Introduction
Selenium and selenocysteine
Selenium (Se) is a broadly essential micronutrient involved in a great number of biological and physiological processes such as immunity or thyroid function. Due to this, selenium deficiency is associated with many physiopathological conditions (i.e. inflammation, cancer, heart disease), but excess can also be damaging to the organism. (1) What is remarkable for us is this chemical element’s implication in the formation of what is considered as the 21st amino acid: selenocysteine (Sec, U). This residue’s structure is almost identical to cysteine’s (Cys) one, with one difference setting them apart: Cys contains sulfur (S) whereas Sec contains Se, and this structural variation is responsible for an improved catalytic activity. Thus, we could say selenoproteins are the more-efficient and oxidation-resistant homologues for proteins containing Cys residues in their catalytic site. (2)

Selenoproteins
Selenoproteins have been identified in the three domains of life (eukarya, archaea and eubacteria), and if we focus on the human selenoproteome, 25 selenoprotein genes have been found. Sec is a residue encoded by the UGA codon, which usually acts as a STOP translation signal, so in order to incorporate Sec in that codon we need two things: specific machinery and SECIS (Cis-acting Sec insertion sequence) elements. When these two factors are present, the organism is able to introduce Sec residues in the amino acid sequence. (3)
Since selenoproteins have a Se in their catalytic redox active site, they are enzymes with oxidoreductase action and play an important role in cell signaling and redox homeostasis. Some examples of selenoproteins are thioredoxin reductases, iodothyronine deiodinases or glutathione peroxidases.(2)
Figure 1. Different amino acids encoded by the diverse codons. The UGA codon can encode both for a STOP codon or a Sec residue. Source: Labunskyy VM, Hatfield DL, Gladyshev VN. Selenoproteins: molecular pathways and physiological roles. Physiol Rev. 2014;94(3):739–777.
BIOSYNTHESIS MACHINERY AND PROCESS
As we have already introduced, both specific machinery and SECIS elements are required for adequate insertion of Sec residues in amino acid sequences. (2) The SECIS element is a cis-acting stem-loop RNA structure of the genome that can be found in the 3’ UTR region (hence, after the encoded protein). Its biological function is indicating to the cell that the sequence corresponds to a selenoprotein and not a regular protein. In a nutshell, it enables the machinery to understand that the UGA codon does not code for a STOP codon, but for a Sec residue. Sec is introduced to the amino acid sequence of selenoproteins by a complex machinery composed by trans-acting factors, a Sec-tRNA[Ser]Sec and a SECIS element. When a ribosome encounters the UGA codon, and only if a SECIS element is found, the Sec machinery will interact with the canonical translation proteins to avoid premature termination. When this occurs, Sec-tRNA[Ser]Sec (a tRNA with an anticodon complementary to UGA) translates UGA to Sec following the steps explained in more detail below. (4) For Sec synthesis, in just a few words, what occurs is that tRNA will incorporate a Ser residue that will posteriorly be phosphorylated and replaced by a Sec residue. However, in much more detail:
- 1. Seryl-tRNA synthesis
- Seryl-tRNA synthetase incorporates a Ser residue in the tRNA[Ser]Sec, which has peculiar characteristics in comparison to other tRNAs (longer sequence, less modified bases, a long arm and a long acceptor stem).
- 2. Phosphorylation of seryl-tRNA[Ser]Sec
- The phosphoseryl-tRNA kinase or PSTK is synthesized and selectively phosphorylates the seryl-tRNA[Ser]Sec to O-phosphoseryl-tRNA[Ser]Sec. This is an eukaryotic-exclusive intermediate step.
- 3. Conversion of Ser-tRNA[Ser]Sec to selenocysteyl-tRNA[Ser]Sec
- SecS (Sec synthetase) catalyzes the conversion of Ser-tRNA[Ser]Sec to selenocysteyl-tRNA[Ser]Sec, which has a selenophosphate (meaning this enzyme incorporates Se's active form). As a result, the final Sec-tRNA[Ser]Sec is generated.
For this recoding, two trans-acting factors are required in eukaryotes: SECIS binding protein 2 (SBP2) and a Sec-specific translation elongation factor (eEFSec). SBP2 associates with ribosomes and contains an L7Ae RBD with high affinity and specificity for SECIS elements and eEFSec, which recruits Sec-tRNA[Ser]Sec and easens Sec’s incorporation to the polypeptide. SBP2 also contains a Sec incorporation domain (SID), which enhances RBD’s activity and interaction. Binding of SBP2 to SECIS elements on selenoprotein mRNA and the cascade events that follow are key for protecting selenoprotein mRNA from premature degradation. Thus, SBP2 is essential for the upstream stabilization of selenoprotein mRNA and selenoprotein translation.
Eukaryotic SECIS elements are formed by two helixes separated by an internal loop, a core structure formed by a GA Quartet structure, and an apical loop or bulge. The core SECIS structure is the part that interacts with SBP2.(2)
Other SECIS-binding proteins, with an identified regulatory function, have also been identified and characterized. Examples of these latter types of proteins are the eukaryotic initiation factor 4a3 (eIF4a3) and nucleolin. The ribosomal protein L30 is not regulatory, but part of the basal Sec insertion machinery. (2)

Figure 2. Schematic representation of Sec residues’ incorporation to a growing polypeptide chain. Source: Labunskyy VM, Hatfield DL, Gladyshev VN. Selenoproteins: molecular pathways and physiological roles. Physiol Rev. 2014;94(3):739–777.
Selenoproteome evolution
Even though selenoproteins can be found in the three domains of life (archaea, eukarya and eubacteria), not every species in each domain contains selenoproteins: some have conserved them, others have lost them and some others have Cys-homologues. The role performed is also variable between life domain: archaea’s selenoproteins are involved in hydrogenotrophic methanogenesis, eubacterias’ in redox homeostasis and compound detoxification and, finally, eukaryota selenoproteins are involved in redox regulation and energy metabolism.As with other aspects, phylogenetic proximity rules how similar or different two selenoproteomes will be, in a way that close species have less differences between them. Aquatic organisms (plants or animals) usually have bigger selenoproteomes in comparison to terrestrial organisms. Mammalians, on the other hand, have reduced their use of selenoproteins, either because they have lost them or substituted the Sec residue for a Cys one, and now have around 25 selenoproteins (21 of them present in all vertebrates).
After many years of research and analyses performed, as it can be inferred, we have enough data to draw some conclusions. Nonetheless, there are still many mysteries surrounding the evolutionary trend of the selenoproteome and further bioinformatic analysis needs to be done.
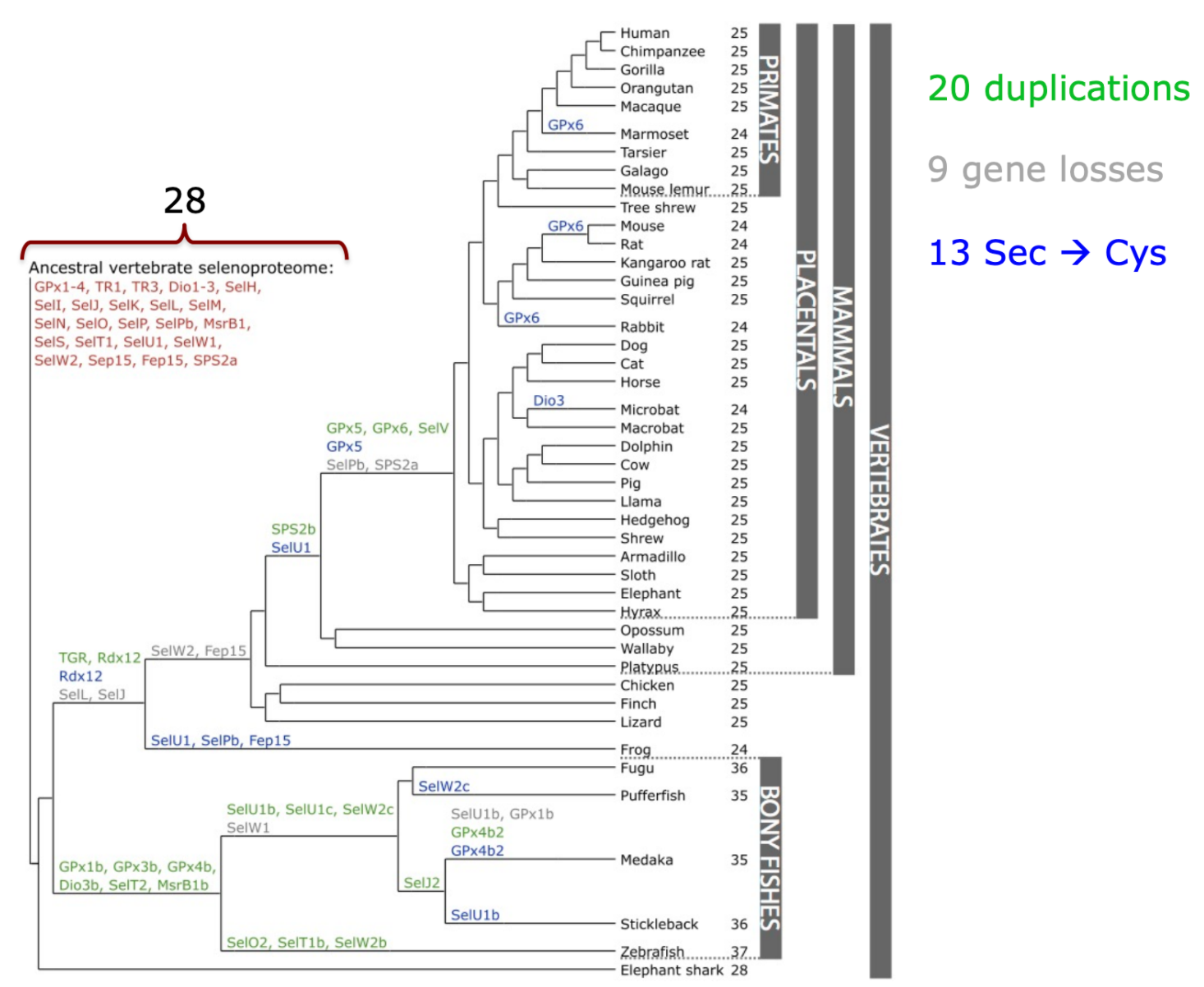
Figure 3. Phylogenetic tree of selenoproteins in mammals / other vertebrates. Source: Mariotti M, Ridge P, Zhang Y, Lobanov A, Pringle T, Guigo R et al. Composition and Evolution of the Vertebrate and Mammalian Selenoproteomes. PLoS ONE. 2012;7(3):e33066
Selenoprotein families
GLUTATION PEROXIDASES (GPX)
The GPx family is a family of enzymes present in the three domains of life (archaea, eubacteria and eukarya) that plays a role in hydrogen peroxide signaling, redox homeostasis and detoxification of hydroperoxides. Mammalians have 8 paralogues present in our genome (GPx1 to GPx8), and they can be categorized in two separate subfamilies according to their structure: GPx1 to GPx4 and GPx6 contain a Sec residue in their active site, whereas GPx5, GPx7 and GPx8 contain a Cys residue, thus making them Cys homologues instead of selenoproteins. Some mammals have GPx6 in the form of a Cys homologue too. (2)
In more detail...
- GPx1: most abundant selenoprotein in mammals, expressed in all cell types (but specially in the liver and kidney). Its role is in protecting cells from oxidative damage, so deficient or augmented activity can affect hydrogen peroxide-mediated responses. This homotetrameric enzyme catalyzes glutathione (GSH)-dependent reduction of hydrogen peroxide to water, decreasing toxic hydrogen peroxide levels.
- GPx2: enzyme with specificity for hydrogen peroxide that is found in the epithelium and GI tract. Its expression is controlled by Nrf2, a transcription factor involved in the antioxidant response.
- GPx3: enzyme with specificity for hydrogen peroxide that constitutes the major GPx form found in plasma.
- GPx4: very remarkable GPx due to a number of reasons. First of all, it is monomeric and present in a great number of cell types and tissues, acting as a housekeeping selenoprotein. It is not as affected by Se availability as its other family proteins. Its gene has three isoforms, encoding for nuclear (nGPx4), mitochondrial (mGPx4) and cytosolic (GPx4) proteins. nGPx4 and mGPx4 are expressed in testes, where it has been suggested to intervene in male gametogenesis. On the other hand, cGPx4 is expressed during embryonic development and in adult organs. This enzyme reduces complex phospholipid hydroperoxides associated with membranes, since its active site lacks a loop structure and this allows access for these kind of molecules.
- GPx5: not a Selenoprotein, is a Cys homologue.
- GPx6: enzyme only expressed during embryonic development, in the olfactory epithelium. As it has already been stated, some mammals have this enzyme in their Cys-homologue version.
IODOTHYRONINE DEIODINASES (DIO)
Deiodinases (DIO) are a family of integral membrane selenoenzymes with a thioredoxin fold,composed of three paralogous proteins in mammals: DIO1, DIO2 and DIO3, which can also be found in other vertebrates and simple eukaryotes. Their role is in thyroid hormone activity regulation, since they are in charge of reductive diodination. We mostly produce T4, the inactive form of the thyroid hormones, and this has to be converted to rT3 (thanks to DIO3) or to T3 via deiodination catalyzed by this family’s DIO1 and DIO2. (2)
- DIO1: mostly found on the plasma membrane. It usually acts in peripheral tissues, like liver and kidney, converting T4 to T3. It is the main regulator of thyroid hormone blood levels.
- DIO2: mostly found on the plasma membrane. Essential for maintaining T3 levels in the brain during the critical period of development. Involved in intracellular roles, specially in skeletal muscle cells during development and muscle regeneration process and brown adipose tissue during adaptive thermogenesis.
- DIO3: found in the endoplasmic reticulum (ER). Critical during embryological development.
THIOREDOXIN REDUCTASES (TXNRDS)
This family is composed of 3 TXNRD flavoenzymes in mammals: TXNRD1, TXNRD2 and TXNRD3, all of which contain a Sec residue in the penultimate position of the COOH-terminal. They are responsible for catalyzing the NADPH-dependent reduction of thioredoxin, as well as have a role in redox homeostasis too. Their differences reside in cell localization and function. (2)
- TXNRD1: mainly located in the cytosol and nucleus. It has 6 isoforms in mammals, all of them involved in different processes including actin and tubulin polymerization, enhancement of estrogen receptor’s transcriptional activity or apoptosis. As a consequence, some isoforms (like isoform 1) will have thioredoxin/glutathione reductase activities and others won’t.
- TXNRD2: located in the mitochondria. It is involved in maintaining thioredoxin in a reduced state, responses against oxidative stress and redox-regulated cell signalling.
- TXNRD3: this enzyme also has both glutaredoxin and glutathione reductase activities. It acts in sperm maturation, promoting the formation of sperm’s structural components. It has been shown to catalyze disulfide bond isomerization and promote disulfide bond formation between sperm proteins and GPx4. (2)
METHIONINE-R-SULFOXIDE REDUCTASE 1
MsrB1, is a zinc-containing selenoprotein of mammals which was previously identified as selenoprotein R. When it was discovered it was a methionine-R-sulfoxide reductase like MsrA, the scientific community changed its name to MsrB1. However, it is worth noting that their structures are not similar. In mammals, this enzyme is the main MSRB protein and can be found in the cytosol and the nucleus, with important activity in the liver and the kidneys. Its two homologues, MsrB2 (located in mitochondrias) and MsrB3 (located ER), are, in comparison to MsrB1, less expressed but equally efficient catalytically. Regarding its function, MsrB1 is involved in cellular oxidation repair via stereospecific reduction of methionine-R-sulfoxide to methionine, which is an oxidative reaction that acts as a post-translational modification able to regulate actin assembly and promote filament repolymerization. (2)
METHIONINE SULFOXIDE REDUCTASES A (MSRA)
MsrA is a highly conserved selenoprotein that, under thioredoxin presence, catalyzes the enzymatic reduction of both free methionine-S-sulfoxide and its protein-based form. Thus, it is a stereospecific reductase. (2)
15-KDA SELENOPROTEIN (SEL15)
This is a thioredoxin-like fold protein located in the ER that contains a NH2-terminal signal peptide with a distinct Cys-rich domain, mandatory in order to interact with UGGT (an ER-resident chaperon involved in the quality control process given to proteins in the organelle). It is present in the liver, kidneys, testes and prostate, and its expression is induced when misfolded proteins accumulate in the ER. Regulation of this selenoprotein is also determined by dietary Se. (2)
SELENOPROTEIN I
This selenoprotein is exclusive of vertebrates, and it is a transmembrane protein composed by 7 transmembrane domains and three conserved Asp residues, essential for its catalytic function related to phospholipids de novo synthesis.
SELENOPROTEIN M
This thioredoxin-like selenoprotein is a distant homologue of Sel15, so it is found in the ER but only in the brain. Its role is in neuroprotection and prevention of hydrogen peroxide-induced oxidative damage, but it has also been linked to a protective inhibition of beta-amyloid protein aggregation.(2)
SELENOPROTEIN N
SelN was the first selenoprotein identified due to bioinformatic tools. It is a transmembrane glycoprotein located in the ER which is highly expressed during embryonic development, but less in adult tissues. It has been linked to skeletal muscle regeneration and maintenance of satellite cells, as well as a role as a ryanodine receptor cofactor, mediating the release of calcium from the sarcoplasmic reticulum during muscle contraction. Mutations in this gene have been associated with musculoskeletal disorders of early onset.(2)
SELENOPROTEIN O
This selenoprotein has not been broadly studied given that we lack structural or biochemical information. We do not know its function but we do know it contains one Sec residue in the antepenultimate position in the COOH-terminal end, and that it has a kinase domain and a mitochondrial targeting peptide. Cys-homologues have been found in other species. (2)
SELENOPROTEINS P
It constitutes almost 50% of the total plasmatic Se, since it contains various Sec residues in its structure. It is mainly synthesized in the liver, but its mRNA can be detected in all tissues. It has been suggested to have a role in transportation of Se to peripheral tissues, specially brain and testis.(2)
SELENOPROTEINS K AND S
These two selenoproteins are considered a family because their topology is very similar, even if their sequences are different: both of them are composed of a transmembrane domain in the NH2-terminal region, a Gly-rich segment and Sec residues in the COOH-terminal. They both also reside in the ER membrane. It is one of the most widespread eukaryotic family, being present in several species of different kingdoms. (2)
Their differences reside in the fact that SelS has an additional coiled-coil domain the cytosolic portion of the protein. They have been suggested to be implicated in the ER-associated degradation (ERAD) of misfolded proteins, inflammation and immune response. Regarding this first function, it has been discovered that their genes have functional ER stress response elements and their expression is upregulated when misfolded proteins are accumulated in the ER.(2)
SELENOPROTEINS H, T, V AND W
These are thiol-based oxidoreductases, which are selenoproteins from the Rdx family. They have a thioredoxin-like fold, a conserved C-x-x-U motif and a conserved stretch of amino acids in the COOH-terminal portion of the protein with the tGXFEI(V) consensus sequence. (2)
SELH
Firstly discovered in fruit flies, this selenoprotein has a conserved nuclear targeting RKRK motif in the N-terminal sequence. Its subcellular localization pattern is very distinctive from other selenoproteins, since it is found in the nucleoli. Furthermore, it is very sensitive to Se dietary intake. With regards to its function, it has a glutathione peroxidase activity and regulates the expression of detoxification enzymes. (2)
SELT
This selenoprotein is localized in the ER and Golgi of all tissues, both during embryonic development and adulthood. Its role is posed to be in the regulation of calcium homeostasis, subcellular organization and neuroendocrine function, but more recent studies have suggested it acts on pancreatic beta-cell function’s regulation and glucose homeostasis. (2)
SELW
Another extremely abundant selenoprotein in mammals, located in the cytosol of cells (especially of muscles, heart and brain). As its sister protein, SelW is very sensitive to Se dietary intake. It has been suggested to have a role in redox regulation of 14-3-3 proteins, which are ubiquitous regulatory molecules, but also in protection of neurons against oxidative stress during their development. It has two paralogues: SELW1 and SELW2. (2)
SELV
This selenoprotein is expressed in the testes and belongs to the SelWTH family, so it does not differ characteristically from SelW (it appears to have evolved from its duplication). It is only found in placental mammals, although it has been specifically lost in some of them. It is larger than SelW because of an additional NH2-terminal domain. About its physiological role, it has been suggested to be involved in male reproduction. (2)
SELENOPROTEINS U
SelU was discovered in fish first, and later in birds and unicellular eukaryotes. However, superior mammal species have the Cys-homologues. For instance, humans have three subfamilies: SelU1, SelU2 and SelU3, and they belong to the thioredoxin-like superfamily. (2) As a consequence, we can determine it has redox activity and a Prx-like2 structure. (10)
SELENOPROTEIN J
SelJ has a very restricted phylogenetic distribution, not even existing in mammalian genomes. It appears to be restricted to actinopterygian fishes and sea urchin and shows significant similarity to the jellyfish J1-crystallins. Moreover, in contrast to other selenoproteins, it does not appear to have any functional role, only a structural one. (2)
SELENOPROTEIN L
SelL belongs to the thioredoxin superfamily. As well as the SelJ it has a restricted phylogenetic distribution, exclusive of aquatic organisms. Besides, it is not found in mammals. (2)
SELENOPROTEIN E
SelE is localized in the ER and its function is yet unknown. It is not relevant for our research as it is only found in fish and our species is a bird. (2)
Pyrgilauda ruficollis
The rufous-necked snowfinch (Pyrgilauda ruficollis) is a species of bird in the sparrow family.
Taxonomy
| Eukaryota | |
| Animalia | |
| Chordata | |
| Aves | |
| Passeriformes | |
| Passeridae | |
| Pyrgilauda | |
| Pyrgilauda ruficollis |
Identification

These birds are around 15cm and are very distinctive in their zone. Quite brightly-colored for a snowfinch, adults and juveniles are distinguished because of their appearance. Adults have black lores and whitish faces except for their chestnut or reddish-brown rear ear-coverts and sides of neck.
The rest of the plumage is light brown, straked darker on mantle and scapulars. The wings have two white wingbars formed by the tips of the coverts. Juveniles are paler than adults, without a well-defined face pattern.
It can be confused with the Blanford's snowfinch, but Pyrgilauda ruficollis has two black stripes on the face. They also have evolved a higher rate of metabolism than most other birds, a better tolerance of low temperatures, and a greater capacity for moving.
Voice
Pyrgilauda ruficollis has a soft voice and a chattering alarm call. It also emits buzzing noises while flying.

Behaviour
It is locally very common, usually found in the breeding season in close association with mouse-hares or pikas, in whose burrows it breeds and hides. When breeding season is over, it is more likely to be found in small flocks, when it ranges over a wide variety of mountainous terrain in company of other finches like Blanford’s snowfinches.
Flight is weak and low, and rarely over long distances. It feeds on the ground, on a variety of small seeds and insects.

Distribution and habitat
It is naturally found in Tibet and adjacent areas of central and western China, in the alpine temperate grassland and barren stony steppes and plateaus; it winters south to Uttarakhand , Nepal, Sikkim and Bhutan, sometimes to lower altitudes. Therefore, it is found on wide, open steppe meadows, pastures and near human settlements.
They are mainly sedentary, only making irregular altitudinal movements in response to particularly bad weather conditions and never doing long-distance movements.
MATERIALS AND METHODS
The aim of this project was to predict and annotate all the selenoproteome regarding proteins including: selenoproteins per se, selenoprotein synthesis machinery and proteins related to selenium metabolism. In order to do so, we searched in the genome of Pyrgilauda ruficollis for the amino acid sequences of all these proteins in three different reference species:
- a)Taeniopygia guttata: the phylogenetically closest species to Pyrgilauda ruficollis.
- b)Gallus gallus: when proteins were not present in the finch’s genome.
- c)Homo sapiens: when proteins were not present in both previous species.
- 1. Name of the file containing all the queries from our reference species.
- 2. Name of the directory where the genome database is stored.
- 3. Name of the folder where the user wants to save all the results.
- a) Tblastn
- The Tblastn program compares all the query proteins of the reference species with the genome of Pyrgilauda ruficollis. We use a threshold of 0.01 for the e-value parameter, in order to avoid obtaining data from non-informative alignments. The output of this command is stored in a tabulated format (using the -outfmt 6 option) in a file named blast.tsv.
Once the Tblastn file is generated, we have devised a method to run all the different commands on each of the hits found from a query protein in each different scaffold.tblastn -query query.fa -evalue 0.01 -outfmt 6 -db genome.fa -out blast.tsv
- b) Fasta fetch
- This command is used to extract the scaffold of the Pyrgilauda’s genome from the hit we are working on.
fastafetch genome.fa genome.index scaffoldname > scaffold.fa
The output of this command is stored in a new text file called scaffold.fa. - c) Fasta subseq
- Once we have obtained the sequence of our scaffold, the script proceeds to obtain the region of interest where our gene is located. The scaffold contains both exons and introns, so it is necessary to take a longer sequence than the length predicted by Tblastn. This longer sequence is what we will call fastasubseq.
In order to avoid missing part of the coding sequence of our gene, we decided to go 50.000 nucleotides back to define the start of our fastasubseq. Then, starting from this defined position, we decided to take a fastasubseq measuring 150.000 nucleotides.
Note: in those cases where the length of the scaffold was a limiting factor to follow the above-mentioned fastasubseq parameters, we decided to introduce a command to automatically adjust these parameters according to the length of the scaffold.
fastasubseq scaffold.fa startposition length > fastasubseq.fa
The output of this command is stored in a new text file called fastasubseq.fa.
- d) Exonerate and egrep
- The exonerate command is a tool for pairwise sequence comparison. It will give a predicted annotation of the query protein in the previously defined fastasubseq. Concretely, the output of this command contains information of interest such as start and end of the predicted coding sequence, score of the obtained alignment or coordinates of the predicted exons.
exonerate -m p2g --showtargetgff -n1 -q proteintosearch.fa -t fastasubseq.fa > exonerate.out
Note: to obtain each of the query proteins individually in every loop of our script, we have designed an awk command that saves this ID and sequence in the proteintosearch.fa file. Regarding the coordinates of the different exons, we need to extract them from the exonerate output in order to assemble the full predicted coding sequence of our gene. To do so, we execute an egrep command, which will extract all those lines in the exonerate.out file that contain the word “exon”.
egrep -w exon exonerate.out > onlyexons.gff
The final output of these two commands is stored in a GFF file named onlyexons.gff. - e) Fastaseq from GFF
- This command will execute what was previously announced. Fastaseq from gff will take all the coordinates of the exons predicted by exonerate (isolated by egrep) and assemble them as a single nucleotide sequence corresponding to the predicted coding sequence of our query protein in Pyrgilauda ruficollis.
fastaseqfromGFF.pl fastasubseq.fa onlyexons.gff > predictedntseq.nuc
The output of this command is stored in a new file named predictedntseq.nuc. - f) Fasta translate
- The fastatranslate command is basically used to translate the nucleotide sequence generated by the fastaseq from gff command into a protein sequence.
fastatranslate predictedntseq.nuc -F 1 > predictedaaseq.fa
The translated amino acid sequence is stored in a new text-file named predictedaaseq.fa. - g) T-Coffee
- T-coffee is a multiple sequence alignment server that we will use to align and compare the query protein with the prediction of that query in Pyrgilauda’s genome.
t_coffee proteintosearch.fa predictedaaseq.fa > tcoffee.fa
The output of this command consists in the alignment of the two protein sequences and the t-coffee score for this alignment, which is stored in the tcoffee.fa file. - h) Genewise
- Genewise is an alternative software for prediction of a query protein into a nucleotide sequence. We specifically use Genewise in those cases where the exonerate prediction is meaningless, which was programmed by a conditional If/Else operation.
As a remarkable trait, Genewise requires the specification of the strand in which our gene is located. To solve this problem we devised a strategy for classifying genes as “Forward” or “Reverse” based on whether the starting genomic coordinate was higher (Reverse) or lower (Forward) than the final genomic coordinate in the Tblastn file.
In this sense, we execute the following command on Forward genes:
genewise -pep -pretty -cdna -gff proteintosearch.fa fastasubseq.fa > genewise.fa
And this other command on Reverse genes:
genewise -pep -pretty -cdna -gff -trev proteintosearch.fa fastasubseq.fa > genewise.fa
As it was mentioned, Genewise gives an alternative protein prediction of the query protein. Then, we perform a T-coffee comparison between the query protein and the Genewise predicted one. - i) SECIS element prediction and Seblastian
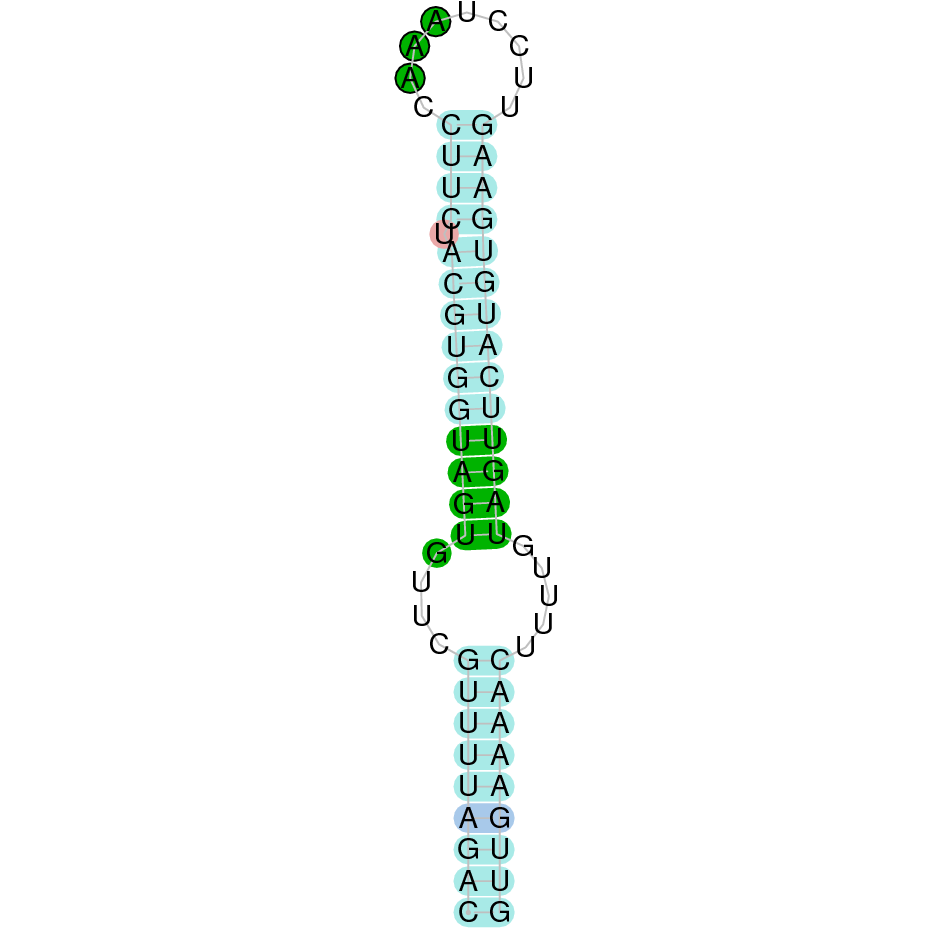
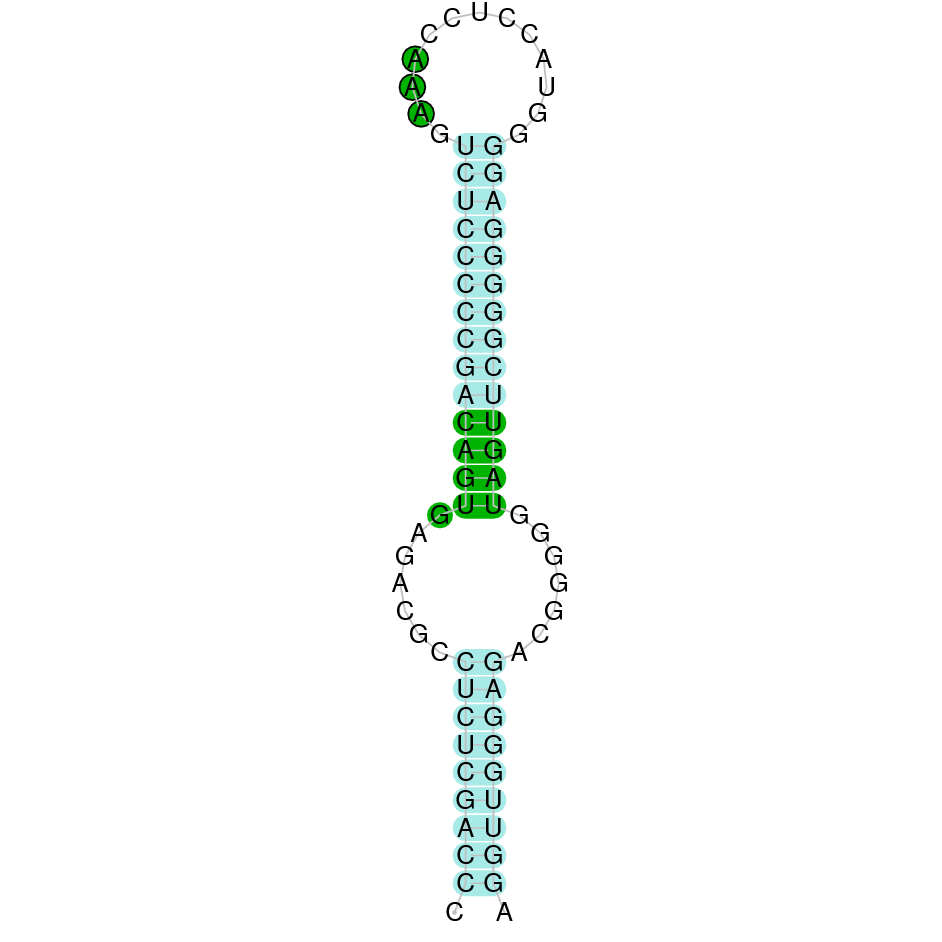
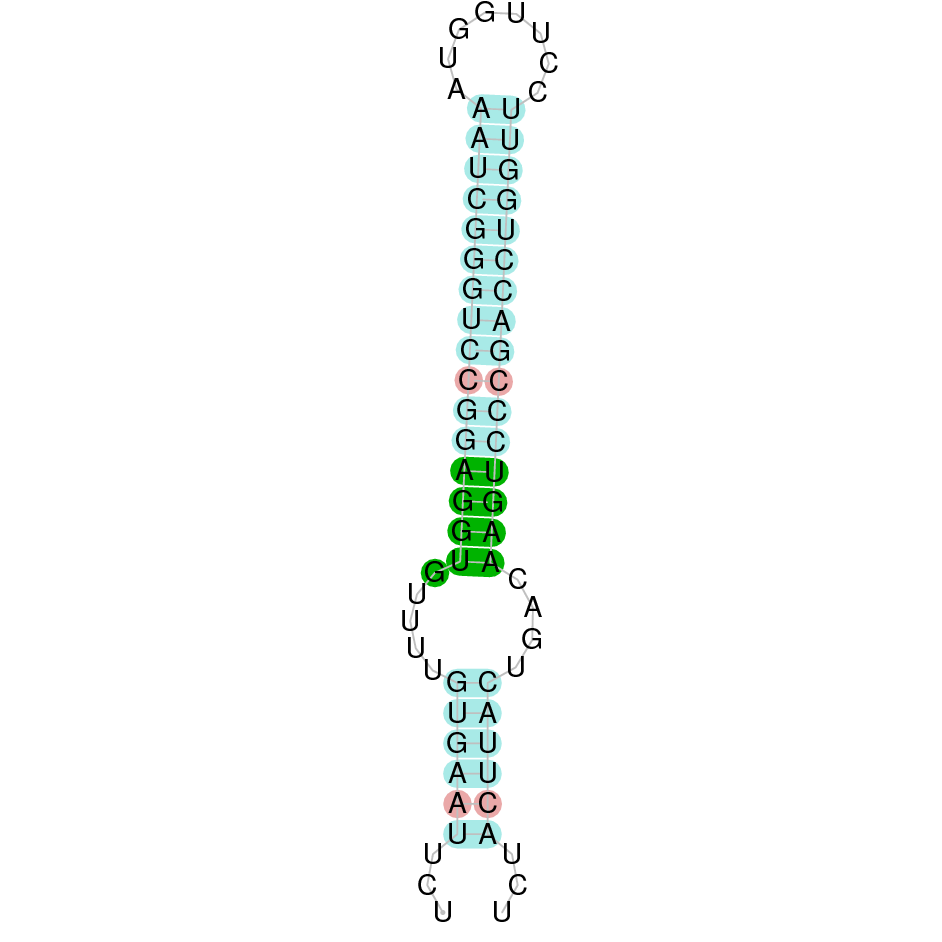
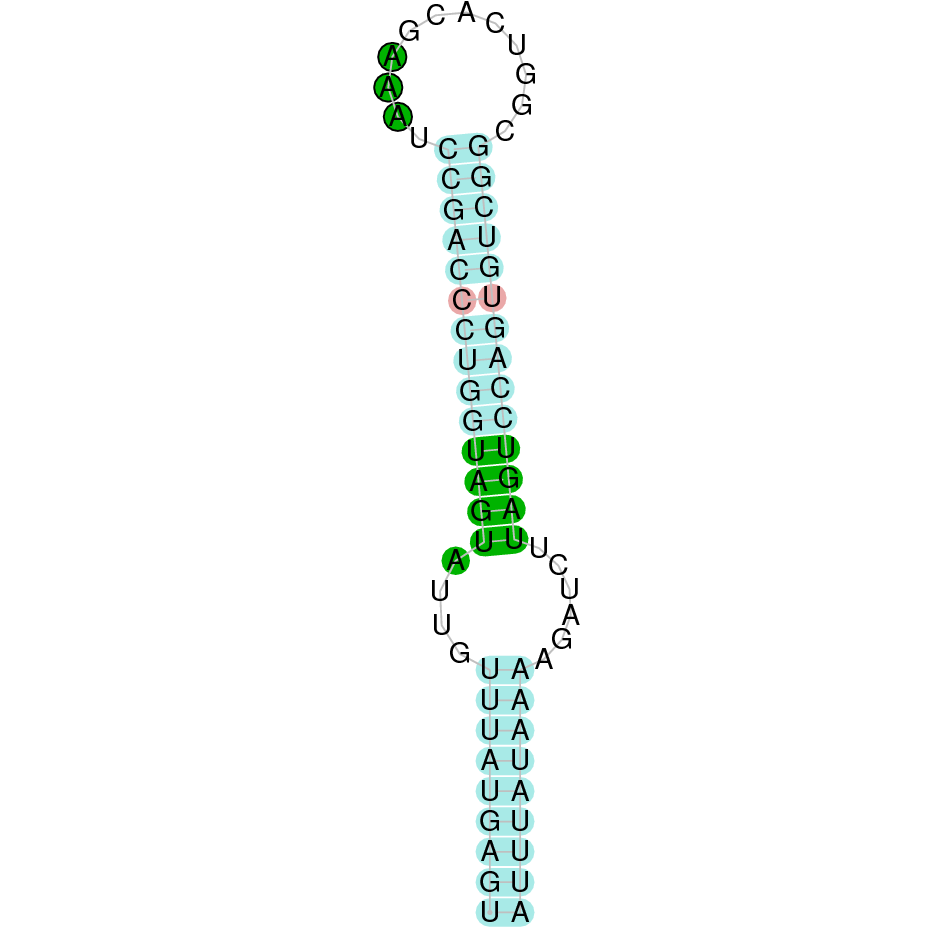
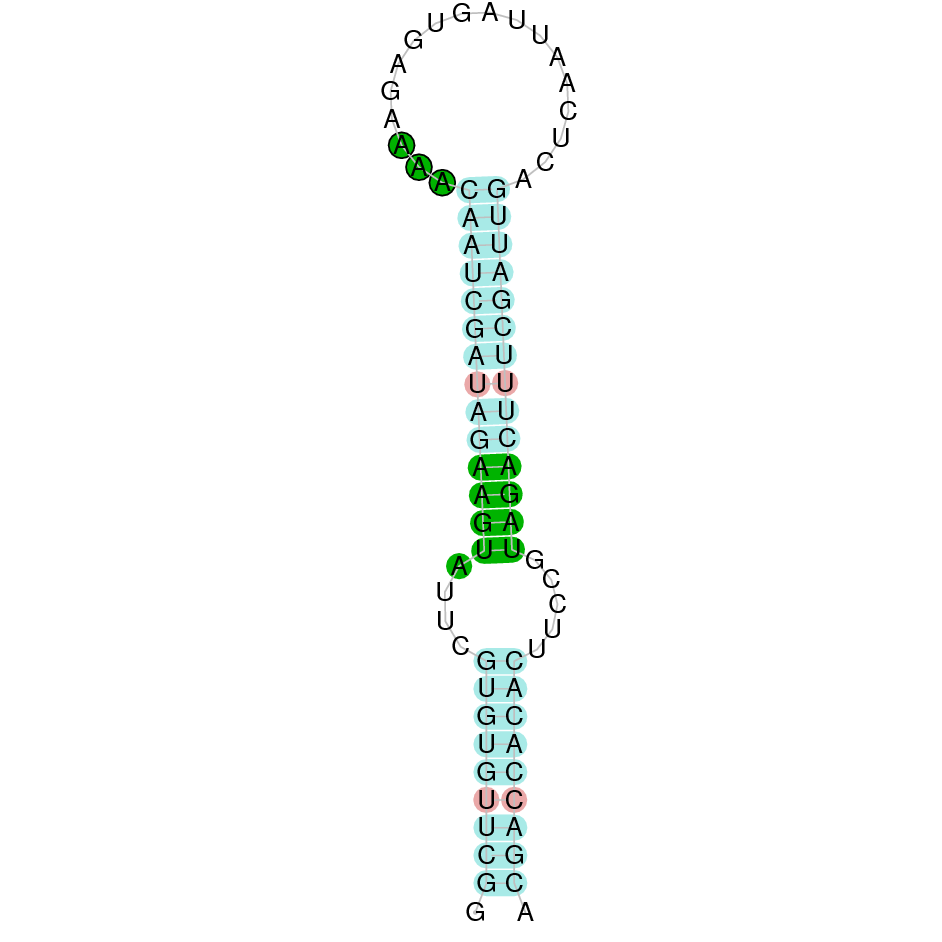
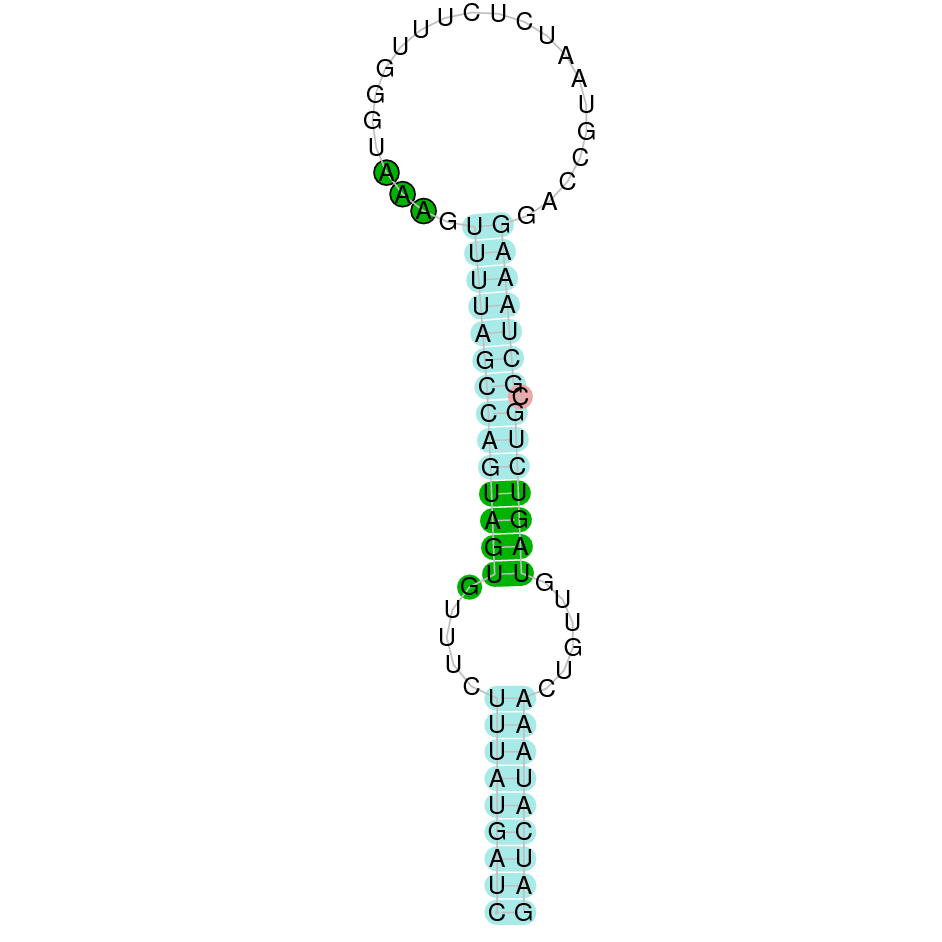
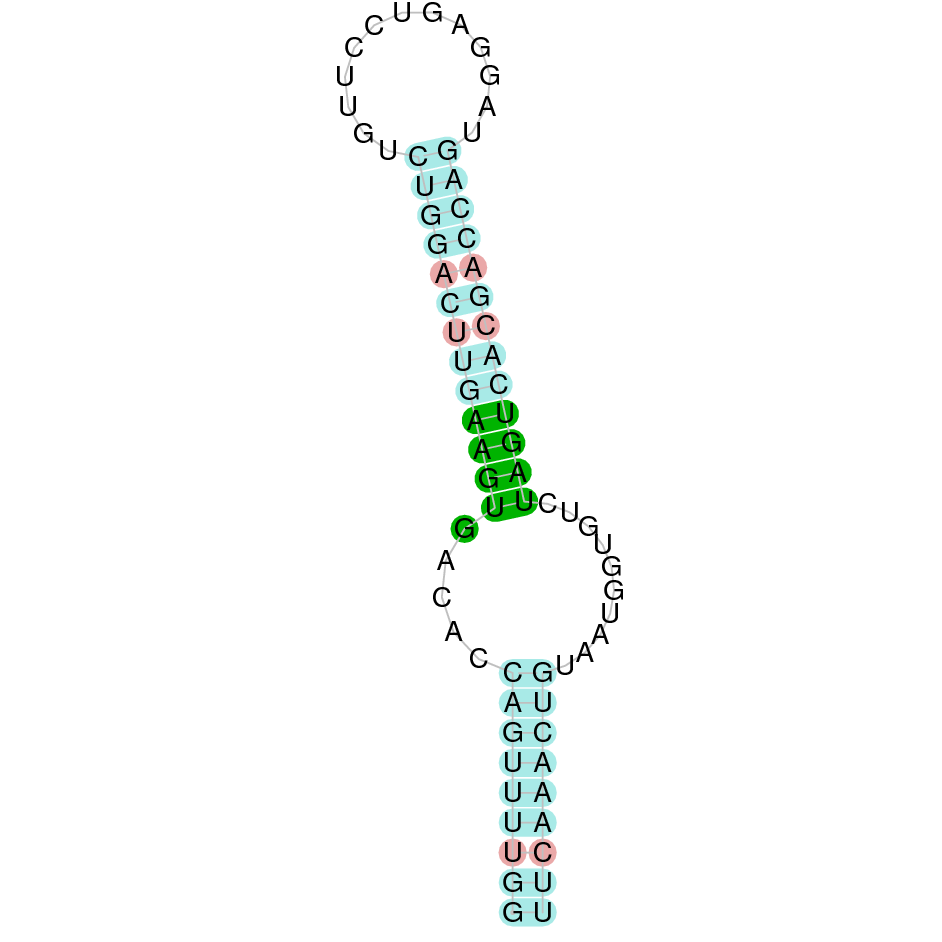
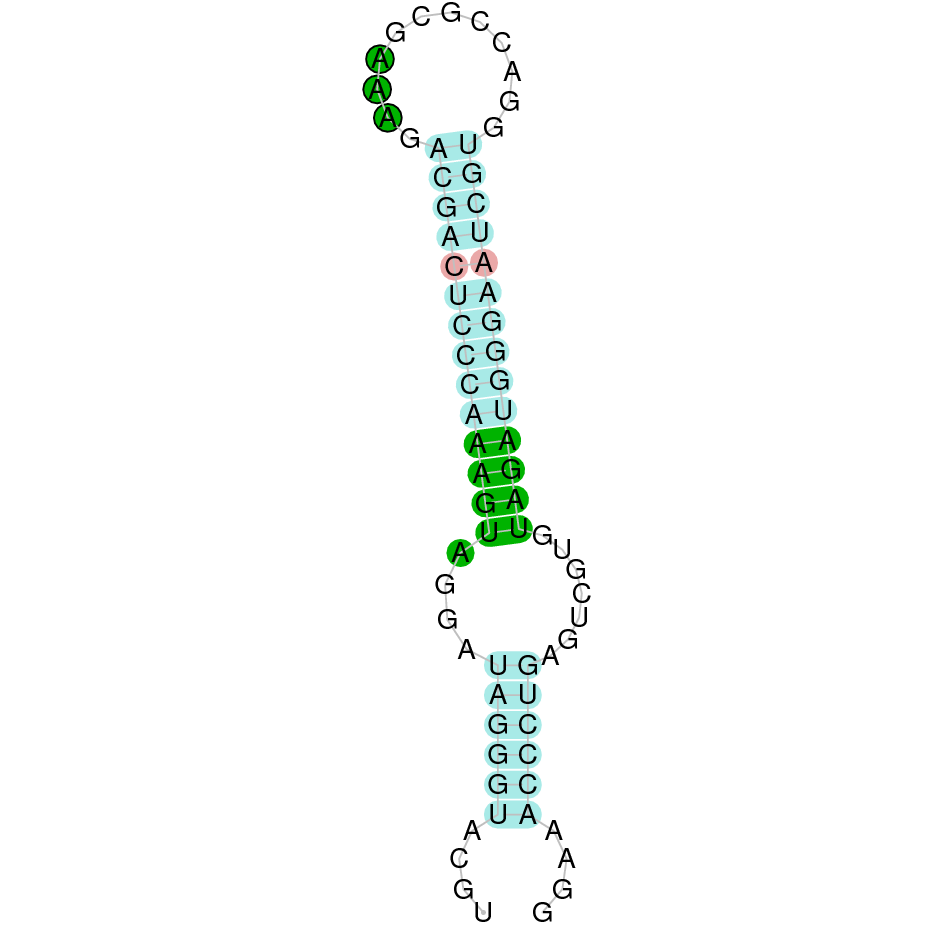
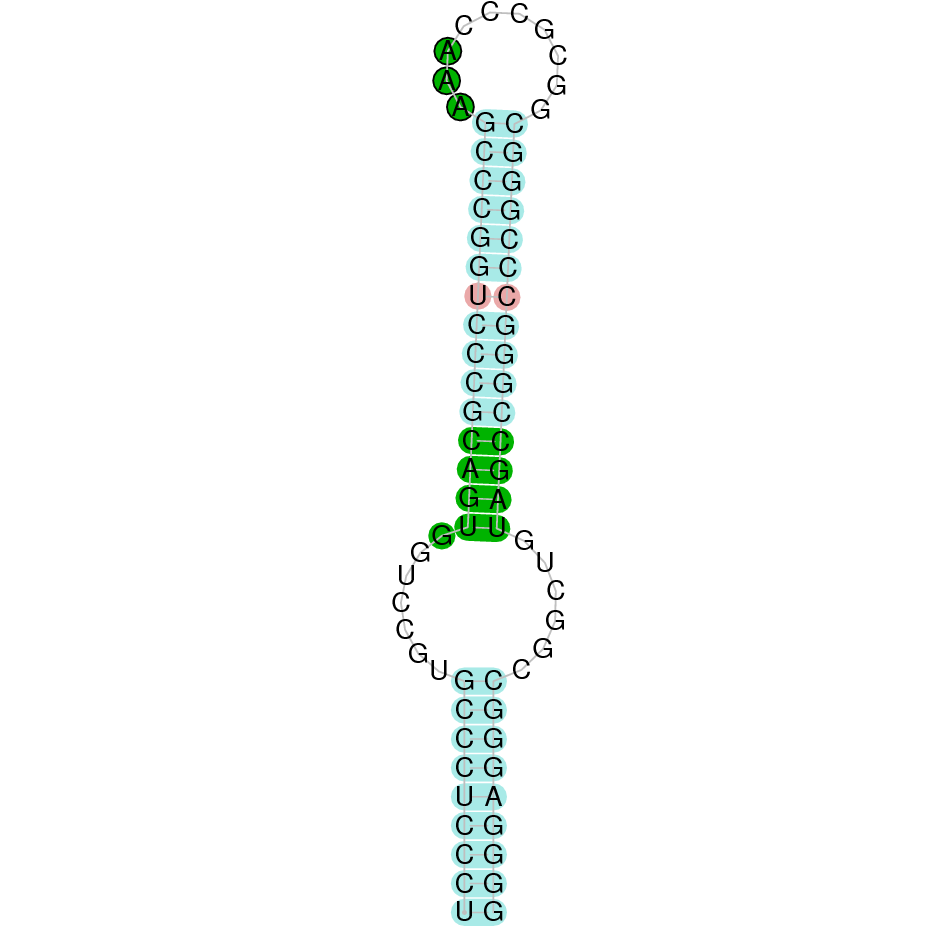
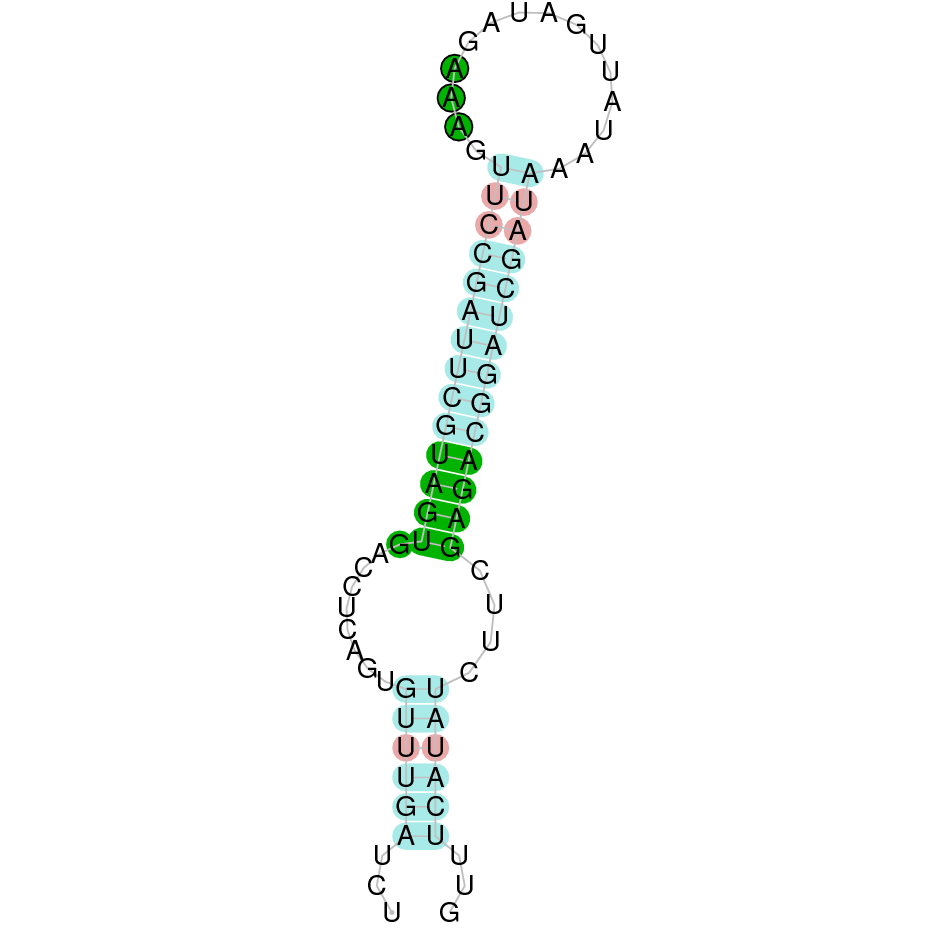
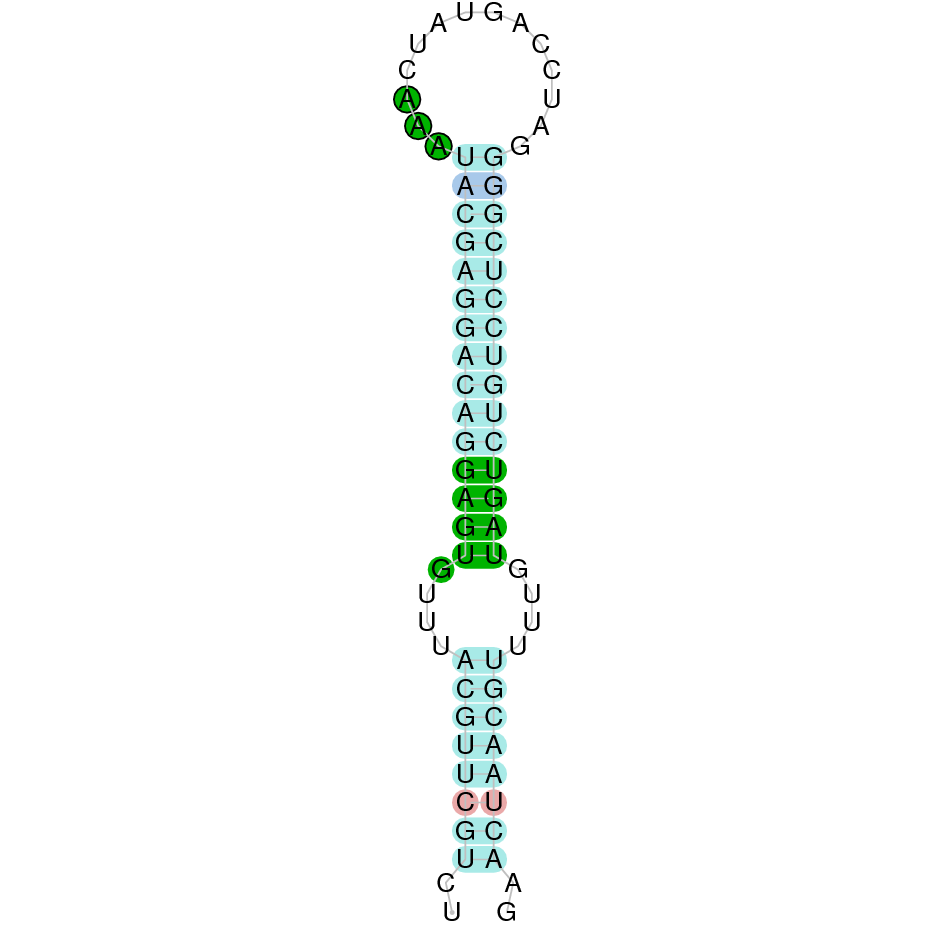
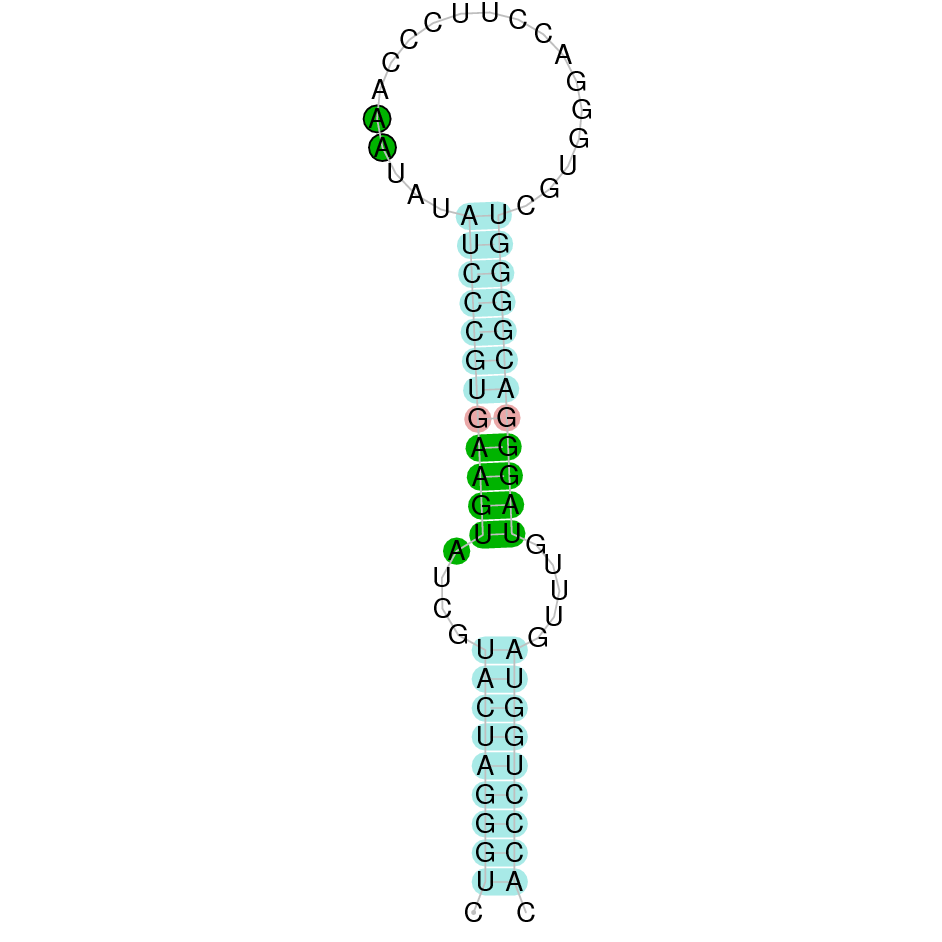
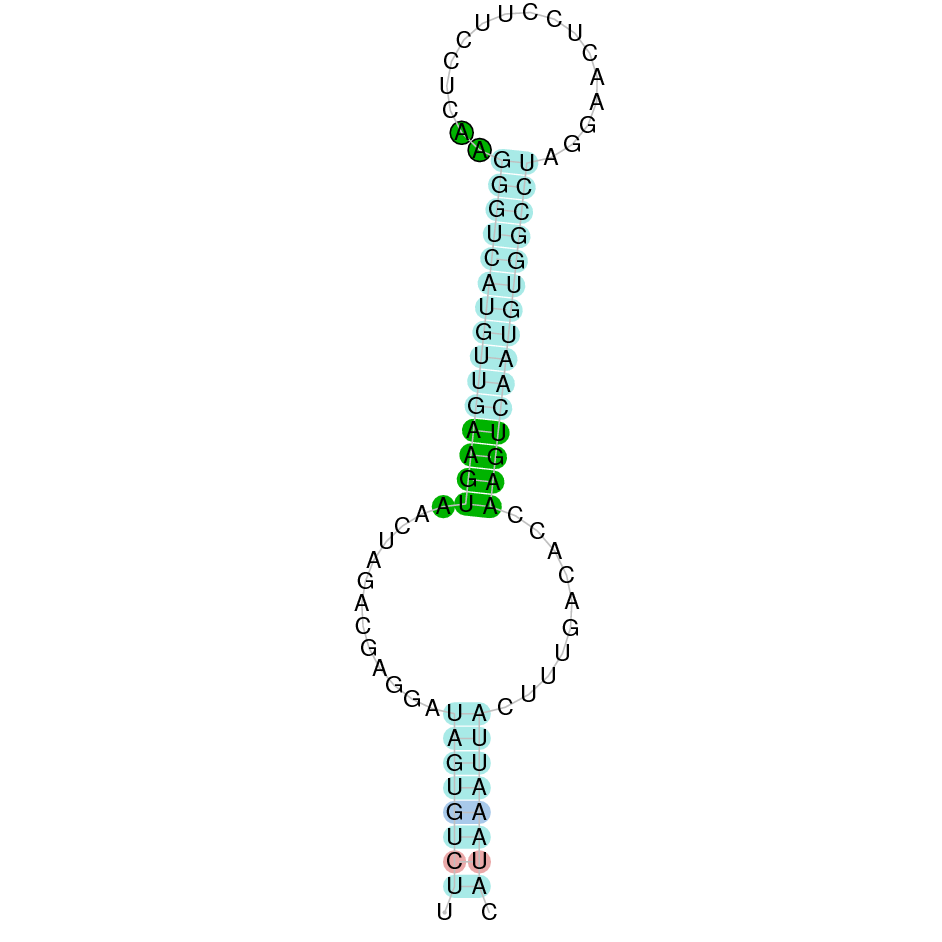
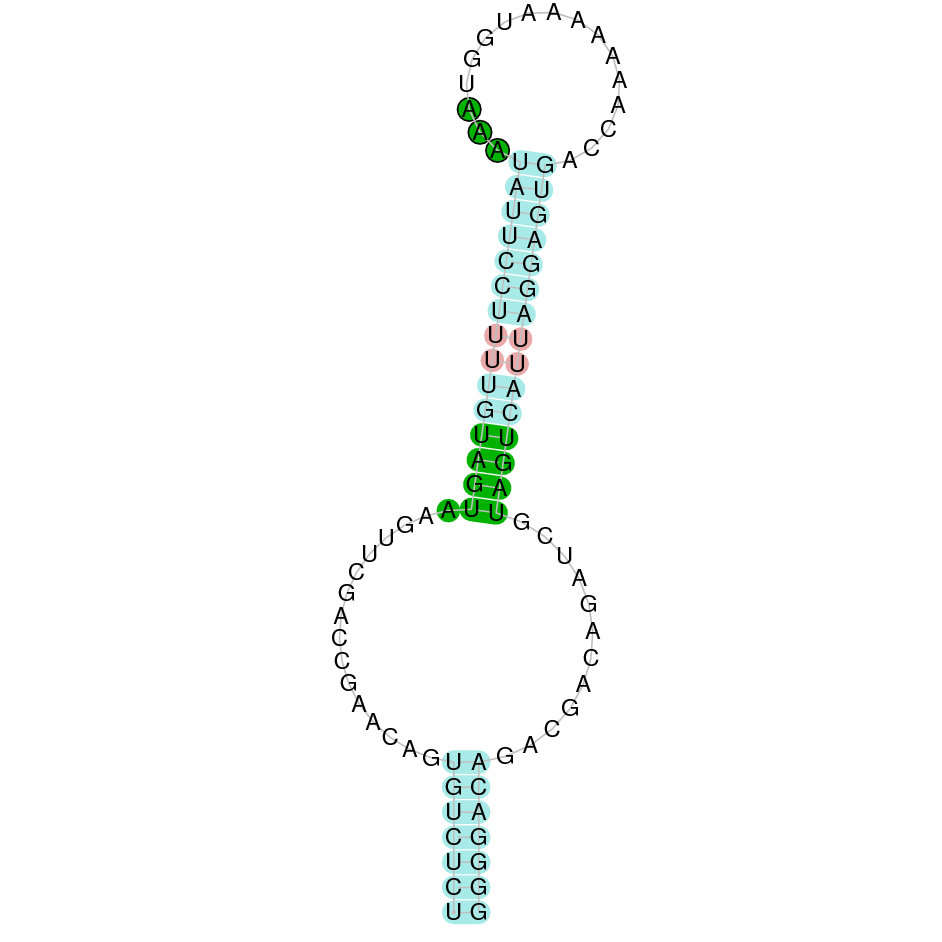
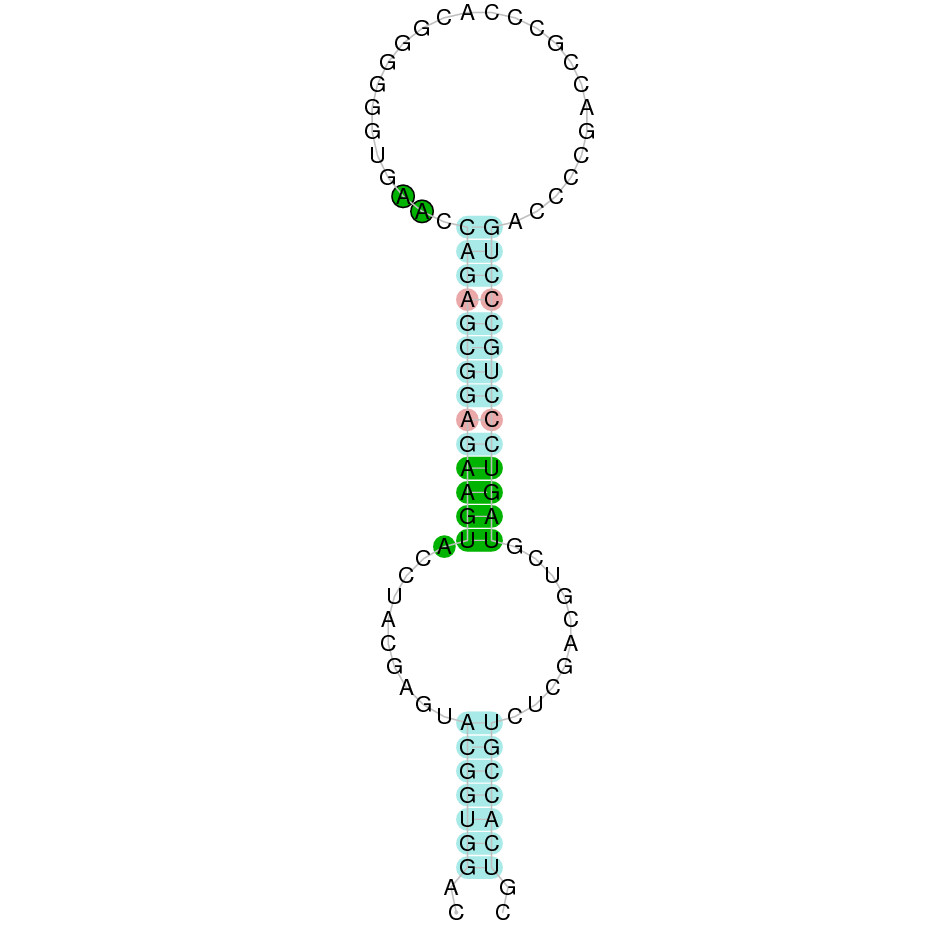
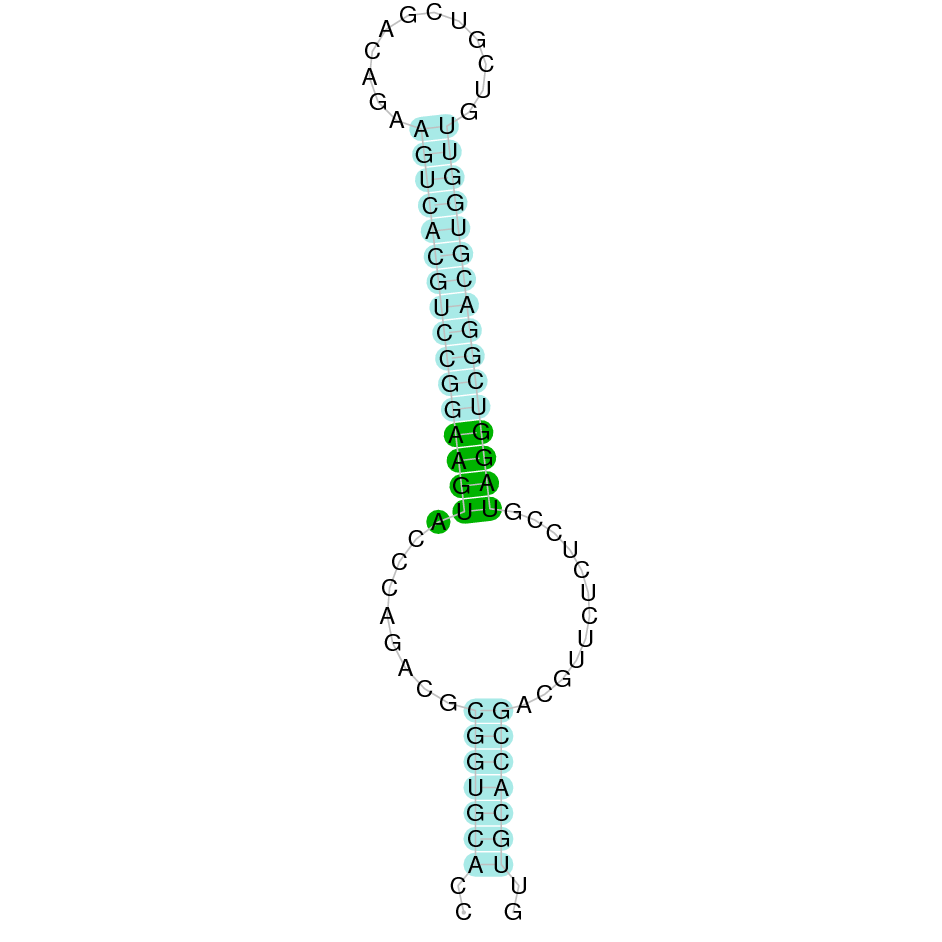
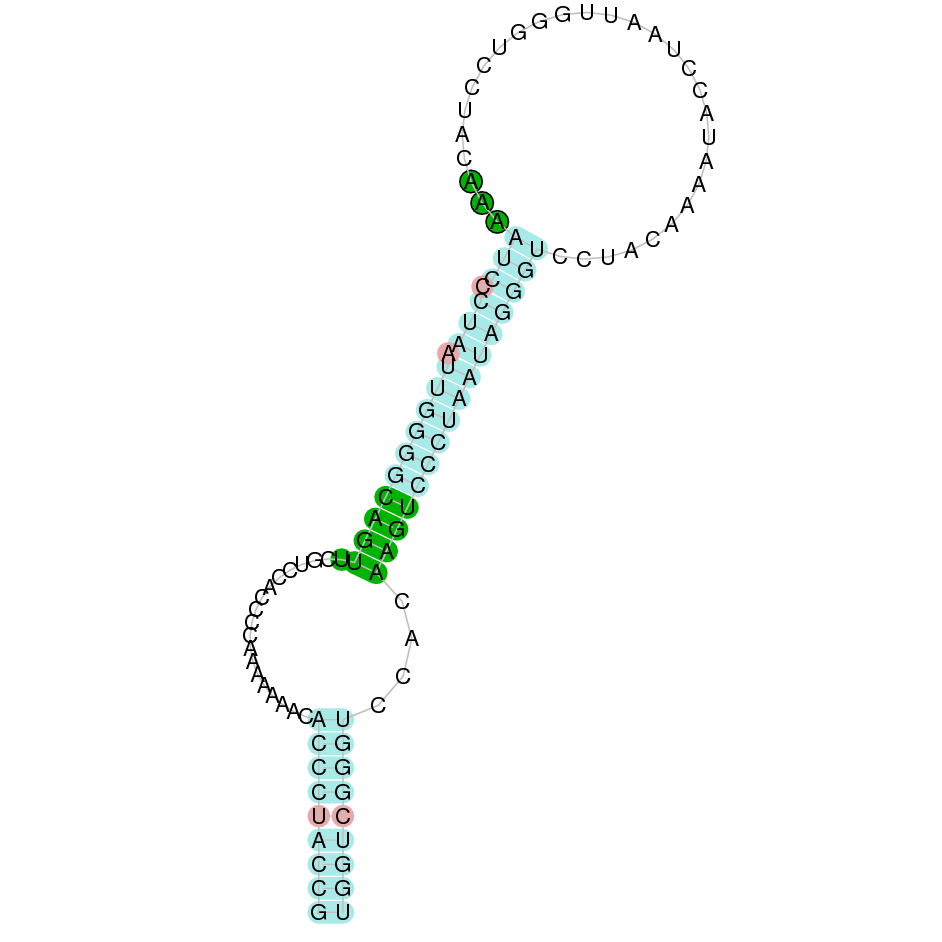
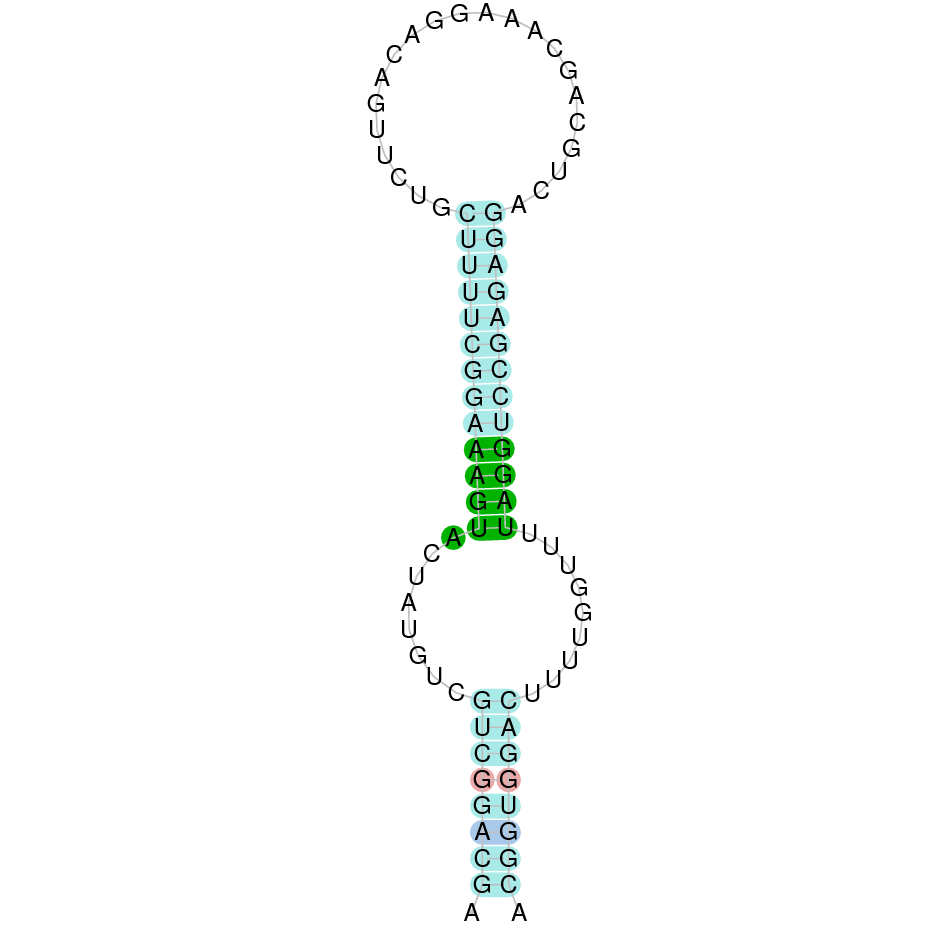
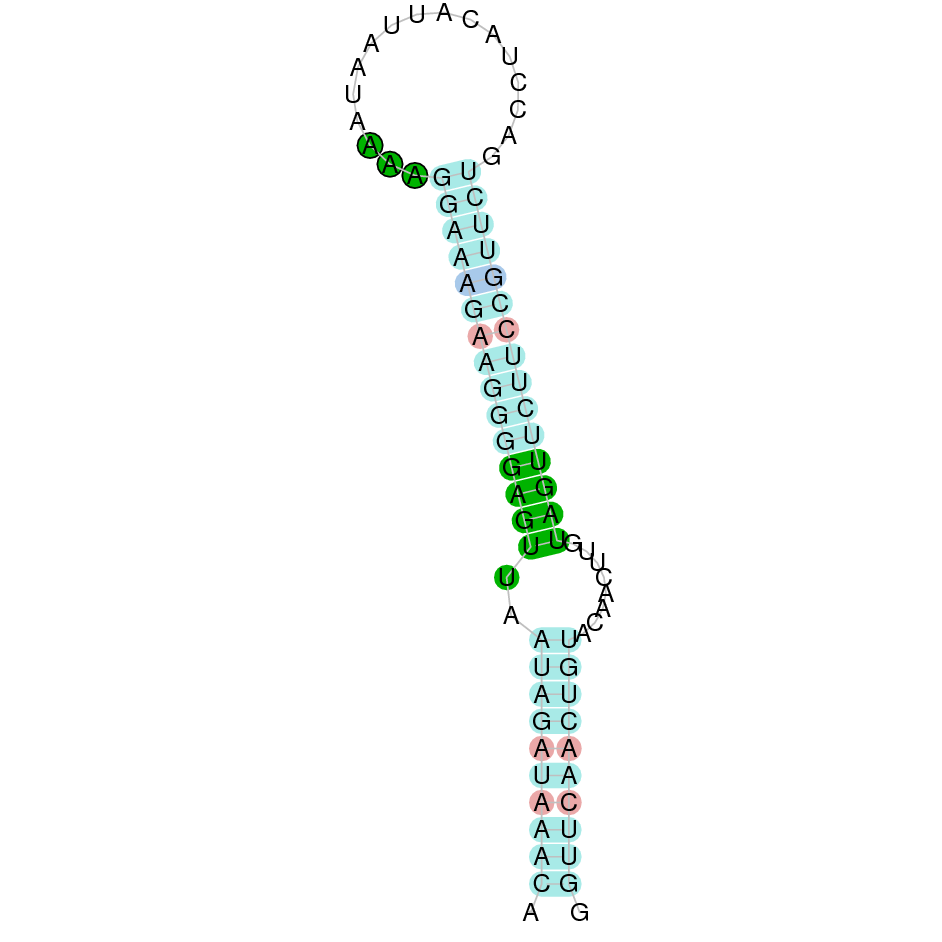
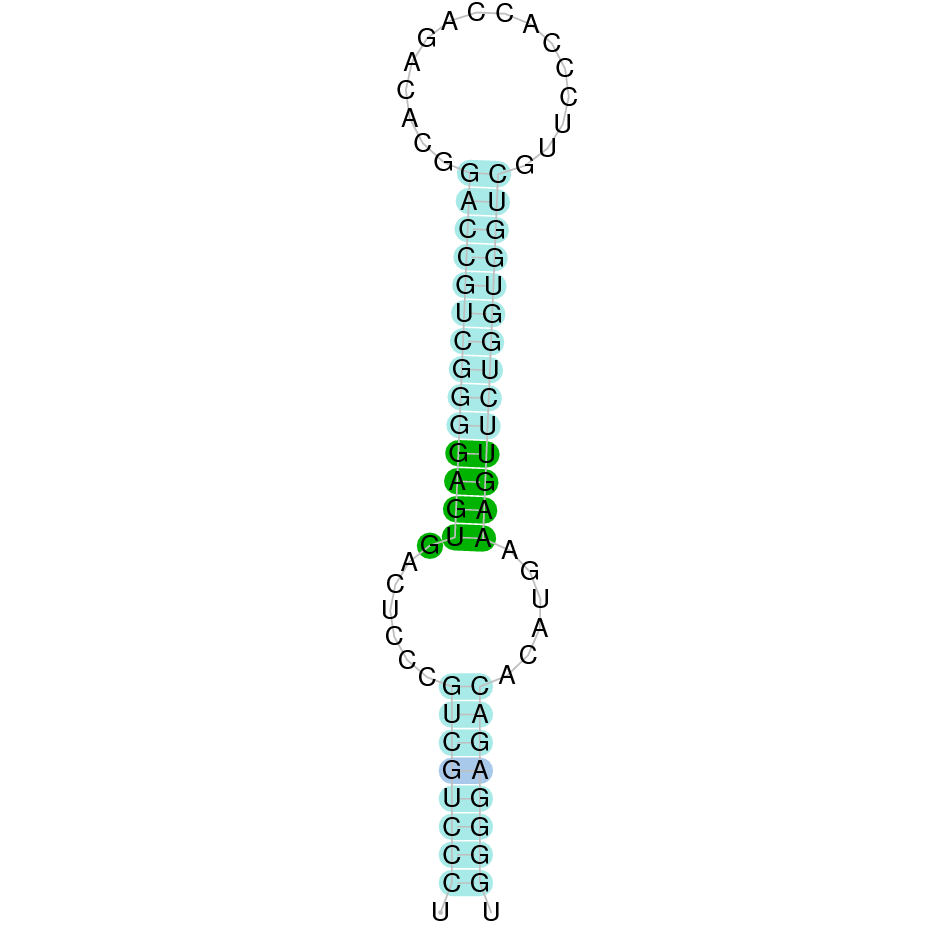
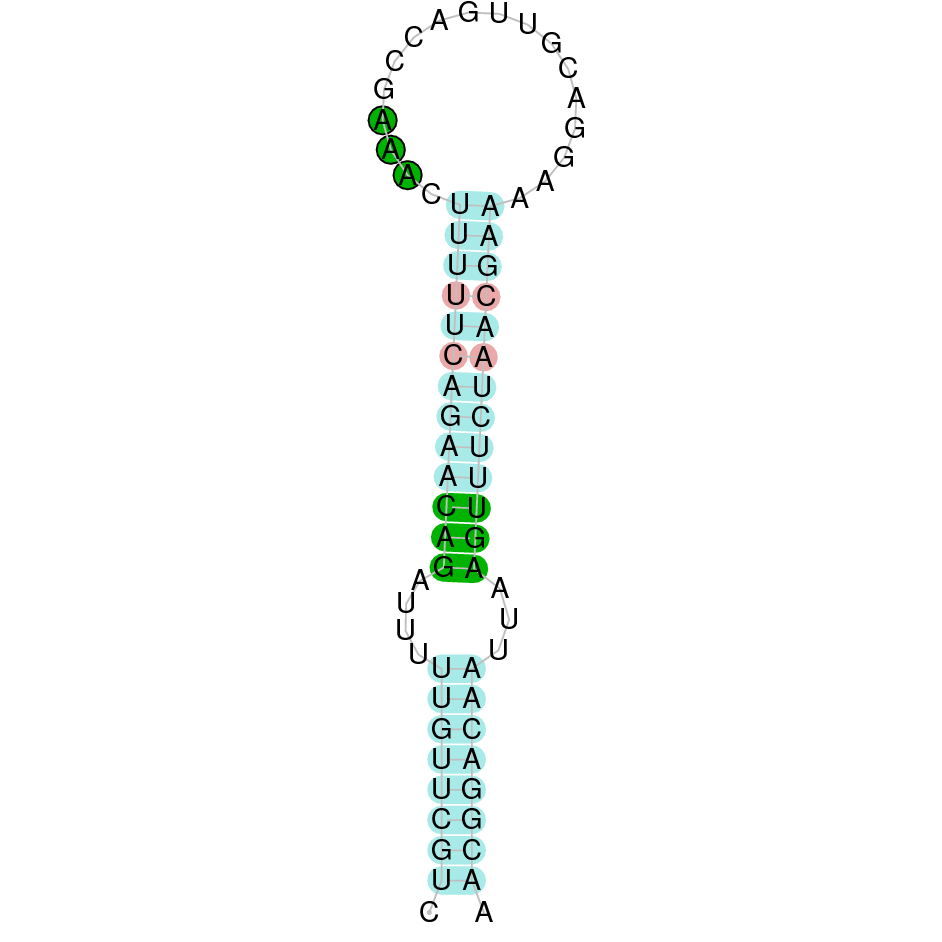
- In the introduction we explained that SECIS elements are necessary for the presence of a selenocysteine residue in those UGA codons that would otherwise be interpreted as STOP codons.
In this sense, we use the software Seblastian to identify the SECIS elements, as well as to predict automatically a hypothetical selenoprotein in our fastasubseq file.
For SECIS element prediction, we introduced manually the content of the fastasubseq.fa file in the Seblastian software leaving the default parameters for searching. Once we obtained the results of the prediction we established some criteria to select our data.
To consider a SECIS element as such, we evaluated its presence in the 3’-UTR region of the gene and whether it was in the same strand as the exons were. - - Protein ID
- - Scaffold ID
- - Start of the gene (genome coordinates)
- - End of the gene (genome coordinates)
- - Strand (Forward/Reverse)
- - Number of exons
- - Presence/Absence of selenocysteine
Once our referent species were chose, the followed analysis is schematized below:
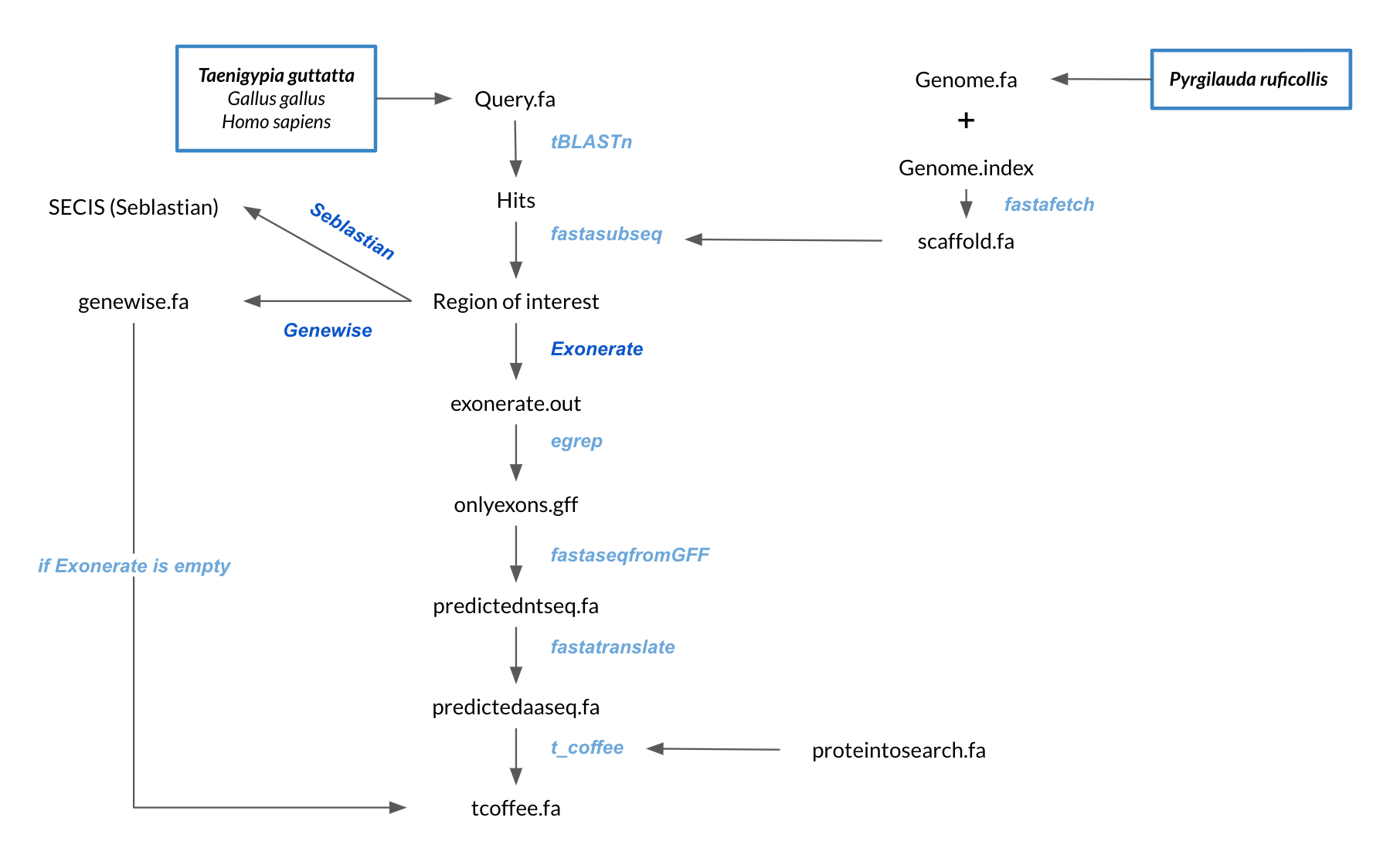
OBTENTION OF Pyrgilauda ruficollis GENOME
The genome of Pyrgilauda ruficollis (genome.fa), which was segmented in scaffolds, was downloaded from the file provided by the UPF Bioinformatics teaching team and was present in the following directory:/mnt/NFS_UPF/soft/genomes/2021/Pyrgilauda_ruficollis OBTENTION OF DIFFERENT SPECIES QUERY PROTEINS
The sequences of the query proteins from Zebra Finch, Chicken and Human were obtained from the selenoprotein database SelenoDB 2.0, where selenoproteins of different reference species can be found well annotated.After obtaining the amino acid sequences of all the queries, we pasted them in the same text-file named referencespecies_proteins.fa, where referencespecies is the common name of our referent species (i.e. finch_proteins.fa).
At this point, all the predicting process was fully automatized by a bioinformatic script written in Bash (Unix) programming language.
Description of the script and the used programs
QUERY PREPARING FOR ANALYSIS
In order to correctly perform the analysis, we change the character “U” (corresponding to Selenocysteine) for an “X”, which represents any possible amino acid, due to the fact that the softwares used do not recognise the “U” as one of them.Moreover, we remove special characters like “#”, “%” and “@” (corresponding to stop codons), as softwares were not able to align them.
These modifications are stored in a new text-file called query.fa.
PREDICTING PROCESS
As it was previously mentioned, our script is able to perform all the needed steps except for selenoprotein annotation, with the exception of SECIS element prediction.To run our script, we only need to call it from the terminal window and give three different inputs in the following order:
DATA ANALYSIS
After collecting all the data from the output of our script, it was the time to analyze all the information.
To do so, we used bash scripting to automatically generate a table containing (you can download an example below):
For more detailed information about the specific commands that we have used, as well as a brief description of all these commands you can download the commented version of our script.
RESULTS
In this section you can see a table summarizing the obtained results after the analysis, regarding selenoprotein prediction in Pyrgilauda ruficollis' genome. The information displayed corresponds to the query of the specie, the selected scaffold for prediction,
gene positions and strand, exonerate prediction and T-coffee alignment for each query of the reference genome (Taenigypia guttata by default, but in some cases it is Homo sapiens or Gallus gallus). Additional information about SECIS elements
(predicted candidates by SECISearch3, if there are any, and images) and selenoprotein prediction via Seblastian is also provided.
Finally, the predicted protein sequence is also available.
The legend used in the table is the following:

DISCUSSION
After aligning Pyrgilauda ruficollis’ with our query proteins, to escape the possibility of considering duplication events when a hit is actually another protein from the same family, exhaustive analysis has taken place to select, for each protein: the hits with the highest identity scores and lowest E-values, as well as the best aligned sequences by the analysis of T-coffee data.
Selenoproteins
Glutation Peroxidases (GPx)
GPx1
GPx1 is a protein responsible for the reduction of hydrogen peroxide to water and oxygen, as well as the reduction of peroxide radicals to alcohols and oxygen. After tBLASTn prediction, 6 scaffolds showed significant hits but the one with the lowest E-value (2,19E-33), highest identity score (96,76%) and better T-coffee alignment (T-coffee score of 995) was selected: JAGGNJ010000002.1. The predicted gene is located between positions 14011422-14012138 in the negative strand, and contains 2 exons, located in positions 14011423-14011770 and 14011896-14012138, both in the negative strand.
It is also worth mentioning that it contains a Sec residue in position 46, the same position as the query protein, taken in this case from the Finch. In addition, the protein starts with Methionine.
SECISearch3 predicted a grade B SECIS element between the positions 14011272-14011336, at the 3’ UTR end of the negative strand, coinciding with the GPx2 selenoprotein in the sequence. Seblastian also predicted a selenoprotein.
The gene that codes for this protein has duplications in the reference species (Zebra finch), and we observe these duplications represented in the Pyrgilauda ruficollis genome. Specifically, we found hits from the three duplications in different scaffolds given their sequence similarity: JAGGNJ010019611.1, JAGGNJ010015723.1, JAGGNJ010009663.1, JAGGNJ010000002.1 and JAGGNJ010001667.1. In more detail:
- - SPP00002524_2.0 and SPP00002526 queries have their best hit on scaffold JAGGNJ010019611 and coinciding in their genomic coordinates, leading us to conclude that one of the two proteins in the Finch is not present in Pyrgilauda ruficollis. It should be noted that this alignment does not have Sec residues neither in the query nor in the predicted sequence.
- - SPP00002525_2.0 query: on scaffold JAGGNJ010000002, so we can consider that this protein has been conserved.
- - SPP00002539 and SPP00002540 were found at the same scaffold (JAGGNJ010002222), being the latter a shorter version of the former, lacking its terminal region.
- - SPP00002541 and SPP00002542 were found at the same scaffold (JAGGNJ010015945) being the latter a shorter version of the former, lacking part of its central region.
Looking at this data, we can conclude that Pyrgilauda ruficollis has conserved two copies from the three than we can observe in Zebra finch. The protein located in scaffold JAGGNJ010000002 conserves the Sec protein and has a SECIS element, so we can assume its role as a selenoprotein in Pyrgilauda ruficollis. Among the second predicted protein, we can say that it is conserved in Pyrgilauda ruficollis but, as it lacks the Sec residue and the SECIS element, the assumption of denying its role as a selenoprotein can be made (it is not either a selenoprotein in Zebra finch).

GPx2
GPx2 is a protein responsible for the reduction of hydrogen peroxide to water and oxygen, as well as the reduction of peroxide radicals to alcohols and oxygen. After tBLASTn prediction, 6 scaffolds showed significant hits but the one with the lowest E-value (1,37E-116), highest identity score (87,26%) and better T-coffee alignment (T-coffee score of 995) was selected: JAGGNJ010009663.1. The predicted gene is located between positions 168504-169140 in the negative strand, and contains 2 exons, located in positions 168928-169140 and 168505-168849, both in the negative strand.
It is also worth mentioning that it contains a Sec residue in position 37, the same position as the query protein, taken in this case from de Finch. In addition, the protein does not start with Methionine, but with Isoleucine, opening the possibility of having missed the beginning of the protein or that this beginning is located more downstream than in the query protein.
SECISearch3 predicted a grade A SECIS element between the positions 168361-168435, at the 3’ UTR end of the negative strand, coinciding with the GPx2 selenoprotein in the sequence. Seblastian also predicted a selenoprotein.
This gene presents duplications in other genomic regions (JAGGNJ010000002, JAGGNJ010001667, JAGGNJ010015723, JAGGNJ010019278 and JAGGNJ010019611), where all of them have a region that, even though it is conserved, lacks N-terminal, C-terminal or both, leading us to conclude that these duplications are not functional.
Looking at this data, we can conclude that Pyrgilauda ruficollis has conserved the GPx2 gene, and it is still a selenoprotein, as it has a Sec residue and a predicted SECIS element.

GPx3
GPx3 is a protein responsible for the reduction of hydrogen peroxide to water and oxygen, as well as the reduction of peroxide radicals to alcohols and oxygen. After tBLASTn prediction, 6 scaffolds showed significant hits but the one with the lowest E-value (2,05E-17), highest identity score (83,04%) and better T-coffee alignment (T-coffee score of 996) was selected: JAGGNJ010019278. The predicted gene is located between positions 4564918-4565968 in the positive strand, and contains 4 exons, located in positions 4564919-4565075, 4565273-4565390, 4565584-4565683 and 4565774-4565968, all located in the positive strand.
It is also worth mentioning that it contains a Sec residue in position 44, the same position as the query protein, taken in this case from the Finch. In addition, the protein does not start with Methionine, but with Glutamine, opening the possibility of having missed the beginning of the protein or that this beginning is located more downstream than in the query protein.
SECISearch3 predicted a grade A SECIS element between the positions 4566312-4566391, at the 3’ UTR end of the positive strand, coinciding with the GPx3 selenoprotein in the sequence. Seblastian also predicted a selenoprotein.
This gene presents duplications in other genomic regions (JAGGNJ010000002 and JAGGNJ010009663), where all of them have a region that, even though it is conserved, lacks N-terminal, C-terminal or both, leading us to conclude that these duplications are not functional.
Looking at this data, we can conclude that Pyrgilauda ruficollis has conserved the GPx3 gene, and it is still a selenoprotein, as it has a Sec residue and a predicted SECIS element.

GPx4
GPx4 is a protein responsible for the reduction of hydrogen peroxide to water and oxygen, as well as the reduction of peroxide radicals to alcohols and oxygen. After tBLASTn prediction, 6 scaffolds showed significant hits but the one with the lowest E-value (4,90E-07), highest identity score (81,90%) and better T-coffee alignment (T-coffee score of 994) was selected: JAGGNJ010001667.1. The predicted gene is located between positions 1331142-1332030 in the positive strand, and contains 4 exons, located in positions 1331143-1331289, 1331370-1331521, 1331649-1331673 and 1331971-1332030, both in the positive strand.
It is also worth mentioning that it contains a Sec residue in position 13, the same position as the query protein, taken in this case from the Finch. In addition, the protein does not start with Methionine, but with an Arginine, opening the possibility of having missed the beginning of the protein or that this beginning is located more downstream than in the query protein. SECISearch3 predicted a grade A SECIS element between the positions 1332187-1332266, at the 3’ UTR end of the positive strand, coinciding with the GPx4 selenoprotein in the sequence. Seblastian also predicted a selenoprotein.
This gene presents duplications in other genomic regions (JAGGNJ010000002, JAGGNJ010009663, JAGGNJ010015723 and JAGGNJ010019611), where all of them have a region that, even though it is conserved, lacks N-terminal, C-terminal or both, leading us to conclude that these duplications are not functional.
Looking at this data, we can conclude that Pyrgilauda ruficollis has conserved the GPx4 gene, and it is still a selenoprotein, as it has a Sec residue and a predicted SECIS element.

GPx5 and GPx6
Gpx5 and Gpx6 are proteins responsible for the reduction of hydrogen peroxide to water and oxygen, as well as the reduction of peroxide radicals to alcohols and oxygen. After tBLASTn prediction, different scaffolds showed significant hits but the one with the lowest E-value, highest identity score and better T-coffee alignment were selected (the others shown very poor alignment significance): both coinciding in JAGGNJ010019278.1, the scaffold where we previously located GPx3. In addition, after deeply investigating this region we noticed that the predicted exons of the three proteins were overlapping.As it has been described in literature, GPx5 and GPx6 were generated by GPx3 duplication in the origin of placental organisms. As Pyrgilauda ruficollis belongs to a previous lineage in evolutionary terms, we do not observe these new proteins in its genome.


GPx7
GPx7 is a protein responsible for the reduction of hydrogen peroxide to water and oxygen, as well as the reduction of peroxide radicals to alcohols and oxygen. After tBLASTn prediction, 5 scaffolds showed significant hits but the one with the lowest E-value (3,11E-17), highest identity score (96,46%) and better T-coffee alignment (T-coffee score of 999) was selected: JAGGNJ0100019611. The predicted gene is located between positions 13898987-13948986 in the positive strand, and contains 2 exons, located in positions 13948987-13949251 and 13951062-13951186, both in the positive same strand.
It is also worth mentioning that the protein does not contain a Sec residue, as we expected since it has been described to be a Cysteine-containing homologous protein. In addition, the protein does not start with Methionine, but with Glutamine, opening the possibility of having missed the beginning of the protein or that this beginning is located more downstream than in the query protein. SECISearch3 did not predict any SECIS element and Seblastian did not predict any selenoprotein.
This gene is found in Chicken’s genome, but not in Zebra finch’s, so we can conclude it has been deleted in the finch. As for Pyrgilauda, it presents duplications in other genomic regions, of which duplication in JAGGNJ010000002 presents a region that, even though it is conserved, lacks N-terminal, C-terminal or both, leading us to conclude that these duplications are not functional. Furthermore, in the scaffold JAGGNJ01001573 we see a very well annotated protein that seems to be a new duplication of this gene in Pyrgilauda’s genome.
Looking at this data, we can conclude that Pyrgilauda ruficollis has conserved the GPx7 protein of the Chicken query and we can confirm that they are both Cysteine-containing homologous proteins. In addition, we can also say that the gene has been duplicated in Pyrgilauda ruficollis, but this duplication is also a Cysteine-containing homologous protein for the same reasons described above.

GPx8
GPx8 is a protein responsible for the reduction of hydrogen peroxide to water and oxygen, as well as the reduction of peroxide radicals to alcohols and oxygen. After tBLASTn prediction, 3 scaffolds showed significant hits but the one with the lowest E-value (1,03E-15), highest identity score (88,69%) and better T-coffee alignment (T-coffee score of 1000) was selected: JAGGNJ010015723. The predicted gene is located between positions 5867100-5869442 in the negative strand, and contains 2 exons, located in positions 5867101-5867204 and 5869178-5869442, both in the positive strand.
It is also worth mentioning that the protein does not contain a Sec residue, as we expected since it has been described to be a Cysteine-containing homologous protein. In addition, the protein does not start with Methionine, but with Lysine, opening the possibility of having missed the beginning of the protein or that this beginning is located more downstream than in the query protein.
SECISearch3 predicted a grade B SECIS element between the positions 5929722-5929788, at the 3’ UTR end of the positive strand. Seblastian didn't predict a selenoprotein.
This gene presents duplications in other genomic regions (JAGGNJ010000002 and JAGGNJ010019611), where all of them have a region that, even though it is conserved, lacks N-terminal, C-terminal or both, leading us to conclude that these duplications are not functional.
Looking at this data, we can conclude that Pyrgilauda ruficollis has conserved the GPx8 gene from the Zebra finch, and it is translated into the Cysteine-containing homologous protein observed in the query.

Iodothyronine deiodinase (DIO)
DIO1
DIO1 is a protein responsible for T4’s deiodination to T3. After tBLASTn prediction, 3 scaffolds showed significant hits but the one with the lowest E-value (1,81E-22), highest identity score (73,46%) and better T-coffee alignment (T-coffee score of 997) was selected: JAGGNJ010019611.1. The predicted gene is located between positions 14665391-14668624 in the positive strand, and contains 3 exons, located in positions 14665392-14665722, 14667231-14667374 and 14668425-14668624, all of them in the positive strand.
It is also worth mentioning that it contains a Sec residue in position 124, the same position as the query protein, taken in this case from the Finch. In addition, the predicted protein starts with Methionine.
SECISearch3 predicted a grade A SECIS element between the positions 14669630-14669697, at the 3’ UTR end of the positive strand, coinciding with the DIO1 selenoprotein in the sequence. Seblastian also predicted a selenoprotein.
As for duplications, this gene is duplicated in Zebra finch but we see the same hits for both queries. This allows us to suppose that our bird has deleted one of the two copies. Since our query SPP00002518_2.0 has one exon less than SPP00002519_2.0 we can infer that the former is the one that has been deleted in Pyrgilauda.
The copy we find is located at scaffold JAGGNJ010019611. Furthermore, it has incomplete duplications in other regions of the genome (JAGGNJ010016056 and JAGGNJ010017168), where all of them have a region that, even though it is conserved, lacks N-terminal, C-terminal or both.
Looking at this data, we can conclude that Pyrgilauda ruficollis has conserved one copy of the DIO1 gene, and it is still a selenoprotein, as it has a Sec residue and a predicted SECIS element.
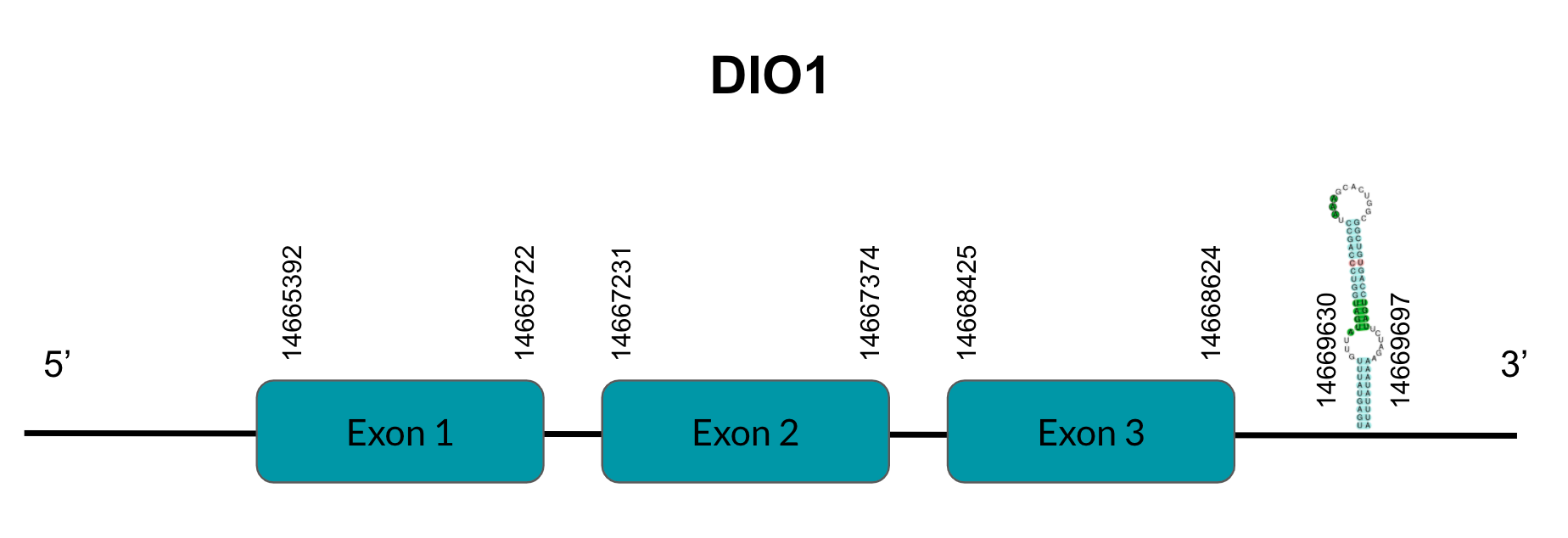
DIO2
DIO2 is a protein responsible for T4’s deiodination to T3. After tBLASTn prediction, 3 scaffolds showed significant hits but the one with the lowest E-value (2,29E-40), highest identity score (99,47%) and better T-coffee alignment (T-coffee score of 996) was selected: JAGGNJ010016056.1. The predicted gene is located between positions 4965319-4977024 in the negative strand, and contains 2 exons, located in positions 4976803-4977024 and 4965320-4965889, both in the negative strand.
It is also worth mentioning that it contains a Sec residue in position 132, the same position as the query protein, taken in this case from the Finch. In addition, the protein starts with Methionine.
SECISearch3 predicted a grade A SECIS element between the positions 4960872-4960946, at the 3’ UTR end of the negative strand, coinciding with the DIO2 selenoprotein in the sequence. On the other hand, Seblastian did not predict any selenoprotein.
As for duplications, they appear in other genomic regions (JAGGNJ010017168 and JAGGNJ010019611), where all of them have a region that, even though it is conserved, lacks N-terminal, C-terminal or both, leading us to conclude that these duplications are not functional.
Looking at this data, we can conclude that Pyrgilauda ruficollis has conserved the DIO2 gene, and it is still a selenoprotein, as it has a Sec residue and a predicted SECIS element.
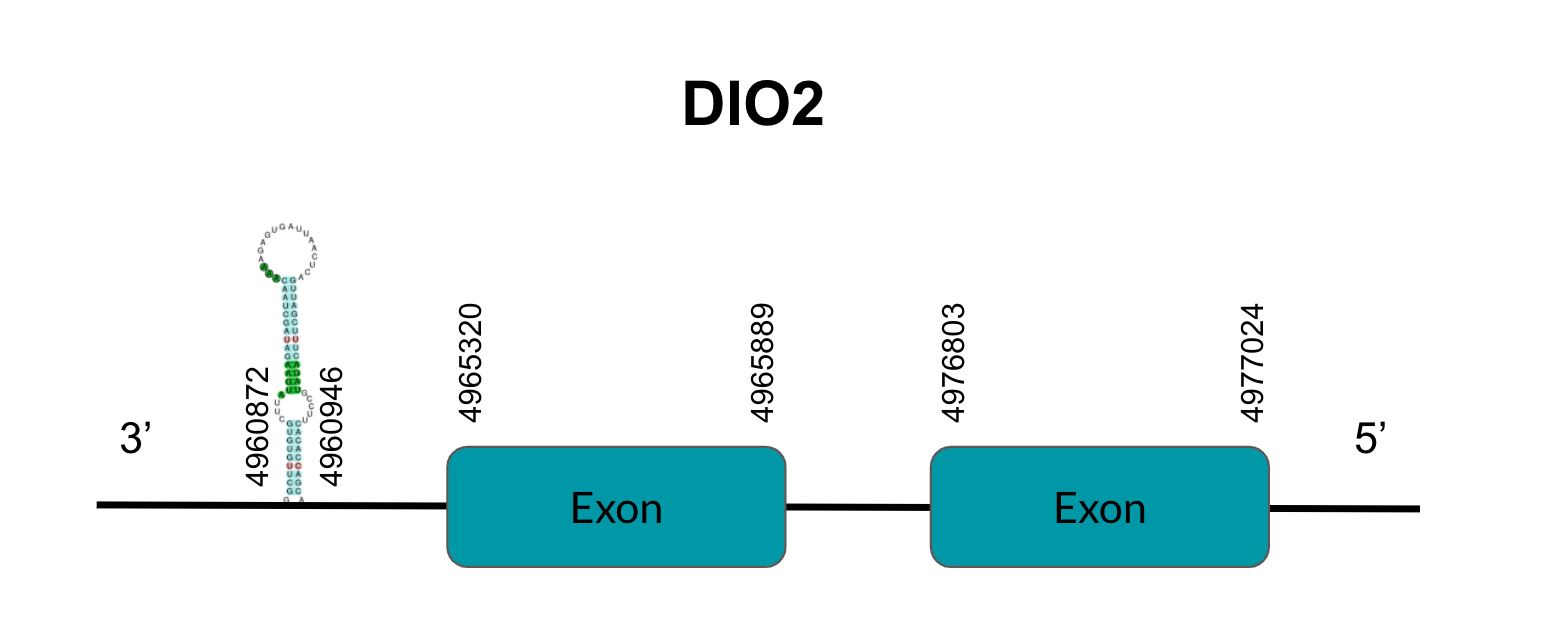
DIO3
DIO3 is a protein responsible for T4’s deiodination to T3. After tBLASTn prediction, 3 scaffolds showed significant hits but the one with the lowest E-value (6,12E-134), highest identity score (98,05%) and better T-coffee alignment (T-coffee score of 995) was selected: JAGGNJ010017168.1. The predicted gene is located between positions 4032271-4032886 in the positive strand, and contains 1 exon, located in positions 4032272-4032886.
It is also worth mentioning that it contains a Sec residue in position 69, the same position as the query protein, taken in this case from the Finch. In addition, the protein does not start with Methionine, but with Valine, opening the possibility of having missed the beginning of the protein or that this beginning is located more downstream than in the query protein.
SECISearch3 predicted a grade B SECIS element between the positions 4033322-4033402, at the 3’ UTR end of the positive strand, coinciding with the DIO3 selenoprotein in the sequence. Seblastian also predicted a selenoprotein.
As for duplications, they appear in other genomic regions (JAGGNJ010016056 and JAGGNJ010019611), where all of them have a region that, even though it is conserved, lacks N-terminal, C-terminal or both, leading us to conclude that these duplications are not functional.
Looking at this data, we can conclude that, although we can assume some issues in the prediction, Pyrgilauda ruficollis has conserved the DIO3 gene, and it is still a selenoprotein, as it has a Sec residue and a predicted SECIS element.
Methionine sulfoxide reductases A (MsrA)
MsrA is a late endosomal enzyme that gives protection to other proteins from oxidative damage. After tBLASTn prediction, 6 scaffolds showed significant hits but the one with the lowest E-value (2,00E-07), the highest identity score (91,99%) and the better T-coffee score (999) was selected: JAGGNJ010018945. However, the gene seems to be truncated compared to the query protein. The predicted gene is located between positions 8146872-8242862 in the negative strand, and contains 3 exons, located in positions 8146873-8147034, 8228143-8228249 and 8242754-8242862, all of them in the negative strand.
It is also worth mentioning that the protein does not contain a Sec residue, as we expected since it has been described to be a Cysteine-containing homologous protein.
In addition, the protein does not start with Methionine, but with Serine, opening the possibility of having missed the beginning of the protein or that this beginning is located more downstream than in the query protein.
SECISearch3 predicted a grade A SECIS element between the positions 8209265-8209345, which is in the negative strand. However, due to the fact that the predicted SECIS element was included between the exon coordinates, we decided to assume that the SECIS prediction was not realistic. Seblastian also predicted a selenoprotein.
Regarding duplication events, we also found a truncated copy of this gene in scaffold JAGGNJ010006888.1.
Finally, as the gene is truncated in both scaffolds, its function is thought to be lost in Pyrgilauda ruficollis.
Methionine sulfoxide reductases B (MSRB)
MSRB1
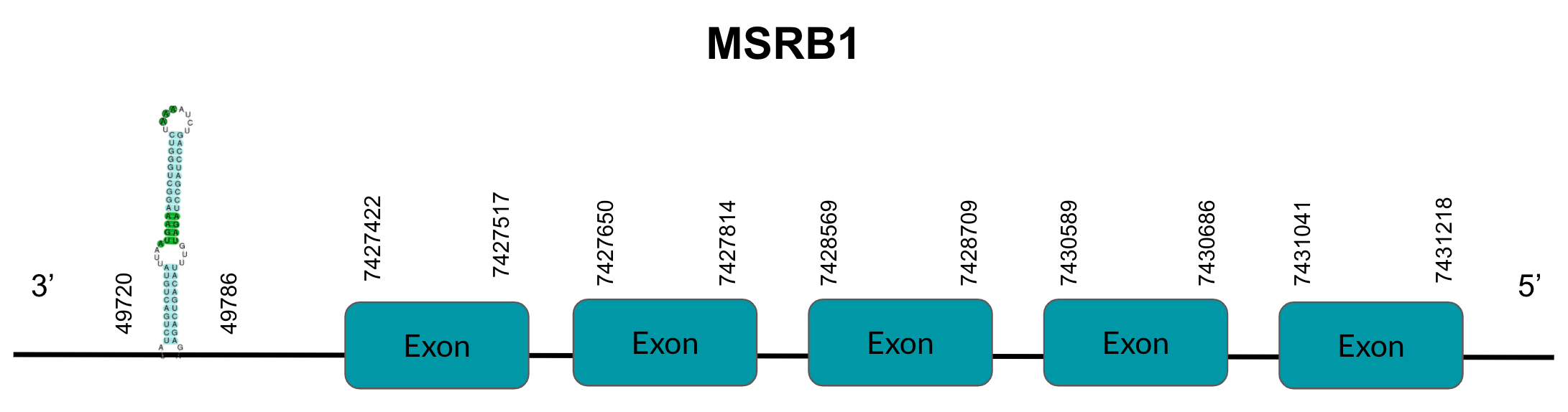
MsrB1, also known as SelX1, is a protein involved in catalyzing the reduction of methionine sulfoxides to methionines. After tBLASTn prediction, only 1 scaffold showed significant hits with a low E-value (4,25E-11), a high identity score (87,19%) and a good T-coffee score (997): JAGGNJ010001111.1.1. The predicted gene is located between positions 7427421-7431218 in the negative strand, and contains 5 exons, located in positions 7427422-7427517, 7427650-7427814, 7428569-7428709, 7430589-7430686 and 7431041-7431218, all of them in the negative strand.
It is also worth mentioning that it does not contain a Sec residue, and the protein does start with Methionine. SECISearch3 predicted a grade A SECIS element between 77427142-77427208 in the negative strand and Seblastian does not predict a selenoprotein.
Looking at this data, we can conclude that Pyrgilauda ruficollis has conserved the sequence coding for MsrB1 that we observe in the Zebra finch. However, neither in Zebra finch nor Pyrgilauda ruficollis MsrB1 is a selenoprotein, as it lacks Sec residues despite the fact that SECISearch3 has predicted a SECIS element.
MSRB2
MsrB2, also known as SelX2 is a protein involved in catalyzing the reduction of methionine sulfoxides to methionines. After tBLASTn prediction, only 1 scaffold showed significant hits with a low E-value (3,30E-09), a high identity score (82,55%) and a good T-coffee score (1000): JAGGNJ010018834.1. The predicted gene is located between positions 1453697-1457567 in the positive strand, and contains 6 exons, located in positions 1453698-1453796, 1454267-1454335, 1455854-1455907, 1456342-1456447, 1456690-1456821 and 1457443-1457567, all of them in the positive strand.
It is also worth mentioning that it does not contain a Sec residue. In addition, the protein does not start with Methionine, but with Arginine, opening the possibility of having missed the beginning of the protein or that this beginning is located more downstream than in the query protein. SECISearch3 did not predict any SECIS element and Seblastian does not predict a selenoprotein.
Looking at this data, we can conclude that Pyrgilauda ruficollis has conserved the sequence coding for MsrB2 that we observe in the Zebra finch. However, neither in Zebra finch nor Pyrgilauda ruficollis MsrB2 is a selenoprotein, as it lacks Sec residues and SECIS elements.

Thioredoxin reductases (TRs)
TR1
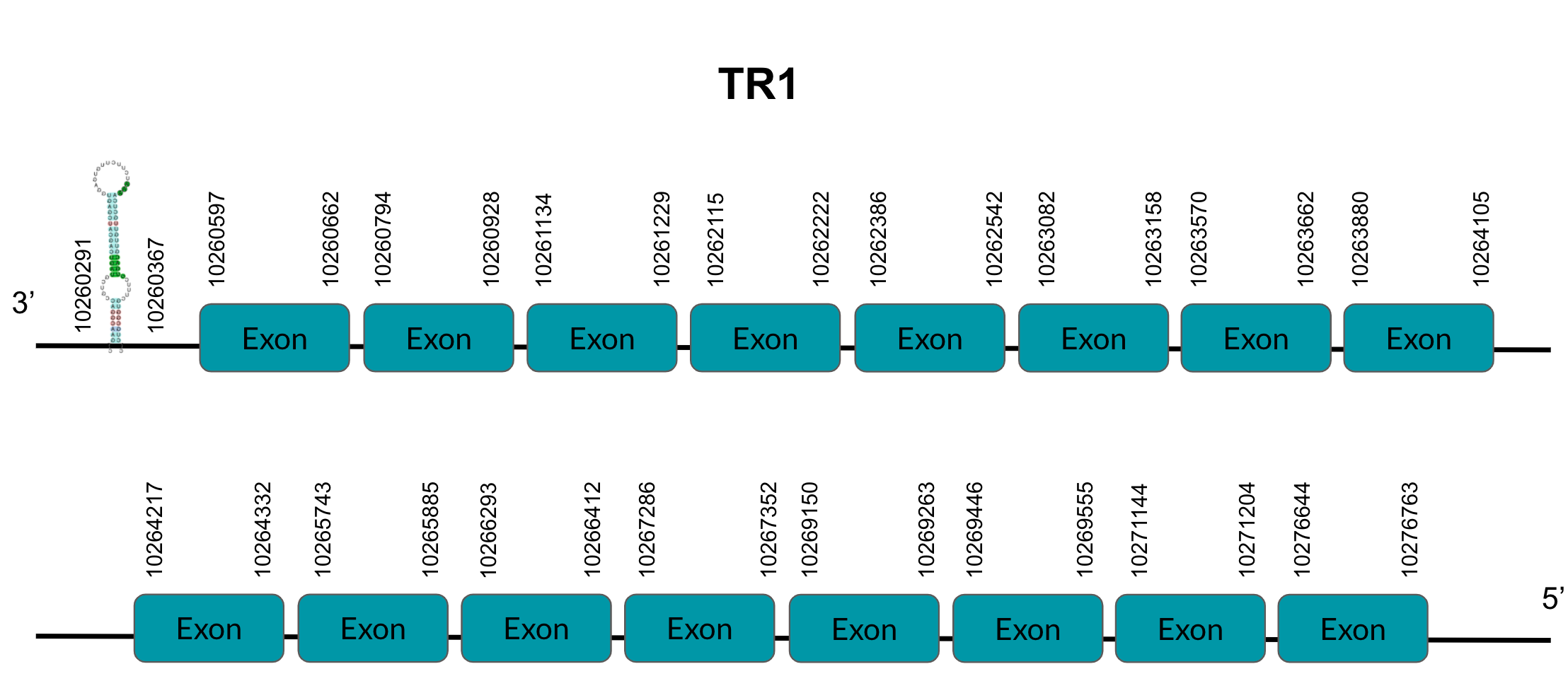
TR1 is a protein responsible for thioredoxin reduction. After tBLASTn prediction, 4 scaffolds showed significant hits but the one with the lowest E-value (1,55E-06), highest identity score (91,60%) and a better T-coffee score (999) was selected: JAGGNJ010000002. The predicted gene is located between positions 10260596-10276763 in the negative strand, and contains 16 exons, located in positions 10260662-10260597,10260928-10260794, 10261229-10261134,10262222-10262115, 10262542-10262386, 10263158-10263082, 10263692-10263570, 10264105-10263886, 10264332-10264217, 10265885-10265743, 10216412-10266293, 10267352-10267280, 10269263-10269150, 10269555-10269446, 10271204-10271144 and 10276763-10276644, all of them in the negative strand.
It is also worth mentioning that it contains a Sec residue in position 611, the same position as the query protein, taken in this case from the Finch. In addition, the protein does not start with Methionine, but with Proline, opening the possibility of having missed the beginning of the protein or that this beginning is located more downstream than in the query protein.
SECISearch3 predicted a grade A SECIS element between the positions 10260291-10260367, at the 3’ UTR end of the negative strand. Seblastian also predicted a selenoprotein.
The gene that codes for this protein has four duplications in the finch, of which we have only found two in Pyrgilauda JAGGNJ010016824 and JAGGNJ010000002.
Therefore, we conclude that two of the duplications were generated in the common ancestor, while the other two additional applications occurred after the divergence of Zebra finch and Pyrgilauda ruficollis, which is why the latter does not present it.
Looking at this data, we can conclude that Pyrgilauda ruficollis has conserved the two copies of the TR1 gene that were present in the common ancestor with Zebra finch. In addition, these two predicted proteins are both selenoproteins, as they have Sec residues and predicted SECIS elements.
TR2
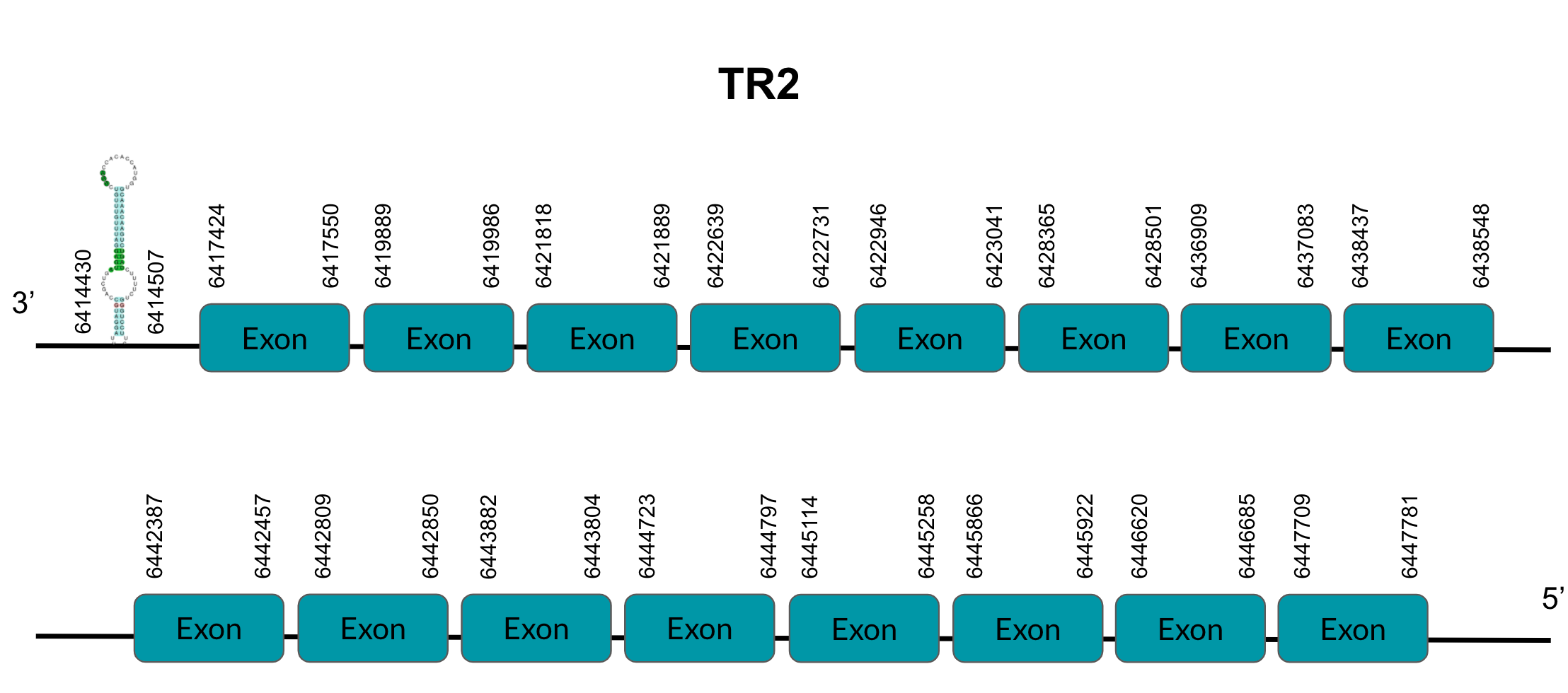
TR2 is a protein responsible for thioredoxin reduction. After tBLASTn prediction, 4 scaffolds showed significant hits but the one with the lowest E-value (2,67E-01), highest identity score (87,80%) and better T-coffee score (999) was selected: JAGGNJ010022944. The predicted gene is located between positions 6417423-6447781 in the negative strand, and contains 16 exons, located in positions 6417424-6417550, 6419889-6419986, 6421818-6421889, 6422639-6422731, 6422946-6423041, 6428365-6428501, 6436909-6437083, 6438437-6438548, 6442387-6442457, 6442809-6442850, 6443804-6443882, 6444723-6444797, 6445114-6445258, 6445866-6445922, 6446620-6446685, 6447709-6447781, all of them in the negative strand.
It is also worth mentioning that it contains a Sec residue 520, the same position as the query protein, taken in this case from the Finch. In addition, the protein starts with Methionine. SECISearch3 predicted a grade A SECIS element between the positions 6414430-6414507, at the 3’ UTR end of the negative strand. Seblastian also predicted a selenoprotein.
This gene presents duplications in other genomic regions (JAGGNJ010000002, JAGGNJ010003778 and JAGGNJ010016824), where all of them have a region that, even though it is conserved, lacks N-terminal, C-terminal or both, leading us to conclude that these duplications are not functional.
Looking at this data, we can conclude that Pyrgilauda ruficollis has conserved the TR2 gene, and it is still a selenoprotein, as it has a Sec residue and a predicted SECIS element.
TR3
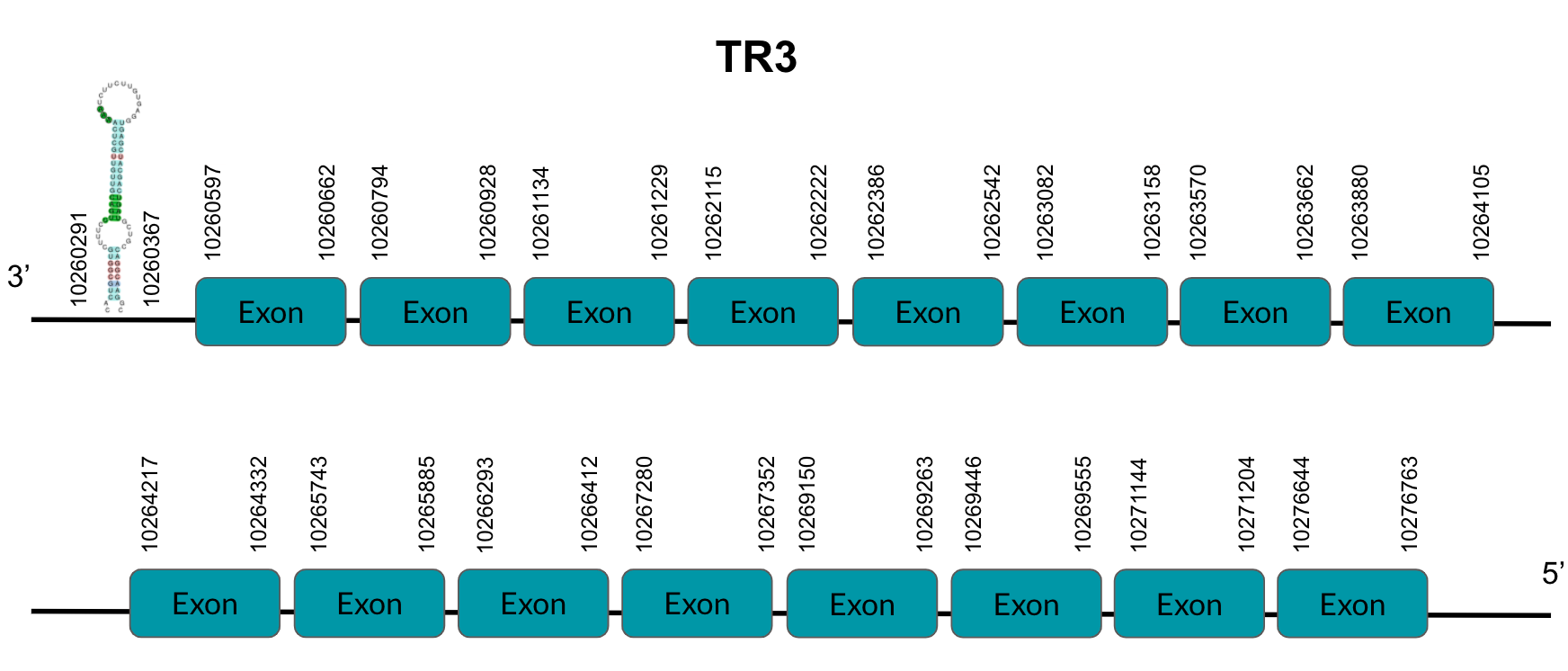
TR3 is a protein responsible for thioredoxin reduction. After tBLASTn prediction, 5 scaffolds showed significant hits but the one with the lowest E-value (3,35E-06), highest identity score (83,53%) and better T-coffee alignment (T-coffee score of 999) was selected: JAGGNJ01000002.1. The predicted gene is located between positions 10260596-10276763 in the negative strand, and contains 16 exons, located in positions 10260597-10260662, 10260794-10260928, 10261134-10261229, 10262115-10262222, 10262386-10262542, 10263082-10263158, 10263570-10263662, 10263880-10264105, 10264217-10264332, 10265743-10265885, 10266293-10266412, 10267280-10267352, 10269150-10269263, 10269446-10269555, 10271144-10271204 and 10276644-10276763, all of them in the negative strand.
It is also worth mentioning that it contains a Sec residue in the position 605, the same position as the query protein, taken in this case from the Chicken. In addition, the protein starts with Methionine.
SECISearch3 predicted a grade A SECIS element between the positions 10260291-10260367, at the 3’ UTR end of the negative strand. Seblastian did not predict any selenoprotein.
Looking at this data, we can conclude that Pyrgilauda ruficollis has conserved the TR3 gene, and it is still a selenoprotein, as it has a Sec residue and a predicted SECIS element.
15-kDa selenoprotein (Sel15)
TSel15 is a protein that interacts with UGGT (an ER-resident chaperon involved in the quality control process given to proteins in the organelle). After tBLASTn prediction, 4 scaffolds showed significant hits but the one with the lowest E-value (2,50E-09), highest identity score (86,179%) and better T-coffee score (995) was selected: JAGGNJ010019611.1. The predicted gene is located between positions 13453840-13472922 in the negative strand, and contains 10 exons, located in positions 13472744-13472922, 13470220-13470338, 13469974-13470132, 13469281-13469434, 13465585-13465687, 13463308-13999437, 13463042-13463133, 13455877-13455970, 13455169-13455259 and 13453841-13454033, all of them in the negative strand.
It is also worth mentioning that it contains a Sec residue in the position 68, the same position as the query protein, taken in this case from the Finch. In addition, the protein doesn't start with a Methionine, but with a Valine, opening the possibility of having missed the beginning of the protein or that this beginning is located more downstream than in the query protein.
SECISearch3 predicted a grade A SECIS element in the positive strand between the positions 13474819-13474888, at the 3’ UTR of the positive strand, differing from the Sel15 selenoprotein in the sequence. Seblastian predicted a selenoprotein.
Looking at this data, we can conclude that Pyrgilauda ruficollis has conserved the Sel15 gene, and it is still a selenoprotein, as it has a Sec residue and a predicted SECIS element.

Selenoprotein I
SelI is a protein involved in the transfer of phosphoethanolamine from CDP-ethanolamine to diacylglycerol and produces phosphatidylethanolamine, which is involved in different functions like in lipid metabolism or protein folding. After tBLASTn prediction, 5 scaffolds showed significant hits but the one with the lowest E-value (3,00E-06), highest identity score (84,9%) and better T-coffee alignment (999) was selected: JAGGNJ010018890. The predicted gene is located between positions 7747828-7772605 in the positive strand, and contains 5 exons, located in positions 7747829-7748101, 7756824-7756932, 7764474-7764501, 7769250-7769451 and 7772414-7772605, all of them in the positive strand.
It is also worth mentioning that it contains a Sec residue in the position 196, the same position as the query protein, taken in this case from the Chicken. In addition, the protein does not start with Methionine, but with a Threonine, opening the possibility of having missed the beginning of the protein or that this beginning is located more downstream than in the query protein.
SECISearch3 predicted a grade B SECIS element between the positions 7836096-7836167, which is in the positive strand. Seblastian did not predict any selenoprotein.
This gene presents duplications in other genomic regions (JAGGNJ010016279, JAGGNJ010023500 and JAGGNJ010021056), where all of them have a region that, even though it is conserved, lacks N-terminal, C-terminal or both, leading us to conclude that these duplications are not functional.
Looking at this data, we can conclude that Pyrgilauda ruficollis has conserved the SelI gene, and it is still a selenoprotein, as it has a Sec residue and a predicted SECIS element.

Selenoprotein M
SelM protein has a role in neuroprotection and prevention of hydrogen peroxide-induced oxidative damage, but it has also been linked to a protective inhibition of beta-amyloid protein aggregation. After tBLASTn prediction, 2 scaffolds showed significant hits but the one with the lowest E-value (4,57E-27), a highest identity score (95,75%) and a better T-coffee score (989) was selected: JAGGNJ010022944.1. The predicted gene is located between positions 98603-99528 in the negative strand, and contains 4 exons, located in positions 99490-99528, 99372-99406, 98816-98894 and 98604-98726, all of them in the same strand.
It is also worth mentioning that it contains a Sec residue in the position 6, the same position as the query protein, taken in this case from the Finch. In addition, the protein does not start with Methionine, but with Glutamine, opening the possibility of having missed the beginning of the protein or that this beginning is located more downstream than in the query protein.
SECISearch3 predicted a grade A SECIS element between the positions 98194-98264, at the 3’ UTR end of the negative strand, coinciding with the SelM selenoprotein in the sequence. Seblastian did not predict any selenoprotein.
Looking at this data, we can conclude that Pyrgilauda ruficollis has conserved the SelM gene, and it is still a selenoprotein, as it has a Sec residue and a predicted SECIS element.

Selenoprotein N
SelN has been linked to skeletal muscle regeneration and maintenance of satellite cells, as well as a role as a ryanodine receptor cofactor, mediating the release of calcium from the sarcoplasmic reticulum during muscle contraction. After tBLASTn prediction, 1 scaffold with significant hits with a low E-value (1,16E-06), a high identity score (87,07%) and a good T-coffee score (998): JAGGNJ010006888.1. The predicted gene is located between positions 54264-66484 in the positive strand, and contains 12 exons, located in positions 54265-54363, 57083-57200, 58207-58337, 58773-58985, 59634-59758, 60470-60607, 61400-61481, 62396-62584, 63487-63592, 64655-64767, 65418-65519 and 66317-66484, all of them in the same strand.
It is also worth mentioning that it contains a Sec residue in the position 400, the same position as the query protein, taken in this case from the Finch. In addition, the protein does not start with Methionine, but with Proline, opening the possibility of having missed the beginning of the protein or that this beginning is located more downstream than in the query protein.
SECISearch3 predicted a grade A SECIS element between the positions 68395-68465, at the 3’ UTR end of the positive strand, coinciding with the SelN selenoprotein in the sequence. Seblastian predicted a selenoprotein.
Looking at this data, we can conclude that Pyrgilauda ruficollis has conserved the SelM gene, and it is still a selenoprotein, as it has a Sec residue and a predicted SECIS element.

Selenoprotein O
We do not know a lot about SelO, but we do know that it has a kinase domain and a mitochondrial targeting peptide. Cys-homologues have been found in other species. After tBLASTn prediction, 1 scaffold showed a significant hit, with a low E-value (4,59E-19), a high identity score (86,03%) and a good T-coffee score (999): JAGGNJ010015945.1. The predicted gene is located between positions 8690962-8690962 in the positive strand, and contains 9 exons, located in positions 8701858-8702270, 8700318-8700521, 8699187-8699367, 8695998-8696128, 8695348-8695628, 8694858-8695008, 8692915-8693100, 8691829-8691985 and 8690963-8691154, all of them in the positive strand.
It is also worth mentioning that it contains a Sec residue in the position 630, the same position as the query protein, taken in this case from the Finch. In addition, the protein does not start with a Methionine, but with Tryptophan, opening the possibility of having missed the beginning of the protein or that this beginning is located more downstream than in the query protein.
SECISearch3 predicted a B SECIS element between the positions 8750327-8750327, at the 3’ UTR end of the positive strand, differing from the SelO selenoprotein in the sequence. Seblastian did not predict any selenoprotein.
The gene that codes for this protein has four duplications in the finch, of which only one (SPP00002541_2.0) conserves selenocysteine. Regarding the annotation of these proteins in the Pyrgilauda’s genome:
Looking at this data, we can conclude that Pyrgilauda ruficollis has conserved two of the four duplication from the Zebra finch, meaning this that two duplications occurred in the Zebra finch posteriorly to the divergence between Zebra finch and Pyrgilauda ruficollis . However, the both predicted copies of SelO in Pyrgilauda ruficollis presented SECIS element prediction and Sec residues, so we can say that they are still selenoproteins.

Selenoproteins P
SelP has been suggested to have a role in transportation of Se to peripheral tissues, specially brain and testis. After tBLASTn prediction, 2 scaffolds showed significant hits but the one with the lowest E-value (3,00E-09), a highest identity score (62,3%) and a better T-coffee score (972) was selected: JAGGNJ010023167.1. The predicted gene is located between positions 304401-308661 in the negative strand, and contains 4 exons, located in positions 308471-308661, 307175-307387, 305476-305593 and 304402-304629, all of them in the negative strand.
It is also worth mentioning that it contains a Sec residue in the position 37, the same position as the query protein, taken in this case from the Chicken. In addition, the protein starts with a Methionine.
SECISearch3 predicted a grade A SECIS element between the positions 303592-303663, at the 3’ UTR end of the negative strand, differing from the SelP selenoprotein in the sequence. Seblastian predicted a selenoprotein.
The gene that codes for this protein has a duplication in the finch, of which SPP00002543 has lost a large part of its selenocysteine content, while SPP00002544 preserves multiple residues of Sec. Regarding the annotation of these selenoproteins in the Pyrgilauda genome, SPP00002543_2.0 is found in scaffold JAGGNJ010019611 and SPP00002544_2.0 is found in scaffold JAGGNJ010023167, both of them well annotated and conserving their selenocysteine content.
Looking at this data, we can conclude that Pyrgilauda ruficollis has conserved the both copies of the SelP gene observed in Zebra finch, and they are still selenoproteins, as they both have Sec residues and a predicted SECIS elements.

Seloproteina R (SelRs)
SelR1
Methionine-R-sufoxide reductase 1 or SelR1 is an antioxidant protein that catalyzes the reduction of methionine sulfoxides in a NADPH-dependent reaction. After tBLASTn prediction, 1 scaffold showed significant hits, with a low E-value (7,07E-40), a high identity score (59,69%) and a good T-coffee score (992): JAGGNJ010002445.1. The predicted gene is located between positions 19819-18733 in the negative strand, and contains 3 exons, located in positions 19756-19810, 18977-19119 and 18734-18850, all of them in the negative strand.
It is also worth mentioning that it contains a Sec residue in the position 92, the same position as the query protein, taken in this case from the Chicken. SECISearch3 predicted a grade A SECIS element between the positions 17208-17280, which is in the negative strand. Seblastian also predicted a selenoprotein.
Looking at this data, we can conclude that Pyrgilauda ruficollis has conserved the SelR1 gene, and it is still a selenoprotein, as it has a Sec residue and a predicted SECIS element.

SelR2
Methionine-R-sufoxide reductase 2 or SelR2 is an antioxidant protein that catalyzes the reduction of methionine sulfoxides in a NADPH-dependent reaction. After tBLASTn prediction, 2 scaffolds showed significant hits but the one with the lowest E-value (9,08E-05), highest identity score (78,97%) and better T-coffee score (998) was selected: JAGGNJ010002222.1. The predicted gene is located between positions 15641230-15649002 in the negative strand, and contains 4 exons, located in positions 15641231-15641323, 15643795-15643942, 15647882-15647958 and 15648931-15649002, all of them in the negative strand.
It is also worth mentioning that the protein does not contain a Sec residue, as we expected since it has been described to be a Cysteine-containing homologous protein. In addition, the protein does not start with Methionine, but with Serine, opening the possibility of having missed the beginning of the protein or that this beginning is located more downstream than in the query protein. SECISearch3 does not predict any SECIS elementsand Seblastian does not predict a selenoprotein.
The prediction of this gene was made from the query protein of the Chicken, as it was not present in Zebra finch. However, the N-terminal region of the query is lost in Pyrgilauda ruficollis, which can lead to a loss of function of this protein.
Looking at this data, we can conclude that Pyrgilauda ruficollis has conserved the SelR2 gene, and it is still a Cysteine-containing homologous protein, as we have seen in the Chicken.

SelR3
Methionine-R-sufoxide reductase 3 or SelR3 is an antioxidant protein that catalyzes the reduction of methionine sulfoxides in a NADPH-dependent reaction. After tBLASTn prediction, only 1 scaffold showed significant hits with a low E-value (3,08E-10), a high identity score (90,72%) and a good T-coffee score (1000): JAGGNJ010009996.1. The predicted gene is located between positions 4567596-4612435 in the positive strand, and contains 5 exons, located in positions 4567597-4567709, 4569521-4569598, 4580214-4580242, 4607194-4607291 and 4612274-4612435, all of them in the positive strand.
It is also worth mentioning that the protein does not contain a Sec residue, as we expected since it has been described to be a Cysteine-containing homologous protein. In addition, the protein does not start with Methionine, but with Alanine, opening the possibility of having missed the beginning of the protein or that this beginning is located more downstream than in the query protein. SECISearch3 does not predict any SECIS elements and Seblastian does not predict a selenoprotein.
Regarding duplication events, this gene presents is found in the scaffold JAGGNJ010002222, lacking N-terminal, C-terminal or both regions, leading us to conclude that this duplication is not functional.
Looking at this data, we can conclude that Pyrgilauda ruficollis has conserved the SelR3 gene, and it is still a Cysteine-containing homologous protein, as we have seen in the Chicken.

Selenoproteins K and S
Selenoprotein K
SelK is a protein that has been suggested to be implicated in the ER-associated degradation (ERAD) of misfolded proteins, inflammation and immune response. Regarding this first function, it has been discovered that their genes have functional ER stress response elements and their expression is upregulated when misfolded proteins are accumulated in the ER. After tBLASTn prediction, only 1 scaffold showed a significant hit, with a very low E-value (7,36E-11), a high identity score (90,62%) and a good T-coffee score (985): JAGGNJ010000002.1. The predicted gene is located between positions 13600790-13602929 in the positive strand, and contains 4 exons, located in positions 13600791-13600809, 13601357-13601447, 13602158-13602238 and 13602836-13602929, all of them in the positive strand.
It is also worth mentioning that it contains a Sec residue in the position 93, the same position as the query protein, taken in this case from the Finch. In addition, the protein starts with a Methionine.
SECISearch3 predicted a grade A SECIS element between the positions 13604233-13601239, at the 3’ UTR end of the positive strand, coinciding with SelK selenoprotein in the sequence. Seblastian predicted a selenoprotein.
Looking at this data, we can conclude that Pyrgilauda ruficollis has conserved the SelK gene, and it is still a selenoprotein, as it has a Sec residue and a predicted SECIS element.

Selenoprotein S
SelS is a protein that is involved in the degradation process of misfolded endoplasmic reticulum (ER) luminal proteins. After tBLASTn prediction, only one scaffold showed a significant hit, with a low E-value (6,00E-05), a high identity score (94,73%) nd a good T-coffee score (984): JAGGNJ010014440.1. The predicted gene is located between positions 2133112-2136337 in the positive strand, and contains 4 exons, located in positions 2136255-2136337, 2134229-2134304, 2134049-2134138 and 2133113-2133220, all of them in the negative strand.
It is also worth mentioning that it contains a Sec residue in the position 20, the same position as the query protein, taken in this case from the Finch. In addition, the protein does not start with Methionine, but with a Glutamic Acid, opening the possibility of having missed the beginning of the protein or that this beginning is located more downstream than in the query protein.
SECISearch3 predicted a grade A SECIS element between the positions 2136894-2136982, at the 3’ UTR end of the positive strand. Seblastian also predicted a selenoprotein.
Looking at this data, we can conclude that Pyrgilauda ruficollis has conserved the SelS gene, and it is still a selenoprotein, as it has a Sec residue and a predicted SECIS element.

Selenoproteins H, T, V and W
SelH
SelH is a protein that acts like an oxidoreductase. After tBLASTn prediction, just 1 scaffold showed a significant hit, with a low E-value (4,81E-38), a high identity score (74,04%) and a good T-coffee score (992): JAGGNJ010009663.1. The predicted gene is located between positions 302816-303252 in the negative strand, and contains 3 exons, located in positions 303206-303252, 302984-303129 and 302817-302911, all of them in the negative strand.
It is also worth mentioning that it contains a Sec residue in the position 19, the same position as the query protein, taken in this case from the Finch. In addition, the protein does not start with a Methionine, but with a Glutamic Acid, opening the possibility of having missed the beginning of the protein or that this beginning is located more downstream than in the query protein.
SECISearch3 predicted a grade B SECIS element between the positions 302612-302703, at the 3’ UTR end of the negative strand, coinciding with SelH selenoprotein in the sequence. Seblastian predicted a selenoprotein.
Looking at this data, we can conclude that Pyrgilauda ruficollis has conserved the SelH gene, and it is still a selenoprotein, as it has a Sec residue and a predicted SECIS element.

SelT
SelT is a protein is a thioredoxin-like enzyme that is found at the endoplasmic reticulum membrane. After tBLASTn prediction, only 1 scaffold showed a significant hit, with a low E-value (2,8E-11), a high identity score (92,76%) and a good T-coffee score (996): JAGGNJ010012219.1. The predicted gene is located between positions 316773-321460 in the positive strand, and contains 5 exons, located in positions 321339-321460, 320551-320638, 319170-319296, 317965-318075 and 316774-316856, all of them in the positive strand.
It is also worth mentioning that it contains a Sec residue in the position 31, the same position as the query protein, taken in this case from the Finch. In addition, the protein does not start with Methionine, but with an Alanine, opening the possibility of having missed the beginning of the protein or that this beginning is located more downstream than in the query protein.
SECISearch3 predicted a grade A SECIS element between the positions 322151-322233, at the 3’ UTR end of the positive strand. Seblastian also predicted a selenoprotein.
Looking at this data, we can conclude that Pyrgilauda ruficollis has conserved the SelT gene, and it is still a selenoprotein, as it has a Sec residue and a predicted SECIS element.

SelW1
SelW1 is a protein involved in redox regulation of 14-3-3 proteins. Since SelW1 is not present at the query protein list available in SelenoDB neither in Finch nor Chicken, we tried to annotate the human SelW1 protein sequence in the genome of Pyrgilauda ruficollis. However, no significant hits were obtained after tBLASTn prediction.
This data seems to indicate that SelW1 was lost in a common ancestor previous to the divergence of the Finch lineage and the Chicken lineage, so it is obvious that Pyrgilauda ruficollis does not either present this selenoprotein.

SelW2
SelW2 is a protein involved in redox regulation of 14-3-3 proteins. After tBLASTn prediction, only 1 scaffold showed significant hits, with a low E-value (1,98E-08), a high identity score (85,39%) and a good T-coffee score (989): JAGGNJ010006777.1. The predicted gene is located between positions 1421184-1422517 in the positive strand, and contains 4 exons, located in positions 1421185-1421207, 1421845-1421942, 1422165-1422241, 1422437-1422517, all of them in the positive strand.
It is also worth mentioning that the protein does not contain a Sec residue, as we expected since it has been described to be a Cysteine-containing homologous protein.. In addition, the protein does not start with Methionine, but with Valine, opening the possibility of having missed the beginning of the protein or that this beginning is located more downstream than in the query protein. In fact, the protein that we have predicted has lost the N-terminal region of the query.
SECISearch3 predicted a grade B SECIS element between the positions 1520618-1520722, at the 3’ UTR end of the positive strand, coinciding with the SelW2 selenoprotein in the sequence. Seblastian did not predict any selenoprotein.
Looking at this data, we can conclude that Pyrgilauda ruficollis has conserved the sequence of the SelW2 gene from the Human, but lacking the N-terminal region, which may be an indication of a loss of function in Pyrgilauda ruficollis. In addition, assuming a not loss of function of the gene it would code for the Cysteine-containing homologous protein.

SelV
SelV is a selenoprotein thought to have evolved from SelW duplication. It has been suggested to play a role in male placental mammal’s reproduction. In this sense, it is quite obvious that neither Finch nor Chicken have SelV in their selenoproteome. Consequently, we tried to annotate the human SelV protein sequence in the genome of Pyrgilauda ruficollis. However, no significant hits were obtained after tBLASTn prediction.
This data seems to indicate that SelV was lost in a common ancestor previous to the divergence of the Finch lineage and the Chicken lineage (probably in the separation of the lineages dividing placental mammals from other species), so it is obvious that Pyrgilauda ruficollis does not present this selenoprotein either.

SEL U family (SelU1, SelU2 and SelU3)
SelU family is a selenoprotein family that belongs to the thioredoxin-like superfamily, and they are implicated in redox homeostasis. Since SelU proteins were not present at the query protein list available in SelenoDB neither in Finch nor Chicken, we tried to annotate the human SelU1, SelU2 and SelU3 protein sequences in the genome of Pyrgilauda ruficollis. However, no significant hits were obtained after tBLASTn prediction.
This data seems to indicate that the SelU family was lost in a common ancestor previous to the divergence of the Finch lineage and the Chicken lineage, so it is obvious that Pyrgilauda ruficollis does not either present these selenoproteins.



Machinery
Eukaryotic elongation factor (eEFsec)
After tBLASTn prediction, only 1 scaffold showed significant hits, with a low E-value (1,86E-07), a high identity score (84,89%) and a good T-coffee score (99): JAGGNJ010000002.1. The predicted gene is located between positions 11183441-1123440 in the positive strand, and contains 7 exons, located in positions 11233441-11233551, 11233672-11233702, 11266143-11266350, 11268363-11268459, 11270032-11270196, 11298532-11299173 and 11313908-11313985, all of them in the positive strand.
It is also worth mentioning that it does not contain a Sec residue, as expected in all machinery proteins. In addition, the protein does not start with Methionine, but with Valine, opening the possibility of having missed the beginning of the protein or that this beginning is located more downstream than in the query protein.
SECISearch3 did not predict any SECIS element, but we ignored them since machinery proteins do not have selenocysteine in their sequence. Seblastian did not predict any selenoprotein.
Looking at this data, we can conclude that Pyrgilauda ruficollis has conserved the sequence of the eEFSec machinery protein.

Phosphoseryl-tRNA kinase (PSTK)
PSTK is a protein responsible for phosphorylating seryl-tRNA[Ser]Sec to O-phosphoseryl-tRNA[Ser]Sec. After tBLASTn prediction, only 1 scaffold showed significant hits, with a low E-value (1,99E-09), a high identity score of (68,80%) and a good T-coffee score (989): JAGGNJ010005555.1. The predicted gene is located between positions 4009476-4012476 in the negative strand, and contains 6 exons, located in positions 4012285-4012479, 4011848-4012106, 4010918-4011126, 4010429-4010524, 4010208-4010301 and 4009477-4009568, all of them in the negative strand.
It is also worth mentioning that it does not contain a Sec residue, as expected in all machinery proteins. In addition, the protein does not start with Methionine, but with Glycine, opening the possibility of having missed the beginning of the protein or that this beginning is located more downstream than in the query protein.
SECISearch3 predicted a grade B SECIS element between the positions 4004199-4004281, at the 3’ UTR end of the negative strand, coinciding with the PSTK selenoprotein in the sequence. However, we ignored this SECIS element since machinery proteins do not have selenocysteine in their sequence. Seblastian did not predict any selenoprotein.
Looking at this data, we can conclude that Pyrgilauda ruficollis has conserved the sequence of the PSTK machinery protein.
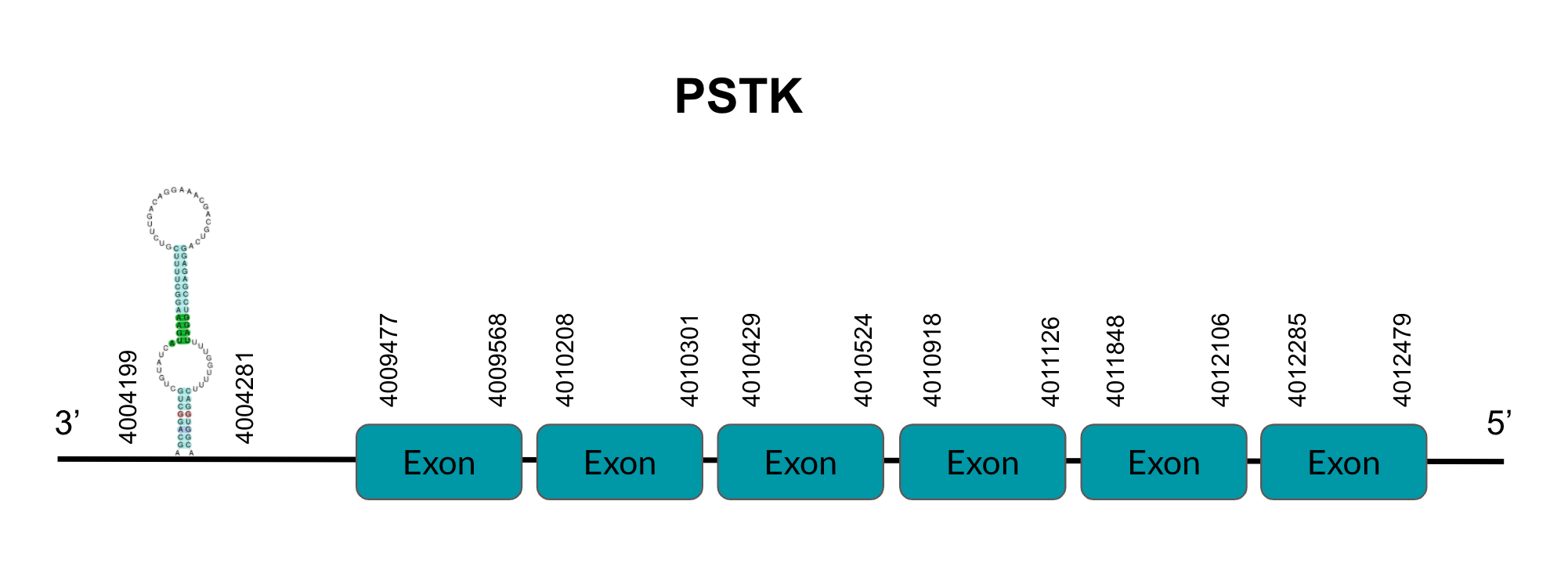
SECIS binding protein 2 (SBP2)
SBP2 is an essential protein for the upstream stabilization of selenoprotein mRNA and selenoprotein translation because of its binding with the SECIS element and the eEFSec. After tBLASTn prediction, 2 scaffolds showed significant hits but the one with the lowest E-value (2,27E-06), highest identity score (89,20%) and the better T-coffee score (1000) was selected: JAGGNJ010014440. The predicted gene is located between positions 9729759-9744646 in the positive strand, and contains 15 exons, located in positions 9729760-9729941, 9730510-9730834, 9730951-9731086, 9731480-9731713, 9732557-9732690, 9734455-9734586, 9735106-9735189, 9735997-9736164, 9736316-9736457, 9738053-9738222, 9739296-9739428, 9740401-9740563, 9741263-9741483, 9742129-9742283, 9742773-9742992 and 9743976-9744646, all of them in the positive strand.
It is also worth mentioning that it does not contain a Sec residue, as expected in all machinery proteins. In addition, the protein does not start with Methionine, but with Lysine, opening the possibility of having missed the beginning of the protein or that this beginning is located more downstream than in the query protein.
SECISearch3 surprisingly predicted a grade B SECIS element between the positions 9828915-9828993, at the 3’ UTR end of the negative strand. However, we ignored this SECIS element since machinery proteins do not have selenocysteine in their sequence. Seblastian did not predict any selenoprotein. Seblastian did not predict any selenoprotein.
The gene that codes for this protein has a duplication in the Zebra finch, but it is observed to have exactly the same hits for both queries in Pyrgilauda ruficollis. For this reason, we assume that one of the two copies of the SBP2 gene has been deleted in the bird. Since the query SPP00002528 is perfectly aligned in the genome, while the SPP00002529 is truncated, it is evident that it is the latter that has been lost in Pyrgilauda ruficollis.
Looking at this data, we can conclude that Pyrgilauda ruficollis has conserved one of the two copies of SBP2 gene observed in Zebra finch and it conserves its role in selenoprotein synthesis machinery.
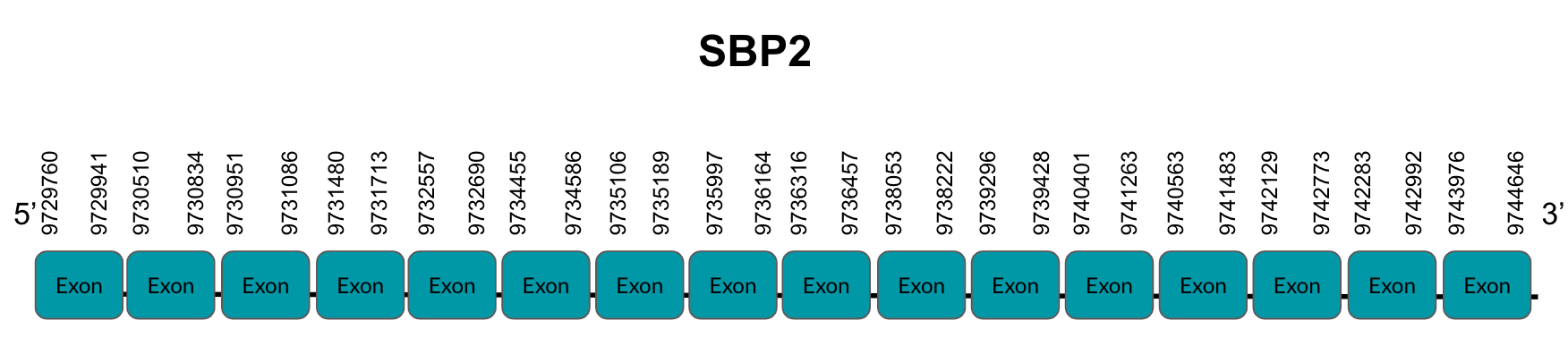
tRNA Sec 1 associated protein 1 (SECp43)
Secp43 or tRNA Sec 1 associated protein 1 is a machinery protein that acts as a cofactor in selenoprotein expression. After tBLASTn prediction, 3 scaffolds showed significant hits but the one with the lowest E-value (6,80E-15), highest identity score (95,51%) and better T-coffee score (1000) was selected: JAGGNJ010018278.1. The predicted gene is located between positions 83818-85294 in the positive strand, and contains 2 exons, located in positions 83819-83939 and 85101-85294.
It is also worth mentioning that it does not contain a Sec residue, as expected in all machinery proteins. the protein does not start with Methionine, but with an Isoleucine, opening the possibility of having missed the beginning of the protein or that this beginning is located more downstream than in the query protein. It is one of the longest proteins with a very good alignment.
SECISearch3 predicted a grade B SECIS element between the positions 176731-176797, at the 3’ UTR end of the positive strand, coinciding with the Secp43 selenoprotein in the sequence. However, we ignored this SECIS element since machinery proteins do not have selenocysteine in their sequence. Seblastian did not predict any selenoprotein.
The gene that codes for this protein is the one found in the JAGGNJ010018278 scaffold. In addition, it presents duplications in other regions of the genome (JAGGNJ010016835 and JAGGNJ010004734) in which all present a region that is conserved but lack the N-terminal region, C-terminal or both, leading us to conclude that these duplications are not functional.
Looking at this data, we can conclude that Pyrgilauda ruficollis has conserved the sequence and the role of the Secp43 machinery protein.
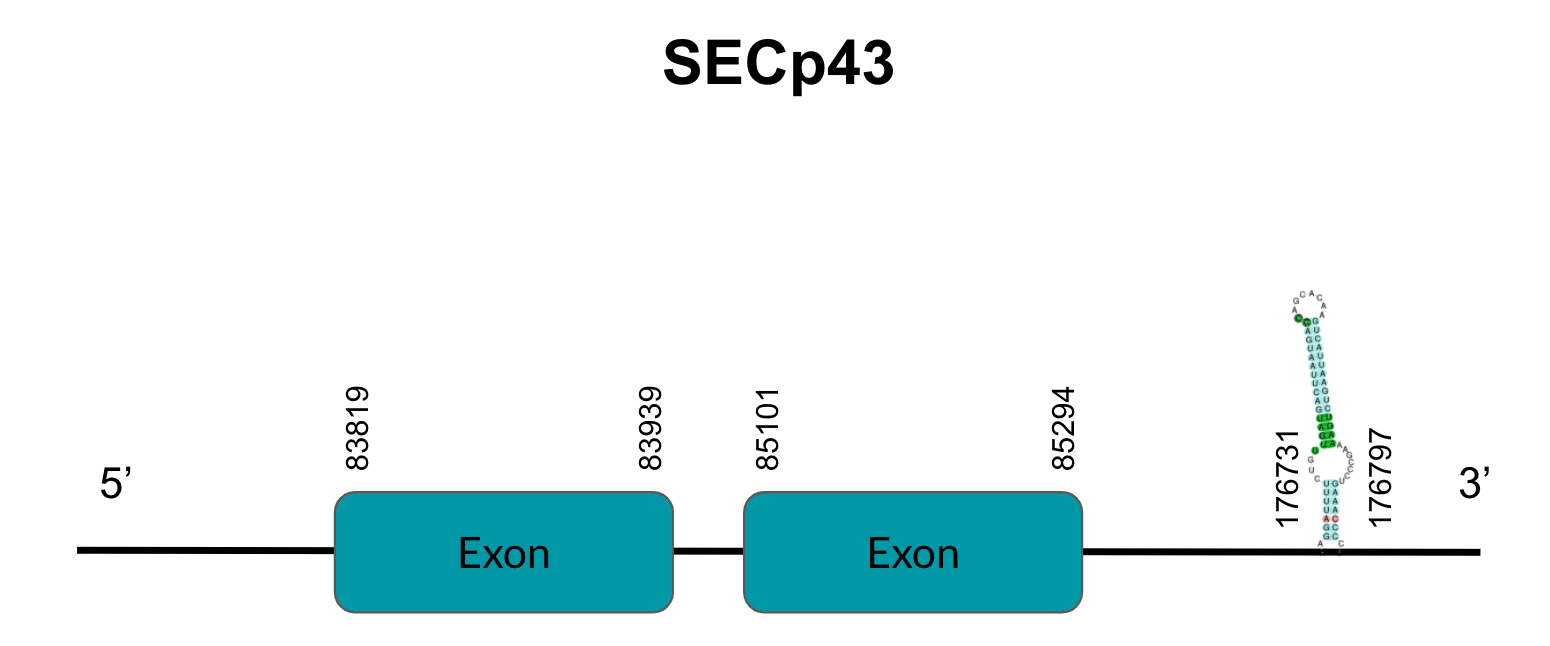
Selenocysteine synthase (SecS)
SecS is a protein which catalyzes the conversion of Ser-tRNA[Ser]Sec to selenocysteyl-tRNA[Ser]Sec, which has a selenophosphate (meaning this enzyme incorporates Se's active form). After tBLASTn prediction, just 1 scaffold showed a significant hit, with a low E-value (4,11E-31), a high identity score (100) and better T-coffee alignment (T-coffee score of 1000): JAGGNJ010000001.1. The predicted gene is located between positions 13472739-13472922 in the negative strand, and contains just one exon, located in positions 50000-50182.
It is also worth mentioning that it does not contain a Sec residue, as expected in all machinery proteins. In addition, the protein does not start with Methionine, but with Isoleucine, opening the possibility of having missed the beginning of the protein or that this beginning is located more downstream than in the query protein. It is one of our longest proteins with one of the best alignments.
SECISearch3 predicted an B grade SECIS element between the positions 13429517-13429591, at the 3’ UTR end of the positive strand. However, we ignored this SECIS element since machinery proteins do not have selenocysteine in their sequence. Seblastian did not predict any selenoprotein. Seblastian did not predict any selenoprotein.
Looking at this data, we can conclude that Pyrgilauda ruficollis has conserved the sequence and the role of the SecS machinery protein.

Selenophosphate synthetase 1 (SPS1)
SPS1 is a selenophosphate protein that depends on a selenium salvage system that recycles selenocysteine. After tBLASTn prediction, 3 scaffolds showed significant hits but the one with the lowest E-value (1,35E-10), highest identity score (86,19%) and better T-coffee score (1000) was selected: JAGGNJ010019723. The predicted gene is located between positions 2894076-2913399 in the negative strand, and contains 8 exons, located in positions 2894077-2894288, 2898938-2899150, 2900674-2900773, 2903693-2903783, 2904593-2904747, 2905511-2905618, 2910027-2910130 and 2913207-2913399, all of them in the negative strand.
It is also worth mentioning that it does not contain a Sec residue, as expected in all machinery proteins. In addition, it starts with Methionine. SECISearch3 surprisingly predicted a grade B SECIS element between the positions 2918285-2918353, at the 5’ UTR end of the negative strand, which makes no sense according to the SECIS element definition. Moreover, we ignored this SECIS element since machinery proteins do not have selenocysteine in their sequence. Seblastian did not predict any selenoprotein.
In addition to our main hit in tBLASTn, this gene presents some duplications in other regions of the genome: scaffolds JAGGNJ010015541 and JAGGNJ010016813. However, all of them have a region that, even though it is conserved, lacks N-terminal, C-terminal or both, leading us to conclude that these duplications are not functional.
Looking at this data, we can conclude that Pyrgilauda ruficollis has conserved the sequence and the role of the SPS1 machinery protein.
Selenophosphate synthetase 2 (SPS2)
SPS2 is a selenophosphate protein that depends on a selenium salvage system that recycles selenocysteine. After tBLASTn prediction, 1 scaffold showed significant hits, with a low E-value (1,77E-05), a high identity score (67,77%) and a good T-coffee score (998): JAGGNJ010019723.1. The predicted gene is located between positions 2894100-2910131 in the negative strand, and contains 7 exons, located in positions 2894101-2894288, 2898938-2899150, 2900674-2900773, 2903693-2903783, 2904593-2904747, 2905511-2905618 and 2910027-2910131, all of them in the negative strand.
It is also worth mentioning that it does not contain a Sec residue, as expected in all machinery proteins. In addition, the protein starts with Methionine. SECISearch3 did not predict any SECIS element, as was also expected in a machinery protein. Seblastian did not predict any protein.
Looking at this data, we can conclude that Pyrgilauda ruficollis has conserved the sequence and the role of the SPS2 machinery protein.
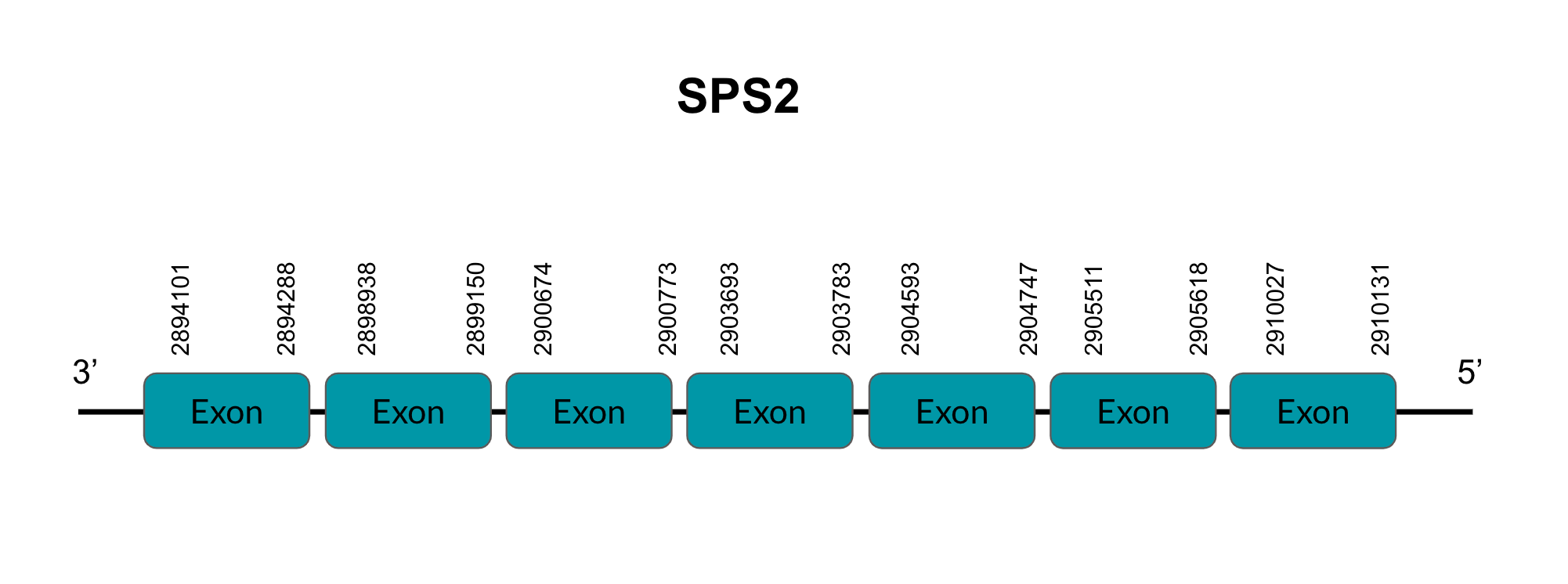
Conclusions
Our project’s main objective was identifying accurately the selenoproteins and associated selenoprotein machinery in Pyrgilauda ruficollis’ genome. As it was previously mentioned, our reference species were chosen according to phylogenetic proximity and quality of the annotation in its genome. With these criteria, we established that our reference species was Taenigypia guttata , commonly known as Zebra finch, but in those cases where the alignment was meaningless, we used Gallus gallus (chicken) or Homo sapiens (human) as the reference species.
Here we report a comprehensive comparative computational analysis of each of these query proteins, that reveal novel insights into the characterization of Sec in this organism. To further extend the reach of our discoveries, we created a webpage in HTML language and updated the Wikipedia webpage for the rufous-necked snowfinch.
We studied 43 proteins in total and the alignment was: 27 with finch, 6 with chicken (PSTK, TR3, SelR1, eEFSec, SelI and GPx7) and 5 with human (GPx5, GPx6, SelW2, SPS2 and SelR2). Others did not align with any of the three. The following characterizations was obtained:
- Selenoproteins: GPx1, GPx2, GPx3, GPx4, DIO1, DIO2, DIO3, TR1, TR2, TR3, Sel15, SelH, SelI, SelK, SelM, SelN, SelO, SelP, SelR1, SelS and SelT.
- Cys-containing homologues: GPx7, GPx8, MsrA, SelR2, SelR3, SelW2.
- Selenoprotein machinery: PSTK, SecS, SBP2, secp43, eEFSec, SPS1, SPS2.
- Proteins that were not predicted: GPx5*, GPx6*, SelV, SelW1, SelU1, SelU2 and SelU3.
- Not expected proteins: MsrB1 and MsrB2.
*GPx5 and GPx6 did align, but it was because their sequence is very similar to GPx3. Actually, Pyrgilauda does not have these genes in their selenogenome.
If we analyze the obtained results, we can determine that we successfully identified and characterized 21 conserved selenoproteins in Pyrgilauda ruficollis , as well as 6 more Cys-containing homolog proteins and 7 machinery proteins.
What we can extract after taking a close look at the analysis’ results would be the following: first of all, that 7 selenoproteins were not found in Pyrgilauda’s genome, implying that their origin occurs later than the species’ divergence from the ancestor. Furthermore, we have not predicted any conversion from selenoprotein to Cys-homologue, which is one of the main contributing processes to the reduction of the selenoproteome.
Additionally, several selenoprotein genes are found duplicated in Pyrgilauda ruficollis, but most of these gene duplications are truncated or incomplete, leading to non functional proteins. Important duplication events were reported in GPx7, SelO, GPx2, etc.
In other cases, gene duplication events occured after the divergence of Pyrgilauda ruficollis and the different reference species, as we see, for instance, in the SBP2 gene.
However, some duplications present in Zebra finch were fully conserved in Pyrgilauda ruficollis, as we can observe in the SelP gene.
We successfully predicted and characterized most of the proteins, but some of them did not start with a Methionine residue, so further research should be conducted regarding this matter. It is possible that this is due to the fact that SelenoDB’s queries proteins do not have a Methionine residue at the beginning either. If this is the case, then a better characterization work of the queries should be done. The fact that some proteins were missing a large number of amino acids made it difficult to obtain a high quality prediction, which decreases the reliability of our results.
We also encountered problems with the Seblastian and SECISearch3 softwares: selenoprotein and SECIS elements’ prediction was not coincident with our results, meaning that there were selenoproteins predicted with our methodology that were not predicted with Seblastian. In light of this, we decided to consider our results trust-worthy because SECISearch3 and Seblastian’s methodology is based on databases of different species to do automatic predictions based on algorithms, and this could lead to not predicting a SECIS or a selenoprotein even if it exists. As a consequence, we have determined that those two softwares’ use would be in corroborating and supporting our own results, obtained with T-coffee.
Regarding further directions of the project, we would like to mention that it is necessary to have more bird species’ genomes well annotated, since the knowledge of this kingdom is much more limited than in others. As beneficial complementary information, more distant species’ genomes should also be annotated since SelenoDB does not have many bird species. This would enable the scientific community to increase the knowledge regarding the evolution of the selenogenome through phylogenetic trees.
Our study suggests that Pyrgilauda ruficollis’ selenoproteome is quite conserved compared with the one of Taenigypia guttata’s . We obtained high identity percentages between sequences, suggesting that the number of mutational changes that cause a change in the amino acid sequence had not been significant, evidencing high homology between both selenoproteomes. Revising our global results, we conclude that our analysis has been successful enough because a high number of proteins have been well predicted in Pyrgilauda ruficollis , and we are thrilled to have contributed, even if only a little, to the current selenoproteome knowledge. We sincerely hope it keeps expanding.
References
(1) Rayman M. The importance of selenium to human health. The Lancet. 2000;356(9225):233-241
(2) Labunskyy V, Hatfield D, Gladyshev V. Selenoproteins: Molecular Pathways and Physiological Roles. Physiological Reviews. 2014;94(3):739-777.
(3) Mariotti M, Ridge P, Zhang Y, Lobanov A, Pringle T, Guigo R et al. Composition and Evolution of the Vertebrate and Mammalian Selenoproteomes. PLoS ONE. 2012;7(3):e33066.
(4) Kasaikina MV, Hatfield DL, Gladyshev VN. Understanding selenoprotein function and regulation through the use of rodent models. Biochim Biophys Acta. 2012;1823(9):1633-42. doi: 10.1016/j.bbamcr.2012.02.018.
(5) Chellan B, Zhao L, Landeche M, Carmean CM, Dumitrescu AM, Sargis RM. Selenocysteine insertion sequence binding protein 2 (Sbp2) in the sex-specific regulation of selenoprotein gene expression in mouse pancreatic islets. Sci Rep. 2020;10(1):18568. doi: 10.1038/s41598-020-75595-4.
(6) Copeland PR, Fletcher JE, Carlson BA, Hatfield DL, Driscoll DM. A novel RNA binding protein, SBP2, is required for the translation of mammalian selenoprotein mRNAs. EMBO J. 2000;19(2):306-14. doi: 10.1093/emboj/19.2.306.
(7) Mariotti M, Lobanov A V., Manta B, Santesmasses D, Bofill A, Guigo R, et al. Lokiarchaeota Marks the Transition between the Archaeal and Eukaryotic Selenocysteine Encoding Systems. Mol Biol Evol. 2016;33(9):2441-53.
(8) Lobanov A V., Fomenko DE, Zhang Y, Sengupta A, Hatfield DL, Gladyshev VN. Evolutionary dynamics of eukaryotic selenoproteomes: Large selenoproteomes may associate with aquatic life and small with terrestrial life. Genome Biol. 2007;8(9).
(9)Papp L, Holmgren A, Khanna K. Selenium and Selenoproteins in Health and Disease. Antioxidants & Redox Signaling. 2010;12(7):793-795.
(10) Jiang L, Ni J, Liu Q. Evolution of selenoproteins in the metazoan. BMC Genomics. 2012;13(1):446.
(11) Clement P, Harris A, Davis A. Finches & Sparrows. London: Christopher Helm; 2013.
Acknowledgements
First of all, we'd like to thank our tutor Valentina del Olmo for her wonderful assistance with many of the coding and approaching challenges that arose.
We are also extremely grateful for Vasilis Ntasis’s help at understanding the magic of computer language, allowing us to save many hours of manual tasks
We'd also want to thank Roderic Guigó, Robert Castelo, Cedric Notredame and Toni Gabaldón, our teachers, for presenting us this challenge: it has made us, undoubtedly, grow as students and future scientists.
Last, but not least, we would like to thank our classmates for their invaluable help. They were the live demonstration of the quote “great minds think alike” , like once said Dr. Francesc Calafell. Our classmates and many teachers are the main reason we are here today.
Again, thanks a lot.