DISCUSSION
Selenoproteins in Nanorana parkeri were characterized by studying its homology with Xenopus tropicalis (frog) and Homo sapiens (human) proteins. Our aim was to focus the analysis principally on Xenopus tropicalis, due to its closer phylogeny to Nanorana parkeri.
The largest and the best-studied selenoprotein families are glutathione peroxidase (GPx), thioredoxin reductase (TR) and iodothyronine deiodinase (Dio) families, with 5, 3, and 3 Sec-containing genes in the human genome, respectively. Though half of mammalian selenoproteins function is still unknown, it has been seen that many of them have a role in redox regulation. Selenoproteins have also been involved in cancer prevention, modulation of the aging process, male reproduction, and immune response. However, selenoproteins with partially characterized biological functions, such as SelH, SelI, SelM, SelN, SelS, SelT, SelW and Sel15; or unknown functions, such as SelK, SelO and SelV, have been less well studied.
The smallest selenoproteome was predicted in frog and in some mammals, with only 24 selenoproteins genes. Nevertheless, 21 selenoproteins were found in all vertebrates: GPx1-4, TR1, TR3, Dio1, Dio2, Dio3, SelH, SelI, SelK, SelM, SelN, SelO, SelP, MsrB1, SelS, SelT1, SelW1 and Sep15. This event shows the high conservation of selenoproteins among vertebrate's evolution. In contrast, some selenoproteins are only found in certain lineages, some were lost during evolution of a particular lineage and some replaced their Sec residue for a Cys residue.
After the split of amphibians, some selenoproteins, like SelU1, Fep15 and SelPb, suffered a Sec- to Cys-containing residue switch. In addition, frog is the only specie that presents both SelW2 and Rdx12 selenoproteins.
From this point, we will describe and discuss the results of our analysis, correlating them with the literature found upon both Xenopus tropicalis and Homo sapiens selenoproteins.

SELENOPROTEINS
Iodothyronine Deiodinase (DI)
The iodothyronine deiodinases family (DI) is the general name for a family constituted of enzymes that catalyze the removal of iodine atoms from various thyroid hormones (Ths) in the thyroid gland and extrathyroidal tissues. As such, they are responsible for both the activation and inactivation of these compounds, and are thus important regulators of TH actions. In mammals, there are three known DI enzymes, all of them containing Sec residues: DI1, DI2 and DI3. It is described that DI1 and DI3 are located in plasma membrane, while DI2 is found in ER.
Particularly, DI3 irreversibly inactivates the thyroid hormone by deiodination of the inner tyrosyl ring, and it is also found that all detected DI3 genes are intronless, while all other genes in vertebrates, apart from SPS2b, consist of multiple exons. In contrast, DI2 is an ER-resistent protein that activates the thyroid hormone by deiodination of the outer tyrosyl ring. A curious characteristic of DI2 is that there is a second in-frame UGA codon in its mRNA. In previous studies, it was observed that the second UGA could insert Sec when the first UGA codon is mutated.
According to this finding explained above, we predicted a gene for DI2 which contains more than one UGA codons (observed as TGA in our gene predict sequence). Thus, it might suggest this codon can have a secondary function introducing Sec residues, when the first UGA codon is mutated, even its main function is to serve as a stop codon.
After our results analysis we could predict DI1, DI2 and DI3 selenoproteins in Nanorana parkeri genome, which concords with the literature. These proteins we predicted contains a Sec residue. In case of DI1 we found a SECIS element predicted in the 3'UTR region of the gene, concording with the Seblastian output. For DI2, two SECIS elements were predicted in 3'UTR region of the gene. DI3 showed the presence of two SECIS elements predicted in the 3'UTR region of the gene, and Seblastian output confirmed this prediction.
Glutathione peroxidase (GPX)
The glutathione peroxidase family (GPx) is the common name for a family of multiple isozymes that catalyze the reduction of H2O2 or organic hydroperoxides to water or corresponding alcohols using reduced glutathione (GSH) as an electron donor (H2O2 + 2GSH fi GS-SG + 2H2O). Aerobic reactions leads to the accumulation of reactive oxygen species that can be toxic to the cells. In this context, aerobic organisms have developed several n-enzymatic and enzymatic systems to neutralize these compounds. Concretely, the enzymatic systems include a set of gene products, such as superoxide dismutases, catalases, ascorbate peroxidases and glutathione peroxidades (GPx), whose principal function is to protect organism from oxidative damage. It is described that GPx1, GPx2, GPx4 and GPx6 are located in the cytosol, begin GPx4 also found in mytochondria nucleus (testis-specific), while GPx3 is located in plasma.
The GPx family is spread over the entire living kingdom and it is the largest selenoprotein family in vertebrates. In humans, eight distinct molecular paralogs coexists, five of which are selenoproteins (GPx1-4 and GPx6), meaning that contain Sec as the active-site residue, and the rest of GPx have evolved from the ancestors just cited. The selenoperoxidases (Sec-GPx) prevail in vertebrates, while GPx homologs having the active-site Sec replaced by Cys (Cys-GPx) are found in terrestrial plants, yeast, protozoa, and bacteria.
It is observed that before the separation of mammals and fishes, Cys-containing GPx7 and GPx8 evolved from a GPx4-like selenoprotein ancestor.
In contrast, the most recently evolved GPx are GPx5 and GPx6, due to a tandem duplication of GPx3 at the basis of placental mammals. As phylogeny reflects, a possible consequence of the origin of GPx5 from a duplication of selenoprotein GPx3, may be the Sec-to-Cys displacement, which must have appeared very early in the GPx5's evolution. For GPx6, several independent Cys conversions were observed: in primate marmoset, in rat and mouse, and in rabbit. It suggests that the Cys-containing GPx6 was not present in the last ancestor of rabbit and rodents because the Sec-containing GPx6 was also observed in other rodents, such as squirrel, guinea pig, and kangaroo rat.
According to these previous findings, we decided to blast Homo sapiens GPx5 and GPx6 against Nanorana parkeri genome. Indeed, due the high homology within the members of the family, we found some hits, though all of them corresponded to other homologous GPx.
As we already said, it is important to highlight that each of the mammalian Sec-containing GPx genes is highly conserved. Four out of five have better than 80% nucleotide sequence identity, while GPx1 has 70% sequence identity within mammalian sequences. GPx4 is one of the most conserved selenoproteins with better than 90% nucleotide sequence identity and considering full length selenoprotein sequences, GPx4 has the highest level of conservation of any selenoprotein.
To confirm the high conservation between the family members, in all cases we blasted Xenopus tropicalis and Homo sapiens protein queries against our studying genome. We found almost the same hits for each GPx in both species, indicating the elevated intrafamiliar relationship. Specially considering GPx1 and GPx4, which were previously defined as the most conserved Sec-containing selenoproteins.
After our results analysis we could predict GPx1, GPx3 and GPx4 selenoproteins in Nanorana parkeri genome. These proteins we predicted contain a Sec residue. In case of Gpx1 we found two SECIS elements predicted in the 3'UTR region of the gene, concording with the Seblastian output. For GPx3, a SECIS element was predicted in 3'UTR region of the gene, and it also corresponded with the Seblastian output. GPx4 showed the presence of two SECIS elements predicted in the 3'UTR region of the gene, and Seblastian output confirmed this prediction.
In case of GPx2, although we could not predict this protein in Sec form in Nanorana parkeri genome, neither from Xenopus tropicalis or Homo sapiens protein queries; we expected it as a Sec-containing selenoprotein, as it is really conserved selenoprotein among all vertebrates. At that point, even it was properly annotated and defined in SelenoDB, we tried to find this protein, as our initial query, in other databases, such as UniGene, but the same results were achieved. However, we could predict three SECIS elements in the 3'UTR region for GPx2 gene.
Due to all literature previously found, that describe GPx1-4 as a selenoproteins, we focused our analysis in predicting these four Sec-containing GPx members. However, once we blasted our studying genome against Homo sapiens GPx protein members, as all of them were well defined in SelenoDB, we found out non-expected Cys-containing selenoproteins, GPx7 and GPx8 members of the family. Comparing these results with many other literature sources, we found that these proteins take part of the ancestral vertebrate Cys-containing homologs, concording with our predictions. Finally, to confirm our results, we blasted both proteins against other vertebrate genomes, using ENSEMBL as the main source. Thus, we found out similar results for GPx7 and GPx8 protein members, which concords with the ancestral vertebrate Cys-containing homologs hypothesis.
Selenoprotein H
The selenoprotein H (SelH) is an ancestral selenoprotein found in all the vertebrates and its function is not completely known. It is regarded as a nucleolar oxide-reductase with an antioxidant function, and it is also related with the gene regulation regarding de novo synthesis of glutation as it contains a DNA-binding-domain. Because of these functions, one of its upregulation consequences is a suppression of the senescence. It is also enrolled in the biogenesis, function and size of mitochondria in the neural cells by activating PKA, PKB, and cAMP.
After our results analysis we could predict SelH selenoprotein in Nanorana parkeri genome. The protein we predicted contains a Sec residue, and a SECIS element was found in the 3'UTR region of the gene, which concord with the Seblastian output. Despite using the human query to predict the selenoprotein, results were correctly obtained due to the high SelH homology in different vertebrates.
Selenoprotein I
The selenoprotein I (SelI) is an ancestral selenoprotein found in all the vertebrates. This protein is one of the more recently discovered selenoproteins. It is characterized by a highly conserved CPD-alcohol phosphatidyltransferase domain that is commonly encountered in choline phosphotransferases (CHPT1) and choline/ethanolamine phosphotransferases (CEPT1). Specifically, CHPT1 catalyzes the transfer of choline to diacylglycerol from CPD-choline and CEPT1 catalyzes and analogous reaction but accepts both choline and ethanolamine. This fact reflects the similarity between these proteins. Besides, SelI has seven predicted transmembrane domains, which correspond to the predicted topologies of CHPT1 and CEPT1. It is observed that the most critical portion of this structure for a correct function are three aspartic acids, which are located between the first and second transmembrane domains. Not only these residues are conserved in all SelI proteins, but also the whole active region is highly closer between SelI and its homologs.
In contrast, the main difference between SelI and its homologs is a Sec containing C-terminal extension in SelI, but its role has yet to be discovered. Moreover, no Cys forms with homology to the SelI C-terminal extension were found. As it is well known, Sec residues often participate in selenenylsulfide bonds with cysteins, thus, it is important to search for cysteines emerged particularly in SelI proteins.
This relevant difference between SelI and its homologous has been seen in our protein query, took from human protein.
In recent studies, a specific ethanolamine phosphotransferase (EPT) activity has been reported while testing human SelI protein for CHPT1/CEPT1 enzymatic activities. Although, they used a bacterial expression system for purification of human SelI, and eukaryotic SECIS elements are not recognized in bacteria, it leads to different possible hypothesis of its function. One possibility is that EPT activity is just the first step in SelI function, with phosphatidylethanolamine further processed in a Sec-dependent step, and another could be that the Sec extension provides totally different substrate specificity to SelI.
After our results analysis we could predict SelI selenoprotein in Nanorana parkeri genome. The protein we predicted contains a Sec residue, but no SECIS elements were found in the 3'UTR region of the gene, which concord with the Seblastian negative output. Despite using the human query to predict the selenoprotein, results were properly obtained due to the high SelI homology in different vertebrates.
Selenoprotein K
The selenoprotein K (SelK) is an ancestral selenoprotein found in all the vertebrates. This protein is a small selenoprotein that resides in the ER and in the plasma membrane. This protein has an unknown biological function and only one published linked this protein to a possible redox function due to the presence of selenocysteine in its active site, protecting cells from oxidative stress.
Some comparative genomics analyses indicate that SelK is the most widespread selenoprotein, found in almost all eukaryotes that use Sec and being replaced with a Cys-containing homolog in some organisms, i.e. nematodes.
It has been also revealed some proteins that interact with SelK, containing ER-associated degradation (ERAD) components. In this complex, SelK showed higher affinity for Derlin-1 what suggest that this selenoprotein could determine the nature of the substrate translocated through the Derlin channel, being also involved in its degradation. SelK gene contains a functional ER stress response element, showing an up-regulation expression in conditions that induce the accumulation of misfolded proteins in the ER. Components of the oligosaccharyltransferase complex an ER chaperone, calnexin, were also found to bind SelK. All these finding suggest that SelK is involved in the Derlin-dependent ERAD of glycosylated misfolded proteins, and that the function defined by the prototypic SelK is the widespread function of selenium in eukaryotes.
It has been found that SelK has more pseudogenes (>27) than any other selenoprotein, being these pseudogenes only described in mammals until now, especially rodent lineages.
In accordance with the information above, no SelK pseudogenes appeared in Nanorana parkeri. As it is already known, it is necessary more than one homologous hit with this protein to define a pseudogene. Thus, only one hit was found out for this protein.
After our results analysis we could predict SelK selenoprotein in Nanorana parkeri genome. The protein we predicted contains a Sec residue, and a SECIS element was found in the 3'UTR region of the gene. As it takes part of the common vertebrate selenoproteins and it was previously well annotated in other databases, this protein was clearly predicted using Xenopus tropicalis as our protein source.
Selenoprotein N
The selenoprotein N (SelN) is an ancestral selenoprotein found in all the vertebrates. This eukaryotic selenoprotein is located basically in the ER membrane. It has a high expression in fetal and growing muscular tissue, skeletal muscle, heart, lung and placenta. It has an important role in the diaphragm, inducing respiratory problems when it is mutated, as it is involved in the intracellular calcium homeostasis. It has been the first selenoprotein directly related with genetic disorders, as it plays an important role in congenital myopathy.
After our results analysis we could predict SelN selenoprotein in Nanorana parkeri genome. The protein we predicted contains a Sec residue, and two SECIS elements were found in the 3'UTR region of the gene. As it takes part of the common vertebrate selenoproteins and it was previously well annotated in other databases, this protein was clearly predicted using Xenopus tropicalis as our protein source.
Selenoprotein 0
The SelO family is the common name for a family of ancestral selenoproteins found in all the vertebrates. These isozymes are localized to mitochondria and expressed in different tissues. This expression is affected by the deficiency of selenium, suggesting that it has a high priority for selenium supply. SelO protein is a redox-active mitochondrial selenoprotein, which seems to have maintained the catalytic phosphotransferase activity in its active site, where it contains a selenocysteine (Sec) residue. The SelO family is composed by two isoforms of the protein, SelO1 and SelO2, both conserved in mice, rats and humans.
This selenoproteins family is found in a wide range of organisms, including eukaryotes, bacteria and yeast, and it is the largest mammalian selenoprotein. Although, there is a majority of eukaryotes and bacteria that have a single-copy protein of SelO, many metazoans have duplicated it. Additionally, this phenomenon is also observed in specific lineages of bony fishes such as zebrafish, in which extra copies of SelO, SelT1 and SelW appeared. Furthermore, several studies found out a duplicated SelO-like gene in the common ancestor of Metazoa, while there was a loss of this duplication in some other lineages like rodents, primates and other mammals.
After our results analysis we could predict SelO selenoprotein in Nanorana parkeri genome. The protein we predicted contains a Sec residue, and a SECIS element was found in the 3'UTR region of the gene. As it takes part of the common vertebrate selenoproteins and it was previously well annotated in other databases, this protein was clearly predicted using Xenopus tropicalis as our protein source. In conclusion, our results may confirm the loss of duplication described for other lineages in the published literature.
Selenoprotein P
The selenoprotein P (SelP) is an ancestral selenoprotein found in all the vertebrates. This protein is a secreted glycoprotein that contains most of the selenium in plasma.
Although its function is unknown, it seems to have antioxidant properties, getting attached to epithelial cells through its heparin-binding properties and protecting the vascular endothelial cells against peroxynitrite toxicity. Recent studies support that due to the high selenium concentration found in SelP, it is probably related to selenium intercellular storage and transport from the liver to the peripheral tissues, which is important to maintain normal brain function. It is also possible that the C-terminal region of this selenoprotein has a redox function.
Moreover, SelP does not have a constant number of Sec residues, and as it has been seen in other studies, it is the unique that hold two SECIS elements. Although, we predicted three SECIS elements, only two of them were properly located, which concord with the literature.
After our results analysis we could predict SelP selenoprotein in Nanorana parkeri genome. The protein we predicted contains a Sec residue. As it takes part of the common vertebrate selenoproteins and it was previously well annotated in other databases, this protein was clearly predicted using Xenopus tropicalis as our protein source.
Selenoprotein R (Msr enzymes)
The Mrs enzymes (methionine sulfoxide reductases) are thiol-dependent enzymes, those function is to catalyze conversion of methionine sulfoxide to methionine. Nowadays, three Msr families are known (MsrA, MsrB, and fRMsr). MsrA and MsrB are responsible for the reduction of methionine-S-sulfoxide and methionine- R-sulfoxide residues in proteins, respectively, whereas fRMsr reduces free methionine-R-sulfoxide. Besides acting on proteins, MsrA can additionally reduce free methionine-S-sulfoxide. In addition, MsrAs and MsrBs evolved to utilize catalytic selenocysteine, which include MrsB1 (SelR), a major MsrB in cytosol and nucleus in mammalian cells. In contrast, MsrA exists like a selenoprotein in some lower organisms, as bacteria or green algae, using a Sec-catalytic residue instead of Cys residue. As we already know, selenocysteine offers catalytic advantage to the protein repair function of Msrs, but also makes these proteins dependent on the supply of selenium and requires adjustments in their strategies for regeneration of active enzymes, meaning that its replacement with cysteine leads to a significant drop or loss of catalytic activity. Interestingly, Msr enzymes are absent in many hyperthermophylic organisms because at higher temperatures, methionine sulfoxide reduction may not require catalysis. A part from that, it has been seen that Msrs have roles in protecting cellular proteins from oxidative stress and through this function they may regulate lifespan in several model organisms.
On one hand, our results showed that the MsrA protein we predicted had no Sec residue, which concords to the literature. On the other hand, we predicted a Sec-containing MsrB1 protein, and also four SECIS elements were found in the 3'UTR region of the gene. Nevertheless, as this protein is annotated in a Cys form in the databases for Xenopus tropicalis and we predicted it in a Sec form, its not a concluding result due to the same discrepancy exposed before in the literature.
Selenoprotein S
The selenoprotein S (SelS) is an ancestral selenoprotein found in all the vertebrates. This enzyme is located in the ER and the cell membrane.
It has an important function in the UPR as it is part of the ERAD (Endoplasmic-reticulum-associated protein degradation) by retrotranslocating the misfolded proteins, and it provides protection against the ROS, concretely it is a thioredoxin-dependent reductase. SelS can reduce hydrogen peroxide but is not an efficient or broad-spectrum peroxidase. It is also enrolled in the inflammatory response, in which the liver secreted it, probably because of its up-regulation by cytokines.
Other recent studies suggest that SelS participates in intracellular membrane transport and maintenance of protein complexes by anchoring them to the ER membrane. Polymorphisms in the SelS gen had been described as a potential risk of inflammatory diseases due the increasing of inflammatory cytokines, such as TNF IL-1. SelS has an anti-apoptotic role and reduces the ER stress in peripheral macrophages and brain astrocytes.
After our results analysis we could predict SelS selenoprotein in Nanorana parkeri genome. The protein we predicted contains a Sec residue, and two SECIS elements were found in the 3'UTR region of the gene, which concord with the Seblastian output. As it takes part of the common vertebrate selenoproteins and it was previously well annotated in other databases, this protein was clearly predicted using Xenopus tropicalis as our protein source.
Selenoprotein T
The selenoprotein T (SelT) is an ancestral selenoprotein found in all the vertebrates. It is localized in the Golgi and the ER. It has an important role in the cell adhesion and enhances the expression of several oxidoreductase genes. In case of a SelT loss, an elevation of SelW1 is found, suggesting that SelW1 may functionally compensate the lack of SelT.
This protein is also important in neuroendocrine processes, as it contributes to the homeostasis of the intracellular calcium and secretion of hormones. The rapid and long-lasting increase of SelT is induced by cAMP and PACAP (neuropeptide pituitary adenylate cyclase-activating polypeptide); moreover it also contributes to the correct folding of the proteins from the RE by its binding to the UDP-glucose-UGTR protein. Finally, SelT is increased upon cell injury, what suggests its important role in oxidoreductase processes.
In previous studies, by building multiple sequence alignment for all vertebrate selenoproteins and analyzing their phylogenetic relationship and sequence features, SelT appeared to be the most conserved selenoprotein. It shows an impressive identity across all mammals, even at the nucleotide sequence level.
After our results analysis we could predict SelT selenoprotein in Nanorana parkeri genome. The protein we predicted contains a Sec residue, and two SECIS elements were found in the 3'UTR region of the gene. As it takes part of the common vertebrate selenoproteins and it was previously well annotated in other databases, this protein was clearly predicted using Xenopus tropicalis as our protein source.
Selenoprotein U
The selenoprotein U family (SelU family) is composed of three members (SelU1, SelU2 and SelU3). The SelU1 selenoprotein is the only member of the family that takes part of the ancestral selenoproteome. Even though SelU function is unknown, the Prx-like2 structure domain presented in these proteins implies their belonging to the thioredoxin-like superfamily.
A relevant finding in prior phylogenetic analysis of Sec- and Cys-containing forms of the SelU family, suggested that all Sec-containing SelU sequences belong to the SelU1 group. Concretely, in mammals there are three Cys-containing SelU proteins (SelU1-3), while in some fishes there are three Sec-containing SelU proteins. It was though that the three Cys-containing SelU proteins in mammals evolved from the three Sec-containing SelU sequences in fish, but there are not enough evidences to determine this Sec-to-Cys early event for SelU2 and SelU3.
Additionally, the SelU proteins of invertebrates diverged into 3 groups, classified into different families in accordance with the proteins of their vertebrate descendants. Sec residues of SelU2 proteins were changed into Cys residues during different moments of these 3 lineages, probably in the early era of invertebrates. The SelU3 lineage represented the most abundant group of invertebrate SelU proteins identified, where 8 SelU3 proteins were found. This suggests a timeframe during which Sec residues were changed into Cys residues during the evolution of the SelU3 family. In SelU1 lineage, this switch between Sec and Cys residues occurred earlier: while all invertebrates of this family were shown to contain Sec, not all mammalian in the SelU1 linage had Sec residues. All this data suggest that the Sec to Cys event occurred during the primitive mammalian stage; nevertheless, a diverged lineage of Sec to Cys was also found in amphibians. Two SelU1 family proteins of two different frogs were found to be Cys-form proteins that potentially changed into Cys-form independently, occurring after the divergence of modern amphibians from a common tetrapoda ancestor.
Even SelU lineage diverged into three families, all retained the Sec-form in the progenitors of the animal kingdom, but this form evolved into the Cys-form in higher mammalian species, without exception. However, Sec to Cys events occurred in different periods of history evolution.
After our results analysis we could predict SelU selenoprotein in Nanorana parkeri genome. According to the information explained above we found for both, SelU1 and SelU2, a Cys residue, which reinforce the potential switch from Sec-to-Cys residue in these selenoproteins. Although we expected a Cys-containing SelU3, our results showed a selenocysteine residue in this protein, even it had not been located in any exon.
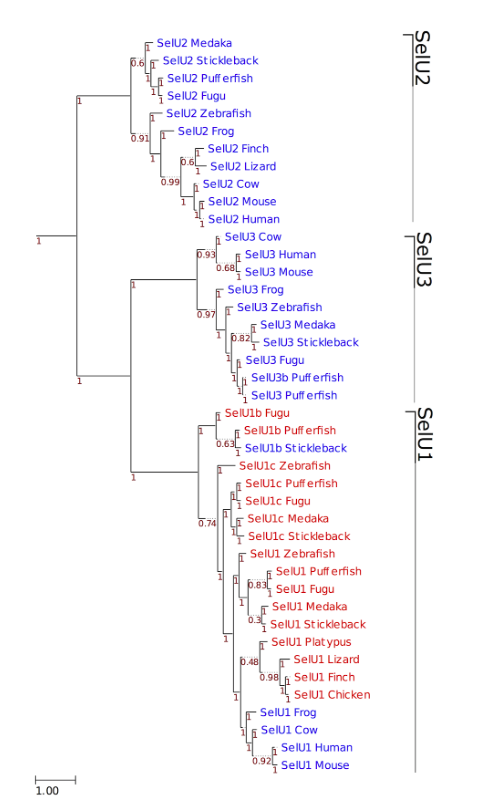
Figure 3. Phylogeny of SelU family in vertebrates. ML tree computed using the JTT substitution model. Sec-containing proteins are shown in red, whereas the Cys-containing homologs are shown in blue. At the bottom left, the distance scale in substitutions per position is shown. Branch support is shown along the tree in red.
Selenoprotein W and Selenoprotein V
The selenoproteins W1 and W2 (SelW1 and SelW2) are members of the ancestral selenoproteome, but only SelW1 takes part of the selenoproteins found in all the vertebrates. Moreover, SelV, which is the least conserved mammalian selenoprotein, appeared at the root of placental mammals by the duplication of SelW.
The functions of SelV and SelW, both located in the cytosol, are not known, but SelV is expressed exclusively in testes, whereas SelW is expressed in a variety of organs. SelW and SelV exhibited the same gene structure; each contained 6 exons with intron locations and phases conserved. Coding regions were within exons 1 - 5.Exon 6 contained only the last portion of the 3'UTR, including the SECIS element. Significant variation between SelW and SelV was found only in exon 1. Translated protein length of this exon has an average length of 261 residues (ranged from 228 amino acids in cat to 334 in dog), in contrast to SelW that had only 9 residues derived from exon 1 in most mammals. Only the last four residues of SelW and SelV in exon 1, which were located immediately upstream of the CxxU motif, were conserved; in contrast, their homology was high in exons 2 - 5 as well as in the SECIS element in exon 6, suggesting that evolution of SelV by SelW gene duplication might have followed up by the addition of N-terminal sequences.
Several SelW homologs were observed across non-mammalian vertebrates. Phylogenetic analysis revealed a distinct group of proteins, SelW2. It was described SelW2 as a selenoprotein in bony fishes, but also in frog and in elephant and shark, which suggests that it was part of the ancestral vertebrate selenoproteome. In mammals, only a remote homolog of SelW2 is present: Rdx12, which is not a selenoprotein and aligns a Cys to the Sec residue of SelW2. Moreover, frog is the only specie in which both selenoproteins SelW2 and Rdx12 have been found. In all other tetrapods, only Rdx12 was found. Thus, it is hypothesized that before the split of amphibians SelW2 duplicated and was immediately converted to a Cys form generating Rdx12, and then SelW2 was lost before the reptile's split.
After our results analysis we could predict two different SelW selenoproteins in Nanorana parkeri genome. One predicted selenoprotein, which contained a Sec residue, could be associated to SelW1, as is described in the literature. In addition, a SECIS element was found. Another predicted selenoprotein that contained a Cys residue might be associated to SelW2, what is also reported in previous studies. Although, four SECIS elements were predicted, only one presented a properly location.
A part from that, due to the importance of Cys-containing SelW2 and Rdx12 selenoproteins in frog, we tried to predict Rdx12 in Nanorana parkeri genome. To achieve this purpose, we looked for the phylogenetic closer organism, Mus musculus, which has this protein form annotated in databases. After all, we could not predict a Cys-containing selenoprotein, though a Sec residue appeared in our protein sequence.
Selenoprotein 15
The selenoprotein 15 (Sel15) protein is an ancestral selenoprotein found in all the vertebrates. It takes part of the Sep15 family, was described for the first time in 1995 by Kacklosch et al., who found the protein in rat prostate. In 1998 the protein was identified in humans, by Gladyshev et al. But, its specific function remains unknown. It has been shown that Sel15 levels specially respond to selenium addition. Moreover, mouse studies suggest that this protein might have redox function and it also may be implicated in the process of posttranslational protein folding, specifically in the quality control. It forms a 1:1 complex with the UDP-glucose: glycoprotein glucosyltransferase (UGGT), an enzyme that is responsible for quality control in the endoplasmic reticulum by oxidative folding and structural maturation of N-glycosylated proteins. It is supposed that Sep15 serves as a disulfide isomerase of glycoproteins targeted by UGGT. Due to redox activity it may also function as an antioxidant. It is highly expressed in prostate, liver, brain, kidneys and testis. The protein expression was shown to be increased in colon cancer and decreased in liver and prostate cancer.
After our results analysis we could predict Sel15 selenoprotein in Nanorana parkeri genome. The protein we predicted contains a Sec residue, and two SECIS elements were found in the 3'UTR region of the gene, which concord with the Seblastian output. As it takes part of the common vertebrate selenoproteins and it was previously well annotated in other databases, this protein was clearly predicted using Xenopus tropicalis as our protein source.
Thioredoxin reductase (TR)
The thioredoxin reductases family (TR) is a protein family composed by flavoproteins, which function as homodimers, actively involved in redox regulation of cellular processes due to their capacity to control the redox status of thioredoxins. It is the only enzyme known to catalyze the reduction of thioredoxin (Trx) and, hence, is a central component in the thioredoxin system. This system is present in all living cells, and it also has an evolutionary history tied to metabolism and redox signaling.
In mammals, there are three known TR isoenzymes: cytosolic TR1, mitochondrial TR3 and TGR, being the last once evolved prior to the split of tethrapodes through a duplication on an ancestral TR1 protein, containing a glutaredoxine domain. Moreover, previous studies have exposed, in mammals, different transcript (splicing forms) and/or protein (isoforms) variants in each mammalian TR. All TR1 alternative splicing is upstream of the first coding exon of the major form of TR1. Among the many splicing forms of TR1, one coded for an N-terminal Grx domain (Grx-TR1). In mammals, the major form of TR1 lacked this domain, but this occurred in TGR and in the TR1 alternative isoform mentioned above. Notably, the Grx-TR1 isoform is absent in rodents. Furthermore, sequence-based phylogenetic analyses suggested that mammalian TGR and TR1 evolved by duplication from the ancestral protein that is similar to fish TR1, and this happened prior to the appearance of amphibians. Some time after the duplication, the Grx domain was retained only in alternative T1-isoform, which was lost in rodents. In addition, sequence analysis highlighted also an important change in the predicted active site of Grx domains of mammalian TGR. In Grx-TR1s, fish TR1 and amphibian, reptile and bird TGRs, a conserved CxxC motif is found.
However, as it was previously described in literature, where only TR1 and TR3 (of TR family) are mentioned as selenoproteins found in all vertebrate, we focused our analysis in these two members of the family, initially not taking into account TGR, also named TR2, because it is not mentioned in literature related to frog lineage. But after our analysis, we were able to find this selenoprotein in Nanorana parkeri genome.
After our results analysis we could predict TR1, TR2 and TR3 selenoproteins in Nanorana parkeri genome, which concords with the literature. These proteins we predicted contains a Sec residue. In case of TR1 we found a SECIS element predicted in the 3'UTR region of the gene, concording with the Seblastian output. For TR3, two SECIS elements were predicted in 3'UTR region of the gene. However, in TR2, no SECIS elements were predicted, concording with the negative Seblastian output.
Selenoprotein Pb
The selenoprotein Pb (SelPb) is an ancestral selenoprotein found in all the vertebrates, and it showed a partial conservation of intron structure with its closest homologs. SelPb function is unknown, but it seems to be responsible for some of the extracellular antioxidant defense properties of selenium.
Although this protein was part of the predicted ancestral selenoproteome, literature suggests that it was generated at the root of vertebrates. During vertebrates evolution a number of different events changed the selenproteome, which implied the loss of SelPb protein in some lineages. However, this selenoprotein was conserved in a Cys form in frog. Although this protein was part of the predicted ancestral selenoproteome, literature suggests that it was generated at the root of vertebrates.
After our results analysis we could predict SelPb selenoprotein in Nanorana parkeri genome. The protein we predicted contains a Cys residue, as it was expected in relation with the literature. As it takes part of the common vertebrate selenoproteins and it was previously well annotated in other databases, this protein was clearly predicted using Xenopus tropicalis as our protein source.
Fep 15 and Selenoprotein M
The Fep15 protein is a relatively recent discovered selenoprotein, described in 2006, which is found to be distantly related to members of the 15 kDa selenoprotein family (Sep15 family). They reported that it could not be found in mammals, and it was only represented in fishes in a Sec-containing selenoprotein form. Nevertheless, a short while ago, it has been found in other species such as cartilaginous fish, elephant, shark, and also found as a Cys homolog in frog. These truths imply that Fep15 formed part of the ancestral vertebrate selenoproteome and it was lost before the reptile's split. In addition, it seems that this selenoprotein was generated during the whole genome duplication occurred at the root of vertebrates, and it specially evolved by duplication of SelM. This last selenoprotein, SelM, which is a selenoprotein found in all vertebrates, shares 31% sequence identity with Sel15 in animals, followed by mutations that resulted in the loss of Cys in the region upstream of the Sec.
As it has been said before, Fep15 is distantly related to Sel15, but various lines of evidence suggest that its function is distinct from those of Sel15 and SelM, two other members of the Sep15 family. Although these parallelism, it is not clear how these differences and similarities within the Sep15 family relate to the physiological functions of these proteins.
In recent studies, neuroprotective functions of SelM and its role in cytosolic calcium regulation have been analyzed suggesting that this selenoprotein may have an important role in protecting against oxidative damage in the brain and may potentially function in calcium regulation.
After our results analysis we could predict Fep15 selenoprotein in Nanorana parkeri genome. The protein we predicted contains a Cys residue, which concord with the explanation above. No SECIS elements were found in the 3'UTR region of the gene, which concord with the Seblastian negative output. Referring to SelM, it was predicted in Nanorana parkeri genome. The protein we predicted contains a Sec residue, and also four SECIS elements were found in the 3'UTR region of the gene.

MACHINERY
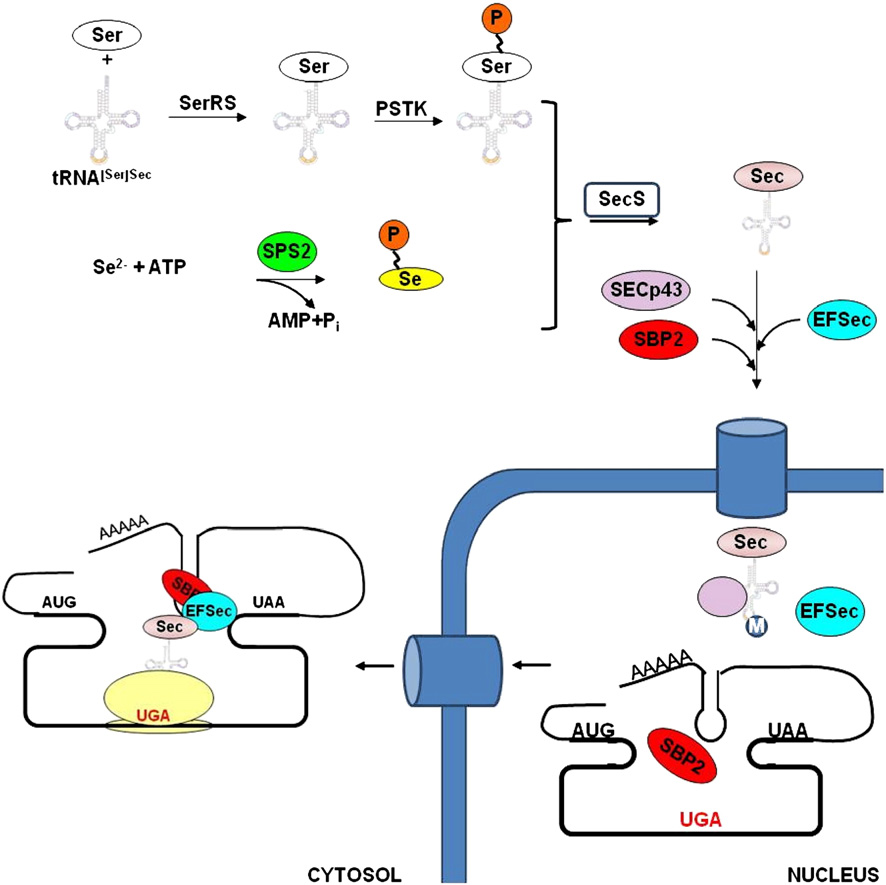
Figure.4.Mechanisms of eukaryotic Sec biosynthesis and incorporation. Sec tRNA[Ser]Sec is initially charged with Ser, which is further phosphorylated by PSTK. SPS2 facilitates the synthesis of selenophosphate, the selenium donor compound. SecS then catalyzes Sec formation. SECp43 may be involved in the methylation of Sec tRNA[Ser]Sec at the A34 position. Protein factors, including SBP2 and EFSec, bind the SECIS element, located in the 3'-UTRs of selenoprotein mRNAs. After translocation to the cytosol, protein factors support interaction with the ribosome and Sec incorporation.
Eukaryotic elongation factor (eEFsec)
Including a Sec amino-acid in a protein requires a selenocysteyl-tRNA[Ser]Sec specific elongation factor in eukaryotes (eEFSec) and in prokaryotes (SELB or Efsec) that suppresses UGA codons that are upstream of Sec insertion sequence (SECIS) elements bound by SECIS-binding protein 2 (SBP2). The EFsec (prokaryotes) is a GTP-dependent RNA binding protein that contains an N-terminal elongation factor domain specific to selenocysteyl-tRNA[Ser]Sec, and a C-terminal selenocysteine insertion (SECIS) binding domain. This protein brings the selenocysteyl-tRNA[Ser]Sec to the ribosomal A site and allows the Sec amino-acid incorporation into the protein, then it dissociates from the ribosome and the SECIS element, and finally reassembles with selenocysteyl-tRNA[Ser]Sec again. In silico analysis of eukaryotic genomes identificated the murine Sec elongation factor, eEFSec which binds to selenocysteyl-tRNA[Ser]Sec; however, it does not associate directly with SECIS elements, but with SECIS binding protein 2 (SBP2) in the presence of selenocysteyl-tRNA[Ser]Sec, thereby forming the Sec decoding apparatus.
After our results analysis we could predict eEFsec machinery protein in Nanorana parkeri genome. The protein we predicted contains a Cys residue, and two SECIS elements were found in the 3'UTR region of the gene.
Phosphoseryl-tRNA kinase (PTSK)
In 1989 selenocysteine (Sec) was identified as the 21st amino acid, and unlike other amino acids, it is synthesized on its transfer RNA, tRNA[Ser]Sec which is initially aminoacylated with serine, then phosphorylated to form phosphoseryl-tRNA[Ser]Sec, and converted to selenocysteyl-tRNA[Ser]Sec. In 1970, a minor species of seryl-tRNA was discovered in rooster liver and was shown to be converted to phosphoseryl-tRNA by a kinase. The seryl-tRNA was subsequently demonstrated to associate with UGA codons and decode UGA in vitro, and was later identified as selenocysteyl-tRNA[Ser]Sec. For years, identification of the phosphoseryl kinase remained unknown; however, recent in silico analyses of the archeal and eukaryotic genomes for novel kinase-like genes that are present within genomes containing the Sec incorporation genes revealed a candidate, phosphoseryl-tRNA[Ser]Sec kinase gene. Phosphoseryl-tRNA[Ser]Sec kinase was subsequently cloned and characterized as a protein that phosphorylates the seryl moiety on seryl-tRNA[Ser]Sec in the presence of ATP and Mg2+. Moreover, the function and homology of this protein is conserved across archaea and eukaryotes that sinthetise selenoproteins, fact that suggests that it plays an important role in selenoprotein biosynthesis and/or regulation.
After our results analysis we could predict PSTK machinery protein in Nanorana parkeri genome. The protein we predicted contains a Cys residue, but no SECIS elements were found in the 3'UTR region of the gene, which concord with the negative Seblastian output.
SECIS binding protein 2 (SBP2)
SBP2 promotes Sec incorporation by associating with SECIS elements and recruiting the eEFSec-selenocysteyl-tRNA[Ser]Sec complex to the ribosome. SBP2 binds to the large ribosomal subunit and the SECIS. The C-terminal half of SBP2 is sufficient to promote Sec incorporation in vitro. SBP2 also contains an RNA binding domain, which works as a suppressor of translational termination. Some reports have identified mutations affecting SBP2-SECIS binding and Sec decoding capacity, and missense mutations in the RNA binding domain of SBP2 identified in patients with abnormal thyroid hormone metabolism confer diminished SECIS binding affinity. Other studies found that the redox state of SBP2 regulates its subcellular localization and Sec decoding function. Oxidative stress induces nuclear sequestration of SBP2, and possibly its associated mRNAs, and results in down-regulation of selenoprotein synthesis.
After our results analysis we could predict SBP2 machinery protein in Nanorana parkeri genome. The protein we predicted contains an initially unexpected Sec residue, and three SECIS elements were found in the 3'UTR region of the gene. The negative Seblastian output might suggest a false positive SECIS prediction.
tRNA Sec 1 associated protein 1 (SECp43)
SECp43 was identified in a degenerate PCR screen for RNA binding proteins, and shown to bind to selenocysteyl-tRNA[Ser]Sec. RNA interference studies targeting SECp43 demonstrated that the protein is required for the Sec-tRNA[Ser]Sec maturation, meaning that it is essencial for methylation of the 2'-hydroxylribosyl moiety in the tremble position of the selenocysteyl-tRNA[Ser]Sec and enhances selenoprotein expression. The result is the formation of methylcarboxylmethyl-5'-uridine-2'-O-hydroxymethylribose. This process is highly sensitive to the primary, seconary and tertiary structure of the tRNA as well as to overall Se status. Mcm⁵U also supports the synthesis of "houskeeping" selenoprotein, as GPx4, TR1 and TR3, whereas the methylated tRNA is needed for expression of "stress-related" selenoproteins, like GPx1, GPx3 and MsrB1. This change in selenoprotein expression pattern is commonly observed during Se defiency, but the precise molecular mechanism reamins unknown. Moreover, a subcellular localization analysis of SECp43 suggest that it may regulate shuttling of the SecS-selenocysteyl-tRNA[Ser]Sec complex between the nucleus and cytoplasm.
After our results analysis we could predict SECp43 machinery protein in Nanorana parkeri genome. The protein we predicted was located in two different scaffolds. The first one presented a Cys residue, without any SECIS neither Seblastian predictions. The second one presented an initially unexpected Sec residue, and one significant SECIS element was found in the 3'UTR region of the gene. The negative Seblastian output might suggest a false positive SECIS prediction.
Selenocysteine synthase (SecS)
Selenocysteine synthase (SecS or SepSecS) was initially established in E.coli in the early 1990s. SecS, a pyridoxal phosphate-dependent enzyme, esterifies the serine moiety of seryl-tRNA[Ser]Sec, forming an aminoacrylyl intermedite which is then exchanged with selenol. In addition, eukaryotic SecS is also a member of the pyridoxal phosphate-dependent transferase superfamily and associates with the supramolecular complex mediating Sec incorporation into selenoproteins.
Recent research demonstrated that mutations in SecS gene are associated with the development of autosomal-recessive progressive cerebellocerebral atrophy and that the observed phenotypes could be partially reproduced in the corresponding KO animal models.
After our results analysis we could predict SecS machinery protein in Nanorana parkeri genome. The protein we predicted contains a Cys residue, but no SECIS elements were found in the 3'UTR region of the gene, which concord with the negative Seblastian output.
Selenophosphate synthetase family (SPS)
The function of selenophosphate synthetase (SPS) is to generate the Se donor compound (selenophosphate) necessary for Sec biosynthesis.
Selenophosphate synthetase (SPS, also called SelD or selenide water dikinase) is unique among the components of the Sec biosynthesis machinery in that it is often a selenoprotein itself. This enzym catalyzes the synthesis of selenophosphate from selenide, ATP, and water, producing the following products, AMP and inorganic phosphate. Selenophosphate is the selenium donor for the synthesis of Sec, which, in contrast to other amino acids, takes place on its own tRNA, tRNAsec. The selenophosphate synthetase enzym (SPS or SelD) is required for Sec synthesis, and it is conserved in all prokaryotic and eukaryotic genomes encoding selenoproteins. This enzyme is described itself as a selenoprotein in most of the species.
As we have already said, this enzyme is itself considered selenoprotein in many species, although functionally equivalent homologs that replace the Sec site with cysteine (Cys) are common. In recent studies, by analyzing diverse organismsIn vertebrates and insects, two paralogous SPS genes have been reported: SPS2, which is a selenoprotein, and SPS1, which is not, and it carries a threonine (Thr) in vertebrates and an arginine (Arg) in insects in place of Sec. In contrast to Cys conversion, Thr or Arg conversion in SPS1 seems to result in the abolishment of the selenophosphate synthase function. Furthermore, it is seen that from an ancestral metazoan selenoprotein SPS2 gene that most likley already carried the SPS1 function, it has been found that SPS1 genes were originated through a number of unrelated gene duplications. Hence, parallel duplications and consequent convergent subfuncionalization in SPS genes have appeared in the segregation to distinct loci of functions originally carried out by a single gene. Thus, it has been found that the presence of Sec/Cys SPS genes, together with a few other gene markers, recapitulates the selenium utilization traits (Sec and SeU) in prokaryotic genomes. Within eukaryotes, specifically within metazoans, we detected a number of SPS homologs with amino acids other than Sec or Cys at the homologous UGA position. We found that Cys- or Sec-containing SPS genes (SPS2) are found in all genomes encoding selenoprot eins, whereas genomes that contain only SPS genes carrying ami no acids other than Sec or Cys at the homologous UGA position (SPS1) do not encode selenoproteins.
After our results analysis we could predict SPS1 and SPS2 machinery protein in Nanorana parkeri genome. Both predicted protein contains a Cys residue. The first one, SPS1, presented a SECIS element in the 3'UTR region of this gene, concording to previous literature. The second one, SPS2, did not present any SECIS elements, which agree with the negative Seblastian output.