RESULTS
Selenoproteins
Photo |
Info |
||||||||||||
| Iodothyronine Deiodinase | |||||||||||||
| Glutathione peroxidase | |||||||||||||
| Selenoprotein O | |||||||||||||
| Selenoprotein P (SelP) | |||||||||||||
| Selenoprotein R (MsrB1) | |||||||||||||
| Selenoprotein U (SelU) | |||||||||||||
| Selenoprotein W (SelW) | |||||||||||||
| Thioredoxin reductase | |||||||||||||
| Sel |

Selenoproteins Machinery
Photo |
Info |
||||||||||||||||||||||||||||||||||
| Eukaryotic elongation factor (eEFsec) | |||||||||||||||||||||||||||||||||||
| Phosphoseryl-tRNA kinase(PSTK) | |||||||||||||||||||||||||||||||||||
| SECIS binding protein 2 | |||||||||||||||||||||||||||||||||||
| tRNA Sec 1 associated protein 1 (SECp43) | |||||||||||||||||||||||||||||||||||
| Selenocysteine synthase (SecS) | |||||||||||||||||||||||||||||||||||
| Selenophosphate synthetase family (SPS) | |||||||||||||||||||||||||||||||||||

ANALYSIS OF SELENOPROTEINS AND MACHINERY:
From our obtained results after running the program, we were able to extract the rellevant information to correctly perform the analysis of each selenoprotein and machinery proteins, following the next steps:
- Protein location and sense strand, in the corresponding scaffold.
- Number of exons of each gene.
- Exons table related with each protein or protein table, which indicates the start and the end position of each exon.
- Origin of the query protein (organism and database), used to predict the final protein.
- SECIS elements obtained from SECIS3Search program.
- Seblastian results from each selenoprotein found.
Selenoproteins
Iodothyronine Deiodinase
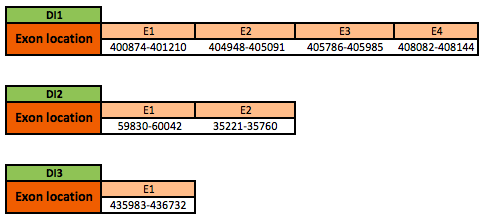
DI1
The DI1 protein is located in the scaffold KN906563.1 between positions 400874 and 408144 in the forward strand. The gene contains 4 exons, with a selenocysteine in exon 1.
This protein was predicted blasting Nanorana parkeri genome against the Xenopus tropicalis DIc protein found in SelenoDB. To find the gene structure we analyzed the exonerate file.
Finally, SECIS element of grade A, predicted in the 3'UTR region, was found. In addition, Seblastian result concorded with SECIS prediction.
DI2
The DI2 protein is located in the scaffold KN908874.1 between positions 35221 and 60042 in the forward strand. The gene contains 2 exons, with a selenocysteine in exon 2.
This protein was predicted blasting Nanorana parkeri genome against the Xenopus tropicalis DIb protein found in SelenoDB. To find the gene structure we analyzed the exonerate file.
Finally, 1 SECIS element of grade A and 2 SECIS elements of grade B, predicted in the 3'UTR region, were found. Though, no Seblastian results were obtained.
DI3
The DI3 protein is located in the scaffold KN906187.1 between positions 435983 and 436732 in the forward strand. The gene contains 1 exon with a selenocysteine.
This protein was predicted blasting Nanorana parkeri genome against the Xenopus tropicalis DIa protein found in SelenoDB. To find the gene structure we analyzed the exonerate file.
Finally, 1 SECIS element of grade A and 1 SECIS element of grade B, predicted in the 3'UTR region, were found. In addition, Seblastian result concorded with SECIS prediction.
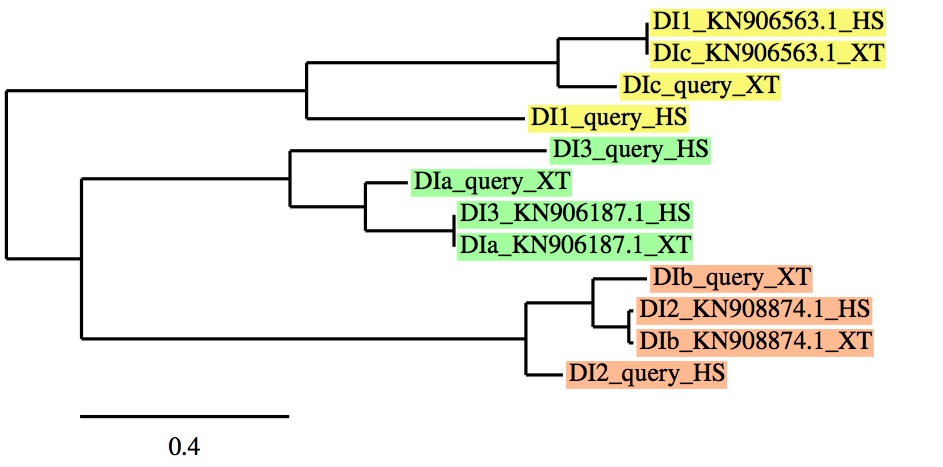
As we have already mentioned, in case of protein families, a phylogenetic tree was performed using a program named phylogeny.fr, to clearly understand and analyze our results. It allows us to correlate: our initial protein queries, both proteins from Xenopus tropicalis (which were not correctly defined in databases -named as "NONE"-;thus, we needed to name as -a, b, c, d, etc...-) and Homo sapiens (which were correctly defined and annotated in databases) with our final protein predictions from the best scaffold in each protein.
Glutathione peroxidase
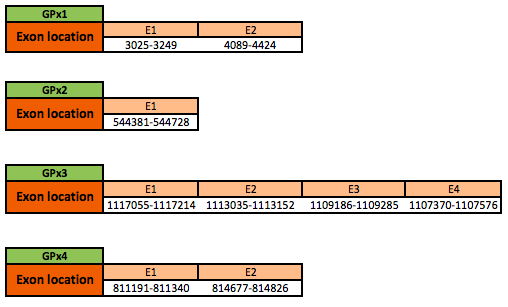
GPX1
The GPx1 protein is located in the scaffold KN910165.1 between positions 3025 and 4424 in the forward strand. The gene contains 2 exons, with a selenocysteine in exon 1.
This protein was predicted blasting Nanorana parkeri genome against the Xenopus tropicalis GPxb protein found in SelenoDB. To find the gene structure we analyzed the exonerate file.
Finally, 1 SECIS element of grade A and 1 SECIS element of grade B, predicted in the 3'UTR were found. In addition, Seblastian result concorded with SECIS prediction.
GPX2
The GPx2 protein is located in the scaffold KN907016.1 between positions 544381 and 544728 in the reverse strand. The gene contains 1 exon, but no selenocysteine was found in the protein predicted.
This protein was predicted blasting Nanorana parkeri genome against the Xenopus tropicalis GPxc protein found in SelenoDB. To find the gene structure we analyzed the exonerate file.
Finally, 1 SECIS element of grade A and 2 SECIS elements of grade B, predicted in the 3'UTR region, were found. Though, no Seblastian results were obtained.
GPX3
The GPx3 protein is located in the scaffold KN906051.1 between positions 1107370 and 1117214 in the reverse strand. The gene contains 4 exons, with a selenocysteine in exon 4.
This protein was not correctly annotated in SelenoDB for Xenopus tropicalis, neither in other protein databases. Thus, this protein was predicted blasting Nanorana parkeri genome against the human GPx3 protein found in SelenoDB. To find the gene structure we analyzed the exonerate file.
Finally, SECIS element of grade A, predicted in the 3'UTR region, was found. In addition, Seblastian result concorded with SECIS prediction.
GPX4
The GPx4 protein is located in the scaffold KN906249.1 between positions 811191 and 814826 in the forward strand. The gene contains 2 exons, with a selenocysteine in exon 1.
This protein was not defined in SelenoDB for Xenopus tropicalis, neither in other protein databases. Thus, this protein was predicted blasting Nanorana parkeri genome against the human GPx4 protein found in SelenoDB. To find the gene structure we analyzed the exonerate file.
Finally, 1 SECIS element of grade A and 1 SECIS element of grade B, predicted in the 3'UTR region, were found. In addition, Seblastian result concorded with SECIS prediction.
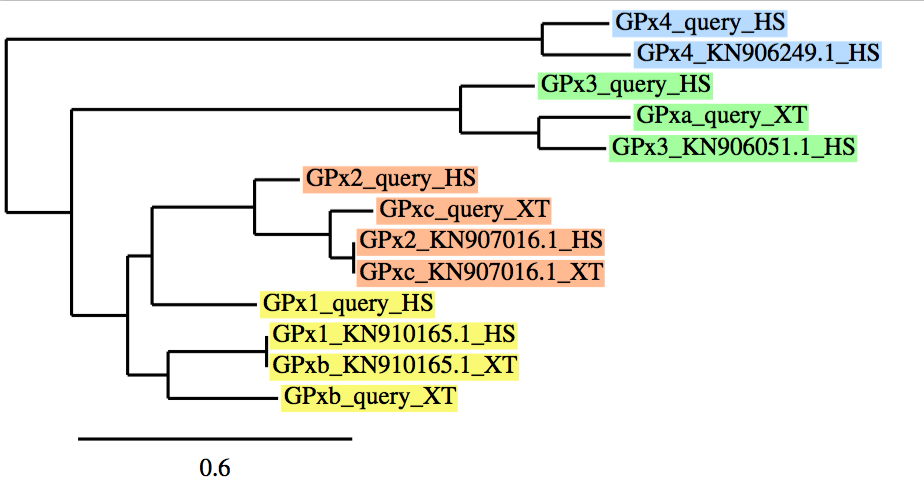
As we have already mentioned, in case of protein families, a phylogenetic tree was performed using a program named phylogeny.fr, to clearly understand and analyze our results. It allows us to correlate: our initial protein queries, both proteins from Xenopus tropicalis (which were not correctly defined in databases -named as "NONE"-;thus, we needed to name as -a, b, c, d, etc...-) and Homo sapiens (which were correctly defined and annotated in databases) with our final protein predictions from the best scaffold in each protein.
Selenoprotein O
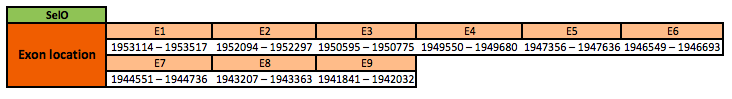
The SelO protein is located in the scaffold KN905923.1 between positions 1941841 and 1953517 in the reverse strand. The gene contains 9 exons, with a selenocysteine in exon 8.
This protein was predicted blasting Nanorana parkeri genome against the Xenopus tropicalis SelO protein found in SelenoDB. To find the gene structure we analyzed the exonerate file.
Finally, SECIS element of grade B, predicted in the 3'UTR region, was found. Though, no Seblastian results were obtained.
Selenoprotein P
The SelP protein is located in the scaffold KN906419.1 between positions 663086 and 664756 in the forward strand. The gene contains 3 exons, with a selenocysteine in exon 1.
This protein was predicted blasting Nanorana parkeri genome against the Xenopus tropicalis SelP protein found in SelenoDB. To find the gene structure we analyzed the exonerate file.
Finally, 2 SECIS elements of grade A and 1 SECIS elements of grade B, predicted in the 3'UTR region, were found. In addition, Seblastian result concorded with SECIS prediction.

Selenoprotein R (MsrB1)
The MsrB1 protein is located in the scaffold KN906356.1 between positions 535896 and 541362 in the reverse strand. The gene contains 3 exons, with a selenocysteine in exon 3.
This protein was predicted blasting Nanorana parkeri genome against the Xenopus tropicalis MsrB1 protein found in HomoloGene. To find the gene structure we analyzed the exonerate file.
Finally, 2 SECIS elements of grade A and 2 SECIS elements of grade B, predicted in the 3'UTR region, were found. Though, no Seblastian results were obtained.

MsrA
The MsrA protein is located in the scaffold KN906068.1 between positions 1116904 and 1135119 in the reverse strand. The gene contains 4 exons. In frog, the selenocysteine (Sec) residue was substituted by a cysteine (Cys) residue.
This protein was predicted blasting Nanorana parkeri genome against the Xenopus tropicalis MsrA protein found in SelenoDB. To find the gene structure we analyzed the exonerate file.
Finally, SECIS element of grade B, predicted in the 3'UTR region, was found. Though, no Seblastian results were obtained.

Selenoprotein U
SelU1
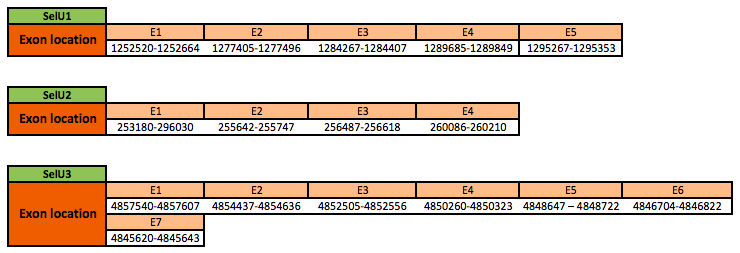
The SelU1 protein is located in the scaffold KN906066.1 between positions 1272520 and 1295353 in the forward strand. The gene contains 5 exons. In frog, the selenocysteine (Sec) residue was substituted by a cysteine (Cys) residue.
This protein was predicted blasting Nanorana parkeri genome against the Xenopus tropicalis SelUc protein found in SelenoDB. To find the gene structure we analyzed the exonerate file.
Finally, SECIS element of grade B, predicted in the 3'UTR region, was found. Though, no Seblastian results were obtained.
SelU2
The SelU2 protein is located in the scaffold KN906045.1 between positions 253280 and 260210 in the forward strand. The gene contains 4 exons. In frog, the selenocysteine (Sec) residue was substituted by a cysteine (Cys) residue.
This protein was predicted blasting Nanorana parkeri genome against the Xenopus tropicalis SelUb protein found in SelenoDB. To find the gene structure we analyzed the exonerate file.
Finally, no SECIS elements were found neither Seblastian results were obtained.
SelU3
The SelU3 protein is located in the scaffold KN905853.1 between positions 4845620 and 4857607 in the reverse strand. The gene contains 7 exons, with a selenocysteine.
This protein was predicted blasting Nanorana parkeri genome against the Xenopus tropicalis SelUa protein found in SelenoDB. To find the gene structure we analyzed the exonerate file.
Finally, SECIS element of grade B, predicted in the 3'UTR region, was found. Though, no Seblastian results were obtained.
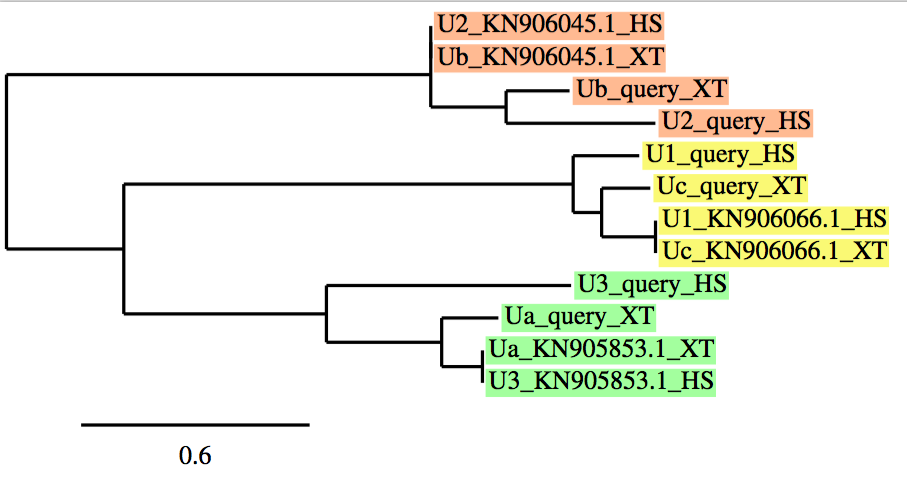
As we have already mentioned, in case of protein families, a phylogenetic tree was performed using a program named phylogeny.fr, to clearly understand and analyze our results. It allows us to correlate: our initial protein queries, both proteins from Xenopus tropicalis (which were not correctly defined in databases -named as "NONE"-;thus, we needed to name as -a, b, c, d, etc...-) and Homo sapiens (which were correctly defined and annotated in databases) with our final protein predictions from the best scaffold in each protein.
Selenoprotein W
SelW1
The SelW1 protein is located in the scaffold KN906069.1 between positions 1346955 and 1348268 in the forward strand. The gene contains 2 exons, but no selenocysteine was found in the protein predicted.
This protein was not correctly annotated in SelenoDB for Xenopus tropicalis, neither in other protein databases. Thus, this protein was predicted blasting Nanorana parkeri genome against the human SelW1 protein found in SelenoDB. To find the gene structure we analyzed the exonerate file.
Finally, 1 SECIS element of grade A and 3 SECIS elements of grade B, predicted in the 3'UTR region, were found. Though, no Seblastian results were obtained.
SelW2
The SelW protein is located in the scaffold KN905922.1 between positions 2394200 and 2396815 in the reverse strand. The gene contains 2 exons, with a selenocysteine in exon 2.
This protein was predicted blasting Nanorana parkeri genome against the Xenopus tropicalis SelWa protein found in SelenoDB. To find the gene structure we analyzed the exonerate file.
Finally, SECIS element of grade B, predicted in the 3'UTR region, was found. Though, no Seblastian results were obtained.
Rdx12
The Rdx12 protein is located in the scaffold KN906527.1 between positions 153842 and 156768 in the forward strand. The gene contains 3 exons, with a selenocysteine in exon 1.
This protein was not defined in SelenoDB for Xenopus tropicalis, neither in other protein databases. Thus, this protein was predicted blasting Nanorana parkeri genome against the Mus musculus Rdx12 protein found in UniProt. To find the gene structure we analyzed the exonerate file.
Finally, 1 SECIS element of grade B, predicted in the 3'UTR region, was found. Though, no Seblastian results were obtained.

Thioredoxin reductase
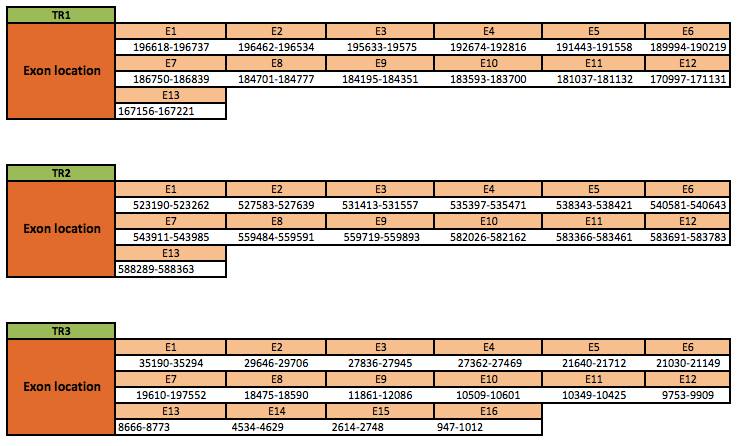
TR1
The TR1 protein is located in the scaffold KN906217.1 between positions 167156 and 2229202 in the reverse strand. The gene contains 13 exons, with a selenocysteine in exon 13.
This protein was predicted blasting Nanorana parkeri genome against the Xenopus tropicalis TRa protein found in SelenoDB. To find the gene structure we analyzed the exonerate file.
Finally, SECIS element of grade A, predicted in the 3'UTR region, was found. In addition, Seblastian result concorded with SECIS prediction.
TR2
The TR2 protein is located in the scaffold KN906108.1 between positions 523190 and 588363 in the forward strand. The gene contains 13 exons, with a selenocysteine in exon 1.
This protein was predicted blasting Nanorana parkeri genome against the Xenopus tropicalis TRc protein found in SelenoDB. To find the gene structure we analyzed the exonerate file.
Finally, no SECIS elements were found neither Seblastian results were obtained.
TR3
The TR3 protein is located in the scaffold KN905983.1 between positions 947 and 35294 in the reverse strand. The gene contains 16 exons, with a selenocysteine in exon 15.
This protein was predicted blasting Nanorana parkeri genome against the Xenopus tropicalis TRb protein found in SelenoDB. To find the gene structure we analyzed the exonerate file.
Finally, 1 SECIS element of grade A and 1 SECIS element of grade B, predicted in the 3'UTR region, were found. In addition, Seblastian result concorded with SECIS prediction.
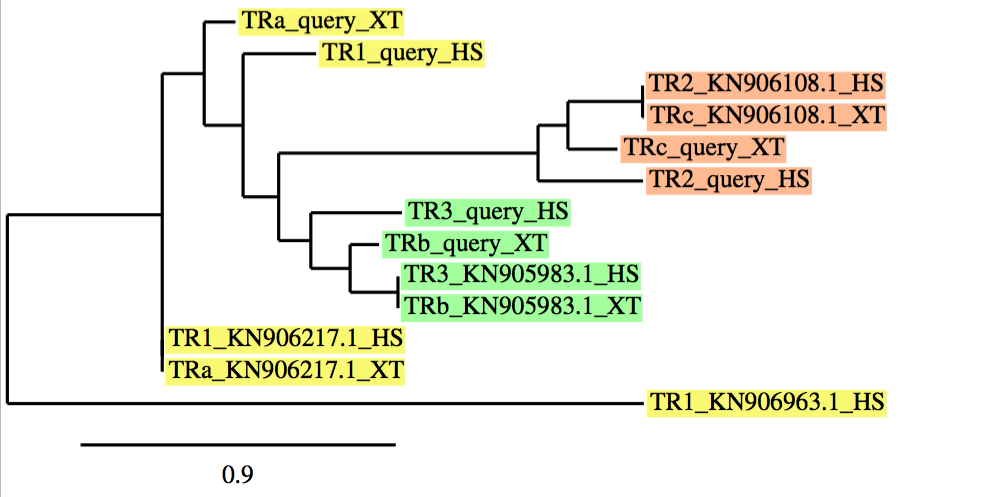
As we have already mentioned, in case of protein families, a phylogenetic tree was performed using a program named phylogeny.fr, to clearly understand and analyze our results. It allows us to correlate: our initial protein queries, both proteins from Xenopus tropicalis (which were not correctly defined in databases -named as "NONE"-;thus, we needed to name as -a, b, c, d, etc...-) and Homo sapiens (which were correctly defined and annotated in databases) with our final protein predictions from the best scaffold in each protein.
Selenoproteins:
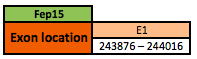
Fep 15
The Fep15 protein is located in the scaffold KN907437.1 between positions 243876 and 244016 in the reverse strand. The gene contains 1 exon. In frog, the selenocysteine (Sec) residue was substituted by a cysteine (Cys) residue.
This protein was predicted blasting Nanorana parkeri genome against the Xenopus tropicalis Fep15 protein found in SelenoDB. To find the gene structure we analyzed the exonerate file.
Finally, no SECIS elements were found neither Seblastian results were obtained.
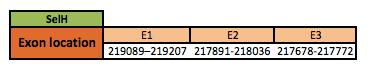
Selenoprotein H
The SelH protein is located in the scaffold KN907381.1 between positions 217678 and 219207 in the reverse strand. The gene contains 3 exons, with a selenocysteine in exon 2.
This protein was not defined in SelenoDB for Xenopus tropicalis, neither in other protein databases. Thus, this protein was predicted blasting Nanorana parkeri genome against the human SelH protein found in SelenoDB. To find the gene structure we analyzed the exonerate file.
Finally, SECIS element of grade A, predicted in the 3'UTR region, was found.In addition, Seblastian result concorded with SECIS prediction.
Selenoprotein I
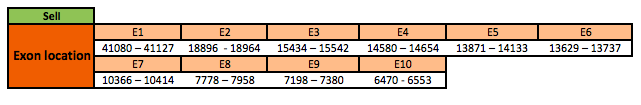
The SelI protein is located in the scaffold KN909114.1 between positions 6470 and 41127 in the reverse strand. The gene contains 10 exons, with a selenocysteine in exon 8.
This protein was not correctly annotated in SelenoDB for Xenopus tropicalis, neither in other protein databases. Thus, this protein was predicted blasting Nanorana parkeri genome against the human SelI protein found in SelenoDB. To find the gene structure we analyzed the exonerate file.
Finally, no SECIS elements were found neither Seblastian results were obtained.
Selenoprotein K
The SelK protein is located in the scaffold KN906037.1 between positions 1366623 and 1369834 in the reverse strand. The gene contains 3 exons, with a selenocysteine in exon 3.
This protein was predicted blasting Nanorana parkeri genome against the Xenopus tropicalis SelK protein found in SelenoDB. To find the gene structure we analyzed the exonerate file.
Finally, SECIS element of grade A, predicted in the 3'UTR region, was found. Though, no Seblastian results were obtained.

Selenoprotein M
The SelM protein is located in the scaffold KN906047.1 between positions 637905 and 654651 in the reverse strand. The gene contains 5 exons, with a selenocysteine in exon 5.
This protein was predicted blasting Nanorana parkeri genome against the Xenopus tropicalis SelM protein found in SelenoDB. To find the gene structure we analyzed the exonerate file.
Finally, 2 SECIS elements of grade A and 2 SECIS elements of grade B, predicted in the 3'UTR region, were found. Though, no Seblastian results were obtained.

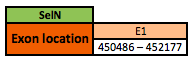
Selenoprotein N
The SelN protein is located in the scaffold KN906994.1 between positions 450486 and 452177 in the forward strand. The gene contains 1 exon with a selenocysteine.
This protein was predicted blasting Nanorana parkeri genome against the Xenopus tropicalis SelN protein found in SelenoDB. To find the gene structure we analyzed the exonerate file.
Finally, 1 SECIS element of grade A and 1 SECIS element of grade B, predicted in the 3'UTR region, were found. Though, no Seblastian results were obtained.
Selenoprotein Pb
The SelPb protein is located in the scaffold KN908356.1 between positions 24006 and 27339 in the forward strand. The gene contains 3 exons. In frog, the selenocysteine (Sec) residue was substituted by a cysteine (Cys) residue.
This protein was predicted blasting Nanorana parkeri genome against the Xenopus tropicalis SelPb protein found in Ensembl. To find the gene structure we analyzed the exonerate file.
Finally, no SECIS elements were found neither Seblastian results were obtained.

Selenoprotein S
The SelS protein is located in the scaffold KN906821.1 between positions 450333 and 465004 in the forward strand. The gene contains 3 exons, with a selenocysteine in exon 1.
This protein was predicted blasting Nanorana parkeri genome against the Xenopus tropicalis SelS protein found in SelenoDB. To find the gene structure we analyzed the exonerate file.
Finally, 1 SECIS element of grade A and 1 SECIS element of grade B, predicted in the 3'UTR region, were found. In addition, Seblastian result concorded with SECIS prediction.

Selenoprotein T
The SelT protein is located in the scaffold KN906746.1 between positions 66024 and 78800 in the forward strand. The gene contains 5 exons, with a selenocysteine in exon 1.
This protein was predicted blasting Nanorana parkeri genome against the Xenopus tropicalis SelT1 protein found in SelenoDB. To find the gene structure we analyzed the exonerate file.
Finally, 1 SECIS element of grade A and 1 SECIS element of grade B, predicted in the 3'UTR region, were found. Though, no Seblastian results were obtained.

Selenoprotein 15
The Sel15 protein is located in the scaffold KN906065.1 between positions 932194 and 946340 in the forward strand. The gene contains 4 exons, with a selenocysteine in exon 1.
This protein was predicted blasting Nanorana parkeri genome against the Xenopus tropicalis Sel15 protein found in SelenoDB. To find the gene structure we analyzed the exonerate file.
Finally, 1 SECIS element of grade A and 1 SECIS element of grade B, predicted in the 3'UTR region, were found. In addition, Seblastian result concorded with SECIS prediction.


Machinery
Eukaryotic elongation factor (eEFsec)
The eEFsec protein is located in the scaffold KN905983.1 between positions 1562965 and 1629305 in the forward strand. The gene contains 5 exons with a cysteine (Cys) residue.
This protein was predicted blasting Nanorana parkeri genome against the Xenopus tropicalis eEFsec protein found in SelenoDB. To find the gene structure we analyzed the exonerate file.
Finally, 2 SECIS elements of grade B, predicted in the 3'UTR region, were found. Though, no Seblastian results were obtained.

Phosphoseryl-tRNA kinase (PTSK)
The PSTK protein is located in the scaffold KN905975.1 between positions 253944 and 260463 in the forward strand. The gene contains 4 exons with a cysteine (Cys) residue.
This protein was predicted blasting Nanorana parkeri genome against the Xenopus tropicalis PSTK protein found in SelenoDB. To find the gene structure we analyzed the exonerate file.
Finally, no SECIS elements were found neither Seblastian results were obtained.

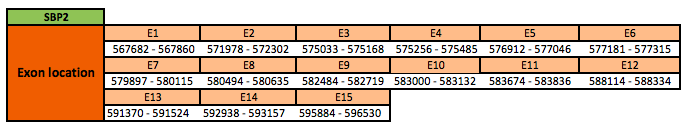
SECIS binding protein 2 (SBP2)
The SBP2 protein is located in the scaffold KN906459.1 between positions 567682 and 596530 in the forward strand. The gene contains 15 exons with a 2 selenocysteine in exon 1, and 1 selenocysteine in exon 2.
This protein was predicted blasting Nanorana parkeri genome against the Xenopus tropicalis SBP2 protein found in SelenoDB. To find the gene structure we analyzed the exonerate file.
Finally, 3 SECIS elements of grade B, predicted in the 3'UTR region, were found. Though, no Seblastian results were obtained.
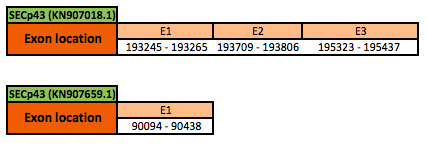
tRNA Sec 1 associated protein 1 (SECp43)
The SECp43 protein is located in the scaffold KN907018.1 between positions 193245 and 195437, and in the scaffold KN907659.1 between positions 90094 and 90438 in the forward strand. The gene contains 3 exons in the first scaffold with a cysteine (Cys) residue, and 1 exon in the second scaffold, with 3 selenocysteine (Sec) residues.
This protein was not defined in SelenoDB for Xenopus tropicalis, neither in other protein databases. Thus, this protein was predicted blasting Nanorana parkeri genome against the human SECp43 protein found in SelenoDB. To find the gene structure we analyzed the exonerate file.
Finally, no SECIS elements were found neither Seblastian results were obtained for the scaffold KN907018.1. However, 1 SECIS element of grade A and 1 SECIS element of grade C, predicted in the 3'UTR region, were found for the scaffold KN907659.1. Though, no Seblastian results were obtained.
Selenocysteine synthase (SecS)
The SecS protein is located in the scaffold KN908153.1 between positions 35129 and 44423 in the reverse strand. The gene contains 3 exons with a cysteine (Cys) residue.
This protein was predicted blasting Nanorana parkeri genome against the Xenopus tropicalis SecS protein found in SelenoDB. To find the gene structure we analyzed the exonerate file.
Finally, no SECIS elements were found neither Seblastian results were obtained.

Selenophosphate synthetase family (SPS)
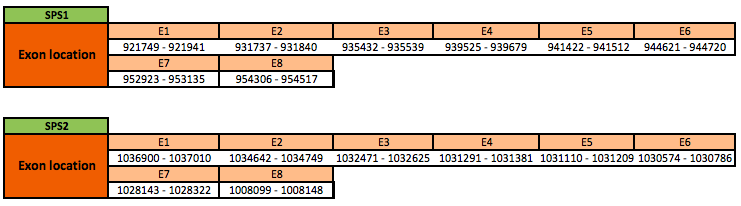
SPS1
The SPS1 protein is located in the scaffold KN905976.1 between positions 921749 and 954517 in the forward strand. The gene contains 8 exons with a cysteine (Cys) residue.
This protein was not defined in SelenoDB for Xenopus tropicalis, neither in other protein databases. Thus, this protein was predicted blasting Nanorana parkeri genome against the human SPS1 protein found in SelenoDB. To find the gene structure we analyzed the exonerate file.
Finally, 1 SECIS element of grade C, predicted in the 3'UTR region, was found. Though, no Seblastian results were obtained.
SPS2
The SPS2 protein is located in the scaffold KN906395.1 between positions 1008099 and 1037010 in the reverse strand. The gene contains 8 exons with a cysteine residue.
This protein was not defined in SelenoDB for Xenopus tropicalis, neither in other protein databases. Thus, this protein was predicted blasting Nanorana parkeri genome against the human SPS2 protein found in SelenoDB. To find the gene structure we analyzed the exonerate file.
Finally, no SECIS elements were found neither Seblastian results were obtained.