: it has been implicated in regulating Ca2+ release from ER calcium stores in neuronal cultures in response to oxygen peroxide [Labunskyy et al., 2014].
The gene in the scaffold PKMU01008734.1 is the only one that was selected from the Tblastn prediction based on our criteria. The gene is located in the forward strand and it contains 5 exons. As expected, as SelM is a selenoprotein, it contains a Sec. T_Coffee shows a good identity. However, the protein we have predicted lacks the first 5 amino acids and 10 of the last ones. This can be because exonerate has not correctly predicted the structure of the gene, and thus the first amino acids are lacking (including the Met in the beginning of every protein). In the sequence studied, as expected, one valid SECIS was confirmed, as it was founded after the gene in the same strand. Moreover, Seblastian predicted this selenoprotein in the sequence. Another grade C SECIS was predicted, however, it was rejected by Seblastian. For all these reasons we can accept our prediction, confirming that Chelonoidis abingdonii has SelM.
SelI
Selenoprotein I (SelI) is a recently evolved selenoprotein, which is found only in vertebrates. It is a transmembrane protein containing seven transmembrane domains and has three conserved aspartic residues that are required for catalytic activity. It also contains a highly conserved CDP-alcohol phosphatidyltransferase domain, which is present in choline (CHPT1) and choline/ethanolamine (CEPT1) phosphotransferases. It is known that CHPT1 and CEPT1 catalyze the last step in de novo synthesis of the two major phospholipids through the transfer phosphocholine and phosphoethanolamine groups to diacylglycerol from CDP-choline and CDP-ethanolamine, respectively. However, the function of SelI remains unknown [Labunskyy et al., 2014].
The gene in the scaffold PKMU01004179.1 is the only one that was selected from the Tblastn prediction based on our criteria. The gene is located in the reverse strand and it contains 10 exons, and it starts with a Met. As expected, because SelI is a selenoprotein, it contains a Sec. T_Coffee shows a good identity. In the sequence studied, as expected, one valid SECIS was confirmed, as it was founded after the gene in the same strand. Moreover, Seblastian predicted this selenoprotein in the sequence. Another grade B was predicted, however, it was rejected by Seblastian. For all these reasons we can accept our prediction, confirming that Chelonoidis abingdonii has SelI.
SelK and SelS
Although selenoprotein K (SelK) and selenoprotein S (SelS) have no significant sequence similarity, they could be assigned to a single SelK/SelS family of related selenoproteins based on their topology, including a single transmembrane domain in the NTD; the presence of a glycine-rich (G-rich) segment that has a high content of glycine, proline, and positively charged amino acids; and a characteristic location of Sec residues in the CTD of the protein. This selenoprotein family is the most widespread in eukaryotic species, being present in nearly all known Se-utilizing organisms ranging from unicellular eukaryotes to humans.
Both SelK and SelS are localized to the ER membrane and belong to a type III group of transmembrane proteins that contain a single transmembrane domain, with the CTD of the protein facing the cytosol. Moreover, as it is shown in the human queries, both proteins contain Sec in the second or third position from the CTD.
Regarding to the function, both SelK and SelS are implicated in ER-associated degradation (ERAD) of misfolded proteins. That means they are involved in recognition, ubiquitination, and retrotranslocation of protein substrates from the ER to the cytosol, and their subsequent degradation by the ubiquitin/proteasome. Moreover, as SelS and SelK deficiency leads to deficient Ca 2+ flux during immune cell activation, they might mediate the anti-inflammatory effects of Se and its role in the immune system [Labunskyy et al., 2014].
SelK
SelK might be involved in binding misfolded proteins and targeting them to the Derlin complex and subsequent proteasome-dependent degradation. It is also involved in the retrotranslocation of only a subset of misfolded proteins from the ER, as knockdown of SelK affected degradation of some misfolded glycoprotein substrates [Labunskyy et al., 2014].
The gene in the scaffold PKMU01003079.1 is the only one that was selected from the Tblastn prediction based on our criteria. The gene is located in the reverse strand and it contains 4 exons, and it starts with a Met. Although the predicted SelK has a very good identity with the human query in T_Coffee, the predicted protein in Chelonoidis abingdonii has lost its last 3 residues, where Sec was found. Both fastatranslate and genewise confirmed that lost. In the sequence studied, as expected, one valid SECIS was confirmed, as it was found after the gene in the same strand. Moreover, Seblastian predicted a selenoprotein in the sequence. Another grade B was predicted, however, it was rejected by Seblastian. For all these reasons we can accept our prediction, confirming that Chelonoidis abingdonii has SelK, but the Sec could not be detected.
SelS
SelS is as a binding partner of p97 and Derlin-1; together with SelS form a retrotranslocation channel. In higher eukaryotes, two additional Derlin-1-related proteins are encoded in the genome (Derlin-2 and Derlin-3). More recently, SelS was found to also bind human Derlin-2 and a long form of Derlin-3. SelS differs from SelK by the presence of an additional coiled-coil domain in the cytosolic portion of the protein, which has been proposed to mediate the interaction with other proteins or oligomerization of SelS [Labunskyy et al., 2014].
The gene in the scaffold PKMU01007956.1 is the only one that was selected from the Tblastn prediction based on our criteria. The gene is located in the reverse strand.
This gene contains 5 exons. It contains a Sec in the same position as human SelS protein does and it is placed in almost the last amino acid. T_Coffee shows a very good identity and similarity, and the predicted protein has almost the same length than the human one. However, the protein we have predicted lacks the first 23 amino acids. This can be because exonerate has not correctly predicted the structure of the gene, and these are lacking (including the Met at the beginning of every protein).
In the sequence studied, one grade A SECIS was found. This SECIS is predicted after the end of our gene, and in the same strand, so we can accept it. Moreover, Seblastian also predicted SelS selenoprotein inside the Chelonoidis abingdonii sequence we have studied.
For all these reasons we can accept our prediction, confirming that Chelonoidis abingdonii has SelS.
Selenoprotein N
Selenoprotein N (SelN) was among the first selenoproteins that were identified through bioinformatic approaches. SelN is an ER-resident transmembrane glycoprotein that is highly expressed during embryonic development and to a lesser extent in adult tissues including skeletal muscle. It’s involved in regulation of intracellular calcium mobilization ans is part of the ancestral vertebrate selenoproteome [Mariotti et al., 2012].
The gene in the scaffold PKMU01005290.1 is the only one that was selected from the Tblastn prediction based on our criteria. The gene is situated in the reverse strand. This gene contains 11 exons, there was an exon in the Exonerate that was misplaced and another that we decided to take out because it had a very low identity and was not included in the genewise file. The gene contains a Sec in the same position as human SelN protein does, but It contains a gap in another position that the human protein has a Sec.
T_Coffee shows a good identity, and the predicted protein has almost the same length that the human one. However, the protein we have predicted lacks the first 11 and the last amino acids, it also has a gap of 42 amino acids in the middle of the sequence. This can be because exonerate has not correctly predicted the structure of the gene, and these are lacking (including the Met in the beginning of every protein). In the sequence studied, one grade A SECIS was found. This SECIS is predicted after the end of our gene, and in the same strand, so we can accept it. However, Seblastian did not predict any selenoprotein inside the Chelonoidis abingdonii sequence we have studied.
For all these reasons we can accept our prediction, confirming that Chelonoidis abingdonii has SelN.
Selenoprotein O
Selenoprotein O (SelO) is one of the least characterized human selenoproteins and is part of the ancestral vertebrate selenoproteome. The majority of SelO homologs contains a Cys residue in place of Sec (Sec-containing SelO sequences are present only in vertebrates). Analysis of vertebrate SelO protein sequences revealed the presence of a mitochondrial targeting peptide and a putative protein kinase but its function remains unknown [Mariotti et al., 2012].
We decided to study three Tblastn predictions for SelO gene based on our criteria. We finally accepted the gene in the scaffold PKMU01009289.1. The gene is located in the forward strand. One of the Tblastn rejected predictions had an extremely short amino acid sequence and the other one had a worse T_coffee alignment. This gene contains 7 exons. It contains a gap in the position that human SelO protein has a Sec. T_Coffee shows a bad alignment with better similarity than identity. The predicted protein has almost the half the length of the human one. The gene we have predicted lacks the first 187 amino acids and the last 52. We tried to make an alignment with lizard’s SelO sequence because the gene may have significatively changed between C. abingdonii and human, but the results obtained were as bad as the ones we got from the human alignment or even worst with gaps in the middle and the end of the sequence. Due to this we kept the H. sapiens-based prediction.
In the sequence studied, we did not find any SECIS and Seblastian did not predict any selenoprotein inside the Chelonoidis abingdonii sequence we have studied.
For all these reasons we cannot be sure that our prediction is correct, but phylogenetically Chelonoidis abingdonii should have SelO.
SelP
Selenoprotein P (SelP) is an abundantly expressed secreted selenoprotein that accounts for almost 50% of the total Se in plasma[Burk and Hill, 2005]. SelP is present in all vertebrates, and contains 10 selenocysteine residues in humans, however this number varies radically from one species to another [Labunskyy et al., 2014]. This protein functions as a Se transporter. The fact that this protein is present in the plasma and that it contains multiple selenocysteine residues indicates that it may serve as a Se supplier to peripheral tissues [Burk et al., 1991].
First, a prediction in scaffold PKMU01001436.1 was selected using the human SelP protein as query. After obtaining the t_coffee file, few similarities and low identity could be observed, therefore, we decided to find another prediction using the lizard protein as a query. The gene in the scaffold PKMU01005800.1 is the only that was finally selected from the Tblastn prediction based on our criteria. The gene is located in the reverse strand and contains 3 exons. It is a selenoprotein in the lizard and the selenocysteine residue is conserved in C. abingdonii. However, no SECIS was predicted and neither did the selenoprotein using the seblastian software. As we have predicted one Sec, the SECIS should be present. It is possible that this part of the genome was not correctly annotated, and so the SECIS cannot be detected.
It is worth mentioning that a protein named Selpb, whose gene can be found in the chicken genome, also gave a prediction in C.abingdonii’s genome, in the same scaffold (and almost same positions) as SelP. Both proteins cannot be codified by the same gene, therefore, one of the predictions was wrong. Taking into account that neither of the predicted proteins contained a SECIS element and based on the fact that the t_coffee file of SelP predicted protein showed better alignment, we decided to choose SelP as the protein present in C.abingdonii’s proteome. Furthermore, this incident could be explained by the fact that Selpb has partial conservation with its closest homolog, which is SelP [Mariotti et al., 2012]. In favor of this decision, Selpb protein transcript could not be found in SelenoDB2, but in Uniprot, which might mean that this protein has indeed disappeared in the reptiles proteome.
Selenoprotein U
Selenoprotein U (SelU) was firstly found in fish and also reported in birds and unicellular eukaryotes, such as Chlamydomonas reinhardtii. In high mammalian species, such as humans and mice, all SelU proteins exist in Cys form, due to the Sec to Cys event that occurred in the early period of mammalian history for the SelU lineage. Three subfamilies of SelU family, SelU1, 2 and 3 are found in humans. The Prx-like2 structure domain presented in these proteins implies that they belong to the thioredoxin-like superfamily [Jiang et al., 2012]. SelU family of proteins deprivation has been linked to induced autophagy, altered the expression of growth factors secreted by stem cells and inhibited apoptosis via PI3K–Akt pathway disruption in rooster stem cells [Sattar et al., 2018]. Their function in other species is yet to be discovered.
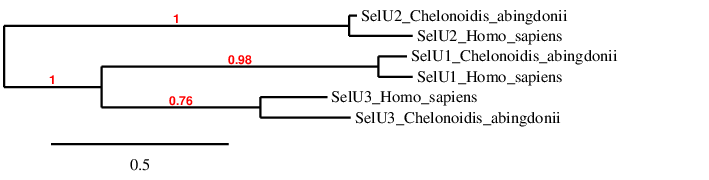
In the phylogeny we see that we have probably predicted the selenoproteins correctly, because each type of SelU clusters directly with its human homolog, and afterwards they are related to each other.
SelU1
The gene in the scaffold PKMU01001923.1 is the only one that was selected from the Tblastn prediction based on our criteria. The gene is situated in the reverse strand. This gene contains 5 exons. The gene contains 2 Cys in the same position as human SelU1 protein does (it is a Cys-containing homolog).
T_Coffee shows a very good identity and similarity, and the predicted protein has almost the same length that the human one. However, the protein we have predicted lacks the last 7 amino acids. This can be because exonerate has not correctly predicted the structure of the gene, and these residues are lacking. The predicted protein starts with a Met, so it is very likely to be a real protein.
In the sequence studied, one grade A SECIS was found. This SECIS is predicted after the end of our gene, and in the same strand, so we can accept it even though without Sec it is not necessary to have one. However, Seblastian did not predict any selenoprotein inside the Chelonoidis abingdonii sequence we have studied. For all these reasons we can accept our prediction, confirming that Chelonoidis abingdonii has SelU1.
SelU2
The gene in the scaffold PKMU01002121.1 is the only one that was selected from the Tblastn prediction based on our criteria. The gene is situated in the reverse strand. This gene contains 6 exons. The gene contains 3 Cys in the same position as human SelU2 protein does (it is a Cys-containing homolog).
T_Coffee shows a good identity and similarity, and the predicted protein has a similar length as the human one. However, the protein we have predicted lacks the first 38 aminoacids. This can be because exonerate has not correctly predicted the structure of the gene, and these are lacking (including the Met in the beginning of every protein).
In the sequence studied, not any SECIS has been found. As we have not predicted any Sec, it is normal that there is not any SECIS present. Seblastian, has not predicted any selenoprotein. For all these reasons we can accept our prediction, confirming that Chelonoidis abingdonii has SelU2.
SelU3
SelU3 protein comes from SelU1 protein, however, unlike SelU1, it is not found in the common vertebrate ancestor, but in some reptiles and amphibians [Mariotti et al., 2012].
The gene in the scaffold PKMU01007067.1 is the only that was selected from the Tblastn prediction based on our criteria. The gene is located in the reverse strand and contains 6 exons. In humans SelU3 is a cysteine-containing homolog, and the cysteine residue has been conserved in C. abingdonii. T_coffee shows a good identity between the human and the tortoise protein, with a small 5-residue gap at the end, which could be due to an incorrect prediction of the structure of the gene in exonerate. Furthermore, the length of the predicted protein is almost the same as the human one (198 amino acids) and the predicted protein starts with a Met, so it is very likely to be a real protein. The C-x-x-C motif usually linked to the thiole-dependant Rdx family of selenoproteins can also be found in the protein’s prediction. In the sequence studied, one grade B SECIS was found. This SECIS is predicted after the end of our gene, in the 3’UTR region, and in the forward strand, therefore it cannot be accepted. However, Seblastian did not predict any selenoprotein in the Chelonoidis abingdonii sequence we have studied. For all these reasons we can accept our prediction, confirming that Chelonoidis abingdonii has SelU3.
Selenoprotein W, SelV, SelT and SelH
Selenoproteins W (SelW), T (SelT), H (SelH), and V (SelV) belong to the Rdx family of selenoproteins. The members of this protein family are characterized by the presence of a conserved Cys-x-x-Sec motif. It was proposed that the Rdx family proteins are thiol-based oxidoreductases, but the exact function of some of these proteins remains unknown [Labunskyy et al., 2014].
The functions of SelV and SelW are not known, however SelV is expressed exclusively in mammal testes, whereas SelW is expressed in a variety of organs. SelW and SelV showed the same gene structure; each contained 6 exons with intron regions and phases conserved. Coding regions were within exons 1-5. Exon 6 contained only the last portion of the 39-UTR, including the SECIS element [Mariotti et al., 2012].
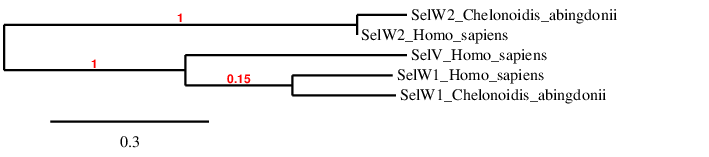
As shown by the phylogenetic tree, our predictions of SelW1 and SelW2 are probably correct, and SelV is mostly related with SelW1 (probably the one from which it emerged).
This family of W selenoproteins can be found in the selenoproteome of the ancestral vertebrate [Castellano et al., 2009]. SelW belongs to the stress-related group of selenoproteins as its expression is highly regulated by the availability of Se in the diet [Howard et al., 2013]. While SelW1 can be found in human with the conserved selenocysteine residue, SelW2 was lost in all tetrapods (but frog), and its homolog can be found under the name Rdx12 with a cysteine residue instead. The hypothesis is that before the split of amphibians SelW2 duplicated and was immediately converted to a Cys form generating Rdx12, and then SelW2 was lost prior to the split of reptiles [Mariotti et al., 2012].
SelW1
The gene in the scaffold PKMU01003401.1 is the only that was selected from the Tblastn prediction based on our criteria. The gene is located in the reverse strand and contains 3 exons. SelW1 is a selenoprotein in humans. We have not been able to find the selenocysteine in C. abingdonii’s protein, either because the protein might be divided in two scaffolds and we could only find the one where there is the biggest part of it or because Exonerate did not correctly predict the initial part of the gene. The Cys-x-x-Sec motif can be found in the human genome (CGAU), but cannot be observed in the tortoise’s genome due to the problems explained before. T_coffee shows a good identity between the human and the tortoise protein, however, there is a 18-residue gap at the beginning, which could be due to an incorrect prediction of the structure of the gene in exonerate. In the sequence studied, two SECIS were predicted. One grade B SECIS was rejected because it was found before the start of the gene, in the 5’UTR region. Thus, we accepted the A grade SECIS that was found after the gene, in the 3’UTR region, and in the same strand. Despite the fact that the tortoise’s protein might or might not have a selenocysteine residue, the presence of a SECIS element can be explained by the fact that this protein is a selenoprotein in the human proteome. However, Seblastian did not predict any selenoprotein in the Chelonoidis abingdonii sequence we have studied. For all these reasons we can accept our prediction, confirming that Chelonoidis abingdonii has SelW1.
SelW2
The gene in the scaffold PKMU01002623.1 is the only that was selected from the Tblastn prediction based on our criteria. The gene is located in the forward strand and contains 3 exons. SelW2 (it is preferable to name it Rdx12) is a cysteine-containing homolog in humans and the cysteine residue has been conserved in C. abingdonii. The Cys-x-x-Sec motif characteristic of SelW family can be found in the human protein, however the selenocystein is replaced by a cysteine (CEPC), and it can also be observed in the tortoise’s genome. T_coffee shows a good identity between the human and the tortoise protein, however, there is a 30-residue gap at the beginning, which could be due to an incorrect prediction of the structure of the gene in exonerate. In the sequence studied, five grade B SECIS were predicted. Two of them were found in the reverse strand and before the start of the gene, therefore were rejected. The three others were found in the forward strand and after the gene, in the 3’UTR region, and the one closer to the end of the gene was chosen. Sattar H., et al state that several selenoproteins were lost across vertebrates after the terrestrial environment was colonized, and that Selenoprotein SelW2 was lost approximately prior to the split of reptiles, as we find it today only in fish and frog. However, this protein is found in mammals in a cys form, under the name Rdx12. It is hypothesized that, before the split of amphibians, SelW2 duplicated and was immediately converted to a Cys form generating Rdx12, and then SelW2 was lost prior to the split of reptiles. This hypothesis would give an explanation of why we are able to predict SECIS elements in its sequence. However, Seblastian did not predict any selenoprotein (as said, Rdx12 does not contain Sec) in the Chelonoidis abingdonii sequence we have studied. For all these reasons we can accept our prediction, confirming that Chelonoidis abingdonii has SelW2.
SelV
SelV is a very low conserved selenoprotein in mammalians and has likely arisen from a duplication of SelW in the placental stem. Then it was modified by addition of N-terminal sequences, whose function is unclear, as well as a deletion of a substantial portion of the 3’-UTR [Mariotti et al., 2012].
Tblastn did not show any prediction of a scaffold that could contain SelV gene in Chelonoidis abingdonii. This is coherent with the literature, as SelV is not supposed to be present in our organism, but it is in Homo sapiens.
SelH
SelH contains a Sec residue within a Cys-x-x-Sec motif, and it localizes specifically in the nucleoli. It is a stress-related selenoprotein. It has an AT-hook motif to bind to DNA, and it binds to sequences containing heat shock and stress response elements [Labunskyy et al., 2014]. Some authors state that this protein also regulates redox homeostasis and suppresses DNA damage [Cox et al., 2016].
The gene in the scaffold PKMU01001079.1 is the one that was accepted from the Tblastn prediction based on our criteria (the other possible prediction was discarded because it was too short). The gene is located in the forward strand.
The Exonerate human-based prediction of Chelonoidis abingdonii SelH showed a gene of 3 exons that coded for a long protein. The predicted protein starts with a Met, but just 10 amino acids after the beginning of it, there are 14 amino acids that cannot be correctly aligned with the human protein. The rest of the protein shows a good similarity and identity. Sec is conserved in the tortoise SelH, but it is not part of a Cys-x-x-Sec motif in the human-based prediction. However, Genewise defined a 2 exon-gene that did not have the beginning of the protein (the initial Met was also discarded) nor the Sec. When predicting this gene with the lizard protein, the results obtained were the similar ones that were stated by Genewise: a short protein that matched with the final part of the human selenoprotein and did not have Met or Sec. Not any viable SECIS was predicted, and Seblastian did not match any protein in the given sequence. The fact that not any viable SECIS could be predicted might be because in some organisms the 3’-UTR is split in two parts and the SECIS is located in the last one [Mariotti et al., 2012], and we may have missed this last element when applying the fastasubseq.
We have accepted the existence of SelH in Chelonoidis abingdonii, but we hypothesise that a part of the gene was lost in reptiles, besides the initial aminoacids and Sec were conserved.
SelT
SelT belongs to the Rdx family of selenoproteins. The members of this protein family are characterized by the presence of a conserved Cysx-x-Sec motif. It was proposed that the Rdx family proteins are thiol-based oxidoreductases, but the exact function of any of these proteins remains unknown [Mariotti et al., 2012].
The gene in the scaffold PKMU01006301.1 is the only one that was selected from the Tblastn prediction based on our criteria. The gene is located in the forward strand.
This gene contains 5 exons. It contains a Sec in the same position as human SelT protein does. T_Coffee shows a very good identity, and the predicted protein has the same length than the human one. Moreover the predicted protein starts with a Met, so it is very likely to be a real protein.
In the sequence studied, two SECIS were predicted. One grade B SECIS was rejected because it was too far of our gene. Thus, we accepted the other SECIS a grade A one. This SECIS is predicted after the end of our gene, and in the same strand. However, Seblastian did not predict any selenoprotein inside the Chelonoidis abingdonii sequence we have studied.
For all these reasons we can accept our prediction, confirming that Chelonoidis abingdonii has SelT.
Selenoprotein machinery
eEFSec
This Sec-specific eukaryotic elongation factor recruits tRNA[Ser]Sec and, together with SBP2, it inserts Sec into nascent protein chains in response to UGA codons. The presence of two separate proteins working as elongation factors (eEFSec, for Sec; and eEF1A, for all the other amino acids) might be dictated by a distant location of SECIS in the 3’-UTR. To decode UGA as Sec, eEFSec undergoes conformational changes upon binding a SECIS element, stimulating functional interactions with the ribosome [Labunskyy et al., 2014]. The gene in the scaffold PKMU01005278.1 is the only one that was selected from the Tblastn prediction based on our criteria. The gene is located in the forward strand. This gene contains 7 exons. As expected, it does not contain a Sec (like human eEFSec), because it is part of the machinery and it is not a selenoprotein itself. T_Coffee shows a good identity and an even higher similarity, and the predicted protein has almost the same length as the human one. However, the protein we have predicted lacks the first 3 amino acids. This can be because exonerate has not correctly predicted the structure of the gene, and thus the first amino acids are lacking (including the Met at the beginning of every protein). In the sequence studied, as expected, not any valid SECIS was found (the predicted secis was in the opposite strand). Moreover, Seblastian did not predict any selenoprotein in the sequence. For all these reasons we can accept our prediction, confirming that Chelonoidis abingdonii has eEFSec.
PSTK
PTSK (phosphoseryl-tRNA kinase) phosphorylates Ser-tRNA[Ser]Sec to produce the phosphorylated intermediate PSer-tRNA[Ser]Sec, serving as a substrate for SecS [Xu et al., 2005]. The gene in the scaffold PKMU01005290.1 is the only one that was selected from the Tblastn prediction based on our criteria, from both human and lizard. We finally selected the lizard query to perform the study, as it predicted one more exon with Exonerate and had a better alignment with T_Coffee compared to the predictions given by the human query. The gene is located in the reverse strand and contains 6 exons. As expected, it does not contain a Sec (as lizard PSTK), because it is part of the machinery and it isnot a selenoprotein itself. T_Coffee shows a good identity and an even higher similarity, and the predicted protein has almost the same length as the human one. In the sequence studied, as expected, not any valid SECIS was found. Moreover, Seblastian did nott predict any selenoprotein in the sequence. For all these reasons we can accept our prediction, confirming that Chelonoidis abingdonii has PSTK.
SBP2
SBP2, also known as SECIS binding protein 2, is stably associated with ribosomes and contains a RNA-binding domain that is known to bind SECIS elements with high affinity and specificity [Copeland et al., 2000]. SBP2 also interacts with eEFSec, which recruits Sec- tRNA[Ser]Sec and facilitates incorporation of Sec into the nascent, growing polypeptide [Tujebajeva et al., 2000]. Furthermore, SBP2 is a limiting factor for selenoprotein synthesis, as knockdown of SBP2 in mammalian cells using siRNA leads to decreased expression of selenoproteins [Papp et al., 2006].
First, a prediction in scaffold PKMU01000862.1 was selected using the human SBP2 protein as query. After obtaining the t_coffee file, few similarities and low identity could be observed, therefore, we decided to find another prediction using the lizard protein as a query. The gene in the scaffold PKMU01000807.1.1 is the only that was finally selected from the Tblastn prediction based on our criteria. The gene is located in the forward strand and contains 16 exons. SBP2is a cysteine-containing homolog or a selenium machinery protein (both things are still considered in the same category in SelenoDB 2) in lizard, however, it is annotated as a selenium machinery protein in human. No residue is highlighted (as residue transformed from selenocysteine) in the transcript of the protein in either of SelenoDB databases (1 or 2), therefore, we cannot know for sure if the residue has been conserved in C. abingdonii’s protein.
T_coffee shows a good identity between the lizard and the tortoise protein, only a few mismatches can be seen and the length of the predicted protein is almost the same as the one in lizard (1062 residues). In the sequence studied, no SECIS was predicted; as this protein has no selenocysteine, not finding any SECIS was expected. Furthermore, Seblastian did not predict any selenoprotein in the Chelonoidis abingdonii sequence we have studied. For all these reasons we can accept our prediction, confirming that Chelonoidis abingdonii has SBP2.
SECp43
SECp43, along with SLA, is specific of eukaryotes. It interacts with the tRNA[Ser]Sec forming a complex. It has a nuclear localization and it may work as a chaperone for SLA and Sec-tRNA[Ser]Sec, being linked to the regulation of the synthesis of selenoproteins through methylation of tRNA[Ser]Sec and the intracellular distribution of SLA [Labunskyy et al., 2014].
We decided to study two Tblastn predictions from human and two Tblastn predictions from lizard for SecS gene based on our criteria. We accepted the gene in the scaffold PKMU01005290.1., obtained from a Tblastn hit from both human and lizard. Finally, we used the human query to align it to the predicted sequence because the T_coffee shows a good identity and a complete alignment, while comparing with the lizard query a gap in the middle of the predicted protein was found. The gene is located in the forward strand and contains 8 exons. Unexpectedly, as it is part of the machinery, it contains a Sec (while the human query contains a tyrosine). It is possible that this is a mistake in the annotation of the genome, because SECp43 has never been reported as a selenoprotein before and it does not have a SECIS element.
T_Coffee shows a good identity, and the predicted protein has almost the same length as the human one. However, the protein we have predicted lacks the first 9 amino acids. This could be because exonerate has not correctly predicted the structure of the gene, and thus the first amino acids are lacking (including the Met at the beginning of the protein). In the sequence studied, as expected, not any valid SECIS was found (the only predicted SECIS was in the opposite strand). Moreover, Seblastian did not predict any selenoprotein in the sequence. For all these reasons we can accept our prediction, confirming that Chelonoidis abingdonii has PSTK.
SecS
SecS (Sec synthase) catalyzes the conversion of the serine moiety on tRNA to selenocysteine-tRNA by incorporating selenophosphate, the active form of Se, into the amino acid backbone and forming Sec-tRNA. In humans, four SecS subunits form a tetramer, which binds two tRNA [Ser]Sec molecules through their long acceptor- TC arms. The interaction of tRNA[Ser]Sec with the active site of SecS induces the enzyme’s conformational change that allows the binding of O-phosphoseryl-tRNA[Ser]Sec , but not free phosphoserine, in order for the reaction to occur [Labunskyy et al., 2014].
We decided to study two Tblastn predictions from human and two Tblastn predictions from lizard for SecS gene based on our criteria. We accepted the gene in the scaffold PKMU01006445.1, obtained from a Tblastn hit from both human and lizard. Finally, we used the lizard query to compare as it had a better alignment with the predicted sequence than the human one. The gene is located in the forward strand. This gene contains 11 exonsand it starts with a Met, so it is probably a real protein. As expected, it does not contain a Sec (as the lizard’s SecS), because it is part of the machinery, but it is not a selenoprotein itself. T_Coffee shows a good identity and an even higher similarity, and the predicted protein has almost the same length as the lizard one, as there is only 1 less amino acid in the predicted sequence compared to the lizard sequence. In the sequence studied, as expected, not any valid SECIS was found (the only predicted SECIS was between the exons). Moreover, Seblastian did not predict any selenoprotein in the sequence. For all these reasons we can accept our prediction, confirming that Chelonoidis abingdonii has SecS.
SPS family
SPS protein family is part of the selenium machinery protein group, therefore, it is involved in the synthesis of selenoproteins. However, the two known members of this family have different functions and characteristics [4]. While SPS2 is required for de novo synthesis of selenophosphate, SPS1 may have a possible role in Sec recycling through a selenium rescue system. Since SPS2 is itself a selenoprotein, it possibly serves as an autoregulator of selenoprotein synthesis [Guimaraes et al., 1996; Kim et al., 1997].
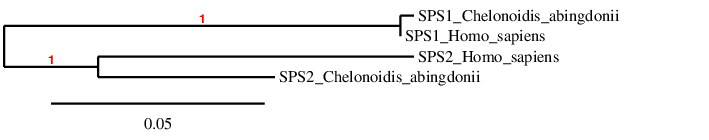
The phylogenetic tree confirms that our predictions are correct, as tortoise SPS1 is firstly related to human SPS1 and the same happens with SPS2.
SPS1
SPS1’s function is not well characterized, though it is believed to play a role in selenoprotein synthesis. Knock-down of this protein’s orthologs in some insects, which lack Sec biosynthesis, has shown to induce growth inhibition and increased intracellular glutamine levels, among other processes [Shim et al., 2009]. That leads to thinking that SPS1 may have a role outside of selenoprotein synthesis [Labunskyy et al., 2014]. On the other hand, both SPS1 and SPS2 interact with SCL (Sec Lyase, which catalyzes degradation of Sec to L-alanine and elemental Se), as shown by an in vitro experiment, supporting the idea of the two proteins’ implication in selenoprotein synthesis processes [Tobe et al., 2009].
Two tblastn predictions were chosen in the beginning, however one of them was rejected based on worse t_coffee alignment and because the scaffold coincided with the one from another protein’s prediction. The gene in the scaffold PKMU01001170.1 is the only that finally was selected from the Tblastn prediction based on our criteria. The gene is located in the forward strand and contains 8 exons. In humans SPS1 is a is a threonine-containing homolog, and this residue has been conserved in C. abingdonii. T_coffee shows a good identity between the human and the tortoise protein, with only 3 mismatches. Furthermore, the length of the predicted protein is the same as the human one (392 amino acids) and the predicted protein starts with a Met, so it is very likely to be a real protein. In the sequence studied, one grade B SECIS was found. This SECIS is predicted before the start of our gene, in the 5’UTR region in the same strand, therefore it cannot be accepted. As this protein has no selenocysteine, not finding any SECIS was expected. Furthermore, Seblastian did not predict any selenoprotein in the Chelonoidis abingdonii sequence we have studied. For all these reasons we can accept our prediction, confirming that Chelonoidis abingdonii has SPS1.
SPS2
Selenophosphate synthetase 2 (SPS2) catalyzes the synthesis of the active Se donor selenophosphate that is necessary for Sec biosynthesis [Xu et al., 2007]. All vertebrates possess Sec-containing SPS2, and that is the reason why it has been proposed to serve an autoregulatory role in selenoprotein synthesis [Guimaraes et al., 1996; Kim et al., 1997]. SPS2 gene was duplicated and the original multiexon gene (SPS2a) was replaced by an intronless gene (SPS2b) in placental mammals [Mariotti et al., 2012]. Because of this event, we expect to find the SPS2a form in C. abingdonii’s genome, as it is not a mammal.
Two tblastn predictions were chosen in the beginning, however one of them was rejected based on worse t_coffee alignment and because the scaffold coincided with the one from another protein’s prediction. The gene in the scaffold PKMU01001233.1 is the only that was finally selected from the Tblastn prediction based on our criteria. The gene is located in the reverse strand and contains 7 exons. SPS2 is a selenoprotein in humans, however, we have not been able to find the selenocysteine in C.abingdonii’s genome, because the protein might be divided in two scaffolds and we could only find the one where there is the biggest part of it. T_coffee shows a good identity between the human and the tortoise protein, however, there is a 111-residue gap at the beginning, which could be due to an incorrect prediction of the structure of the gene in exonerate. In the sequence studied, one grade A SECIS was found. This SECIS is predicted after the end of our gene, in the 3’UTR region in the same strand, therefore it can be accepted. Despite the fact that the tortoise’s protein might or might not have a selenocysteine residue, the presence of a SECIS element can be explained by the fact that this protein is a selenoprotein in the human proteome. Furthermore, Seblastian did not predict any selenoprotein in the Chelonoidis abingdonii sequence we have studied. For all these reasons we can accept our prediction, confirming that Chelonoidis abingdonii has SPS2.