



This project would not had been
possible without the help of our
assigned tutor Diego Garrido
(diego.garrido@crg.eu).
The aim of the present study is the characterization and annotation of selenoproteins, as well as the machinery required for their synthesis, present in Ophiodon elongatus genome, a fish unique to the west coast of North America. In order to map its selenoprotome, we used an homology-based approach using the Danio renio (or zebrafish) genome as reference because of its phylogenetic proximity to the given sequence.
Selenoproteins are a particular type of proteins that contain selenocysteine (Sec) residues in their catalytic redox-active site, which is encoded by the UGA codon that usually acts as a stop signal. To decode UGA as a Sec (and not as a stop codon) it is necessary the presence of the Sec insertion sequence (SECIS) elements, which we will predict. Specifically in our study case, 25 selenoproteins, including 7 Cys-homologues and 6 components of the required machinery proteins, were found and analyzed with bioinformatic tools (tBLASTtn, FastaFetch, FastaSubSeq, Exonerate/Genewise/Seblastian, FastaSeqFromGFF, FastaTranslate and T-coffee).
Our results suggest that Ophiodon elongatus presents a similar selenoproteome to Danio rerio’s genome, but we hypothesize that there is a duplication in GPx seleprotein.
This work intends to contribute to the study of selenoproteome evolution.
Selenium is an essential trace element with important physiological functions in vertebrates, including fish. Nevertheless, selenium is toxic when in excess. Studies pointed out selenium functioned through selenium-containing proteins (selenoproteins) in the form of selenocysteine (Sec) Sec is the 21st natural amino acid and encoded by a UGA codon which normally acts as a stop signal.[1]. To decode UGA as a Sec and not as a stop codon it is necessary the presence of the Sec insertion sequence (SECIS) elements which have a cis-acting stem-loop structure.[2]
SECIS element in eukaryotes is located in the 3’-untranslated region (UTR) of the mRNAs which in some cases is several kilobases away from the UGA codon. SECIS elements have low sequence similarities, however, their secondary structures are highly conserved and contain consensus se- quences which are indispensable for Sec incorporation. The features required of SECIS element for Sec incorporation are a conserved AAR motif with two consecutive unpaired AA or CC residues located within an apical loop, a stem structure containing a quartet of non–Watson-Crick G to A base-pairing referred to as the SECIS core, and an A or G residue preceding the SECIS core. According to their structure, SECIS elements are divided into two categories which differ in the existence of an additional internal loop. A single SECIS element is sufficient for determining a Sec incorporation in all selenoproteins with the exception of SelP, which contains two SECIS elements that direct the decoding of 10 UGA-Sec codons in mammals and up to 17 in fish.
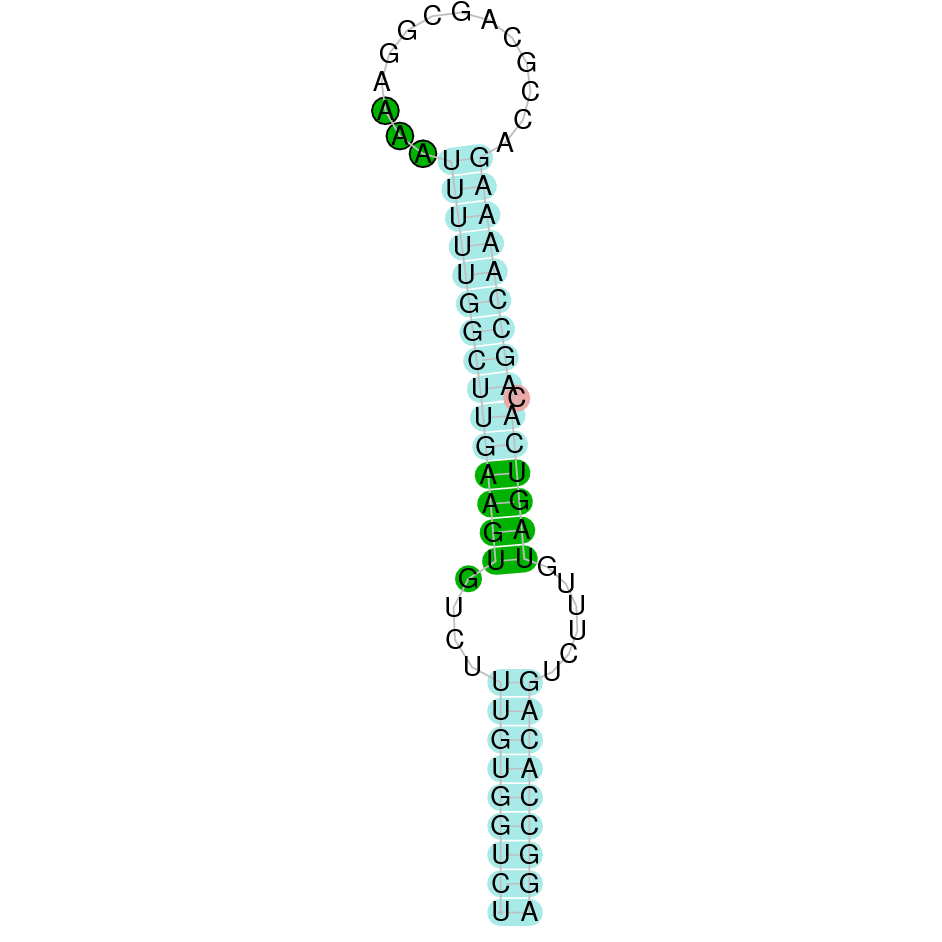
Figure 1: SECIS element (DI)
21 common selenoproteins have been found in all vertebrates: GPx1-4, TR1, TR3, DIO1, DIO2, DIO3, SelH, Sell, SelK, SelM, SelN, SelO, SelP, MsrB1, SelS, SelT1, SelW1, Sep15. Other proteins are only found in certain lineages. Most new selenoproteins are generated by duplication, and others have replaced their Sec with a cysteine.
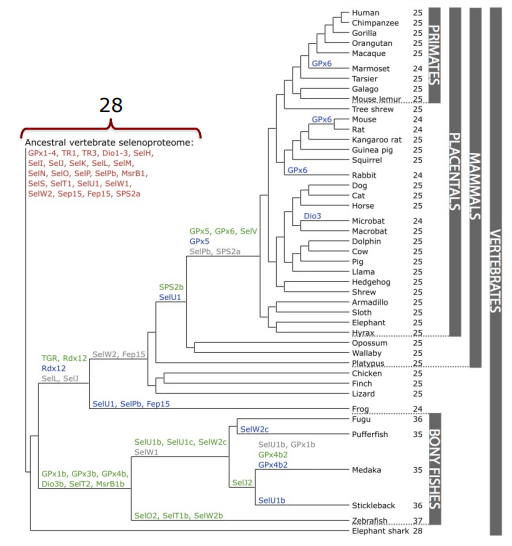
Figure 2: Phylogenetic tree of selenoproteins' evolution
In the following list we mention the main families of selenoprotein:
Deiodinases catalyze activation or inactivation (or both) of thyroid hormones (T3 and T4). Deiodinases are integral membrane selenoproteins with a single transmembrane domain at the NH2-terminal region, where it is also found the active-site Sec residue. DI1 and DI3 are found mostly on the plasma membrane, whereas DI2 is localized to the endoplasmic reticulum (ER).
Glutathione peroxidases are the largest selenoprotein family. These proteins are expressed in all tissues and have an antioxidant function: the degradation of hydroperoxides by a glutathione dependent catalysis. This family is formed for 8 GPx homologs (GPx1-GPx8). GPx1-4 and GPx6 are selenoproteins in humans and in macaque, whereas GPx5, GPx7 and GPx8 are cysteine-containing homologues.
- GPx1 is the most abundant selenoprotein in mammals and catalyzes glutathione (GSH)-dependent reduction of hydrogen peroxide to water.
- GPx2: : It is found on the gastrointestinal epithelium and it acts as a barrier for avoiding the toxicity of hydroperoxides ingested through the diet.
- GPx3: GPx3 is the major GPx form in plasma. It has specificity for hydrogen peroxide.
- GPx4: An ubiquitously expressed protein, it has a protective function against oxidation of lipid peroxides. It appears to be essential for development in vertebrates, and specially crucial to rain development and maintenance during maturity. Unlike other proteins of the GPx family, GPx4 has a structural function as well as an enzymatic one, as it becomes part of the mitochondrial sheath in spermatozoa.
- GPx5: is secreted into the lumen of the epididymis and interacts with spermatozoa.
- GPx6: expression of this gene is restricted to embryos and adult olfactory epithelium.
- GPx7: it has an ubiquitous expression, with a marked overexpression in tumoral tissues.
- GPx8: Besides the enzymatic activity common to the GPx family, Gpx8 takes part in regulation of calcium fluxes in the cytosol and mitochondria.
Methionine sulfoxide reductase (MsrA) is the main peptide in the methionine sulfoxide reduction pathway. MsrA catalyzes thioredoxin-dependent methionine sulfoxide reduction. This enzyme (as well as the MsrB or SelR) is present in all cell types, and have shown to be regulating life spans in mammals, insects, and yeast.
Selenoproteins W, T, H, and V
Selenoprotein W (SelW), T (SelT), H (SelH), and V (SelV) belong to the Rdx family of selenoproteins and are characterized by the presence of a conserved Cys-x-x-Sec motif. All of them are considered thiol-based oxidoreductases.
Selenoprotein P
SelP is an unusual selenoprotein located in human plasma. It contains a total of 10 Sec residues in humans. SelP is responsible for the transport and delivery of selenium to body tissues from the liver, where it is secreted. It has also been associated with antioxidant functions.
15 kDa selenoprotein (Sep15/Sel15)
Protein implicated in disulfide bond assisted protein folding. Fep15 is a distantly related protein of this family, which is absent in mammals. Fep15 can be detected only in fish, and is present in these organisms only in the selenoprotein form. In contrast with other members of the Sep15 family, which contain a putative active site composed of Sec and cysteine, Fep15 has only Sec.
Selenoprotein I
Selenoprotein I is one of the latest discovered selenoproteins. It is only found in vertebrates. SelI is located in the endoplasmic reticulum membrane due to its 7 transmembrane domains. It may be associated with phospholipids synthesis.
Selenoprotein J
SelJ is unusual so it has a highly restricted phylogenetic distribution; being found only in actinopterygian fishes and sea urchin, but not in other vertebrates, including mammals, birds and amphibians. SelJ is preferentially expressed in the eye lens in early embryogenesis, and shares significant similarity with jellyfish crystallins, which act to maintain transparency and proper light diffraction in the eye lens.
Selenoprotein K and S
Selenoproteins K and S are both located in the endoplasmatic reticulum membrane and the cell membrane SelS and SelK are related to the ER-associated degradation of misfolded proteins. Despite they do not share any sequence similarity, they have structural similarities: a single transmembrane domain in the NH2-terminal sequence (transmembrane proteins III), positively charged amino acids, a high content of glycine and Sec residues located in the COOH-terminal of the protein.
Selenoprotein L
Selenoprotein L contains two Sec residues and is present only among aquatic eukaryotes such as fish, invertebrates and marine bacteria.
Selenoprotein M
This protein presents functional and structural homology with SEL 15, with a thiol-disulfide activity and a signal peptide at the N-terminal end. Thus, it has also been proposed as a protein with an important role in disulfide bond formation in the ER. Redox-active motifs observed in SEL M structure indicate that this protein has a redox function.
Selenoprotein N
is expressed in the membrane of the endoplasmic reticulum of several tissues. Different mutations in this gene lead to different congenital muscular disease, named in general SEPN1-related myopathies. Its function is not yet clear, but it is thought to have a role in myogenesis. Animal models and cellular studies have provided advances towards unveiling its function.
Selenoprotein O
Selenoprotein O contains a Sec residue located in C-terminal end. Homologs of human Selenoprotein O have been detected in a wide variety of species, even though the majority of homologs contain a Cys residue in place of a Sec. Its function is yet unknown.
Selenoprotein R
elenoprotein R, also known as MsrB1, is a zinc-containing mammalian selenoprotein. It was firstly designed as selenoprotein R (SelR) but later on, it was found that its function was methionine-R-sulfoxide reductase, like MsrA, so finally it was called MsrB1. Its main functions include cellular oxidation repair, e.g. transcription factors or cytoskeleton.
Selenoproteins U
In superior mammal species selenoprotein U is found in the Cys form. Nevertheless, SelU were found in fish first, as well as in birds and unicellular eukaryotes. Three subfamilies have been reported in humans (those present below). They have a Prx-like2 structure, which means that they belong to the thioredoxine-like superfamily.
They are involved in a sequence of reactions which ultimately leads to the formation of reduced disulfide bonds. TRs reduce thioredoxin, taking its electrons in order to give them to a protein and transform it from the S2 (oxidized) form to the SH2 (reduced) form. This process is an antioxidant mechanism present in all cells. TR1 is cytosolic, TR2 mitochondrial and TR3 is testes specific.
- TR1: Protein with 6 different isoforms formed by alternative splicing and extension of NH2 terminal extreme. Its main function is to reduce NADPH Trx1 dependence but it is also important in regulating different transcription factors, apoptosis, etc. It is located in the cytoplasm and nucleus.
- TR2: Really important in the compaction on reactive oxygen.
- TR3: Protein with multiple isoforms important in the reduction of the tioredoxina mithocondrial (Trx2), that is why it is placed on the mithocondria.
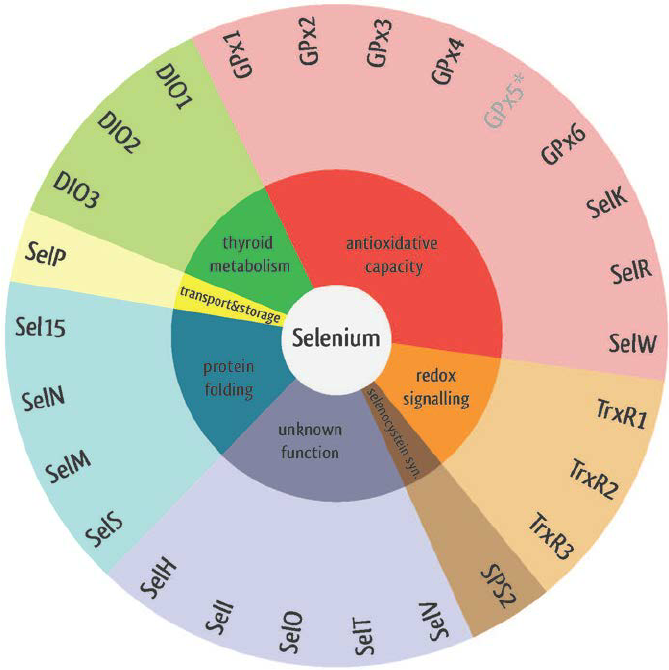
Figure 3: Classes of selenoproteins and their functions
Sec is the only known amino acid in eukaryotes whose biosynthesis occurs on its own tRNA, named Sec-tRNA[Ser]Sec.
Seryl-tRNA synthetase (SerS)
Initially, tRNA[Ser]Sec is aminoacylated with a serine in a reaction catalyzed by seryl-tRNA synthetase (SerS) to form Sec-tRNA[Ser]Sec, which provides the backbone for Sec biosynthesis.
Phosphoseryl-tRNA[Ser]SecKinase (PSTK):
Phosphorylates the serine of Ser-tRNA[Ser]Sec. The phosphate is then replaced by the selenium donor selenide, a selenophosphate (H2Se-PO3-).
Sec Synthase (SecS)
Catalyzes the conversion of phosphorilated Ser-tRNA[Ser]Sec to Sec-tRNA[Ser]Sec or Cys-tRNA[Ser]Sec. It can incorporate a selenophosphate, the active form of selenium, into the backbone forming a Sec-tRNA[Ser]Sec or a phosphte forming a Cys-tRNA[Ser]Sec.
Selenophosphate Synthase 2 (SPS2)
SPS2 is an enzime that converts selenide intoH2Se-PO3-, the active form of selenide and is itself a selenoprotein.
Moreover, SPS2 is also implicated in Cys bioshyntesis. When selenium is in deficiency, SPS2 can use sulfide instead of selenide and generate thiophosphate, an active donor for the SecS-catalyzed reaction making Cys-tRNA from the thiophosphate inserting a Cys into selenoproteins at the UGA-encoed sites instead of Sec.
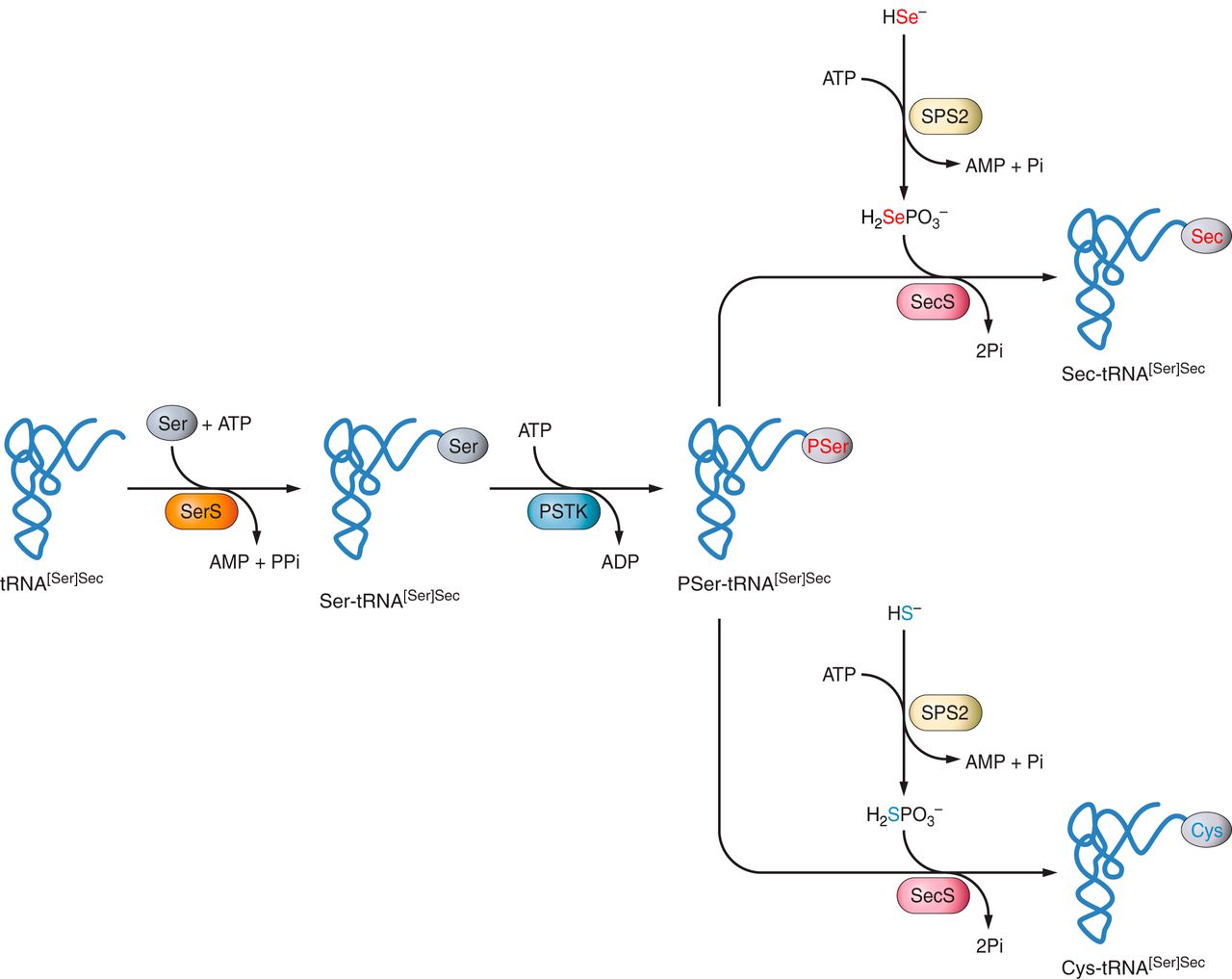
Figure 4: Selenoprotein biosynthesis
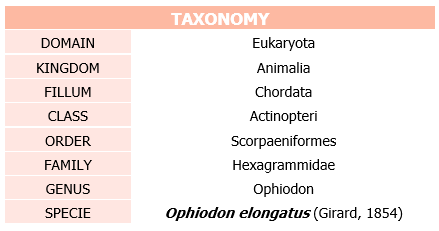

Ophiodon elongatus is a fish of the greenling family Hexagrammidae. It is the only extant member of the genus Ophiodon[5].
Ophiodon elongatus is native to the North American west coast from Shumagin Islands in the Gulf of Alaska to Baja California, Mexico. Specimens upto a size of 152 centimetres (60 in) and a weight of 59 kilograms (130 lb) has been observed.[6]
It is spotted in various shades of grey, however, around 20% of lingcods have blue-green to turquoise flesh. The colour is destroyed by cooking [7]. The lingcod is a popular eating fish, and is thus prized by anglers. Although it is not closely related to either ling or cod, the name "lingcod" originated because it resembles those fish [8]
Maturation of the females is around the age of three and five (61–75 centimetres ) and maturations of the males arroung two years of age (45 centimetres). Adult males can be distinguished externally from females by the presence of a small, conical papilla behind the anal vent. [6]
Lingcod are voracious predators, feeding on nearly anything they can fit in their mouths, including invertebrates and many species of fish. [8]
The aim of our project was to identify and annotate Ophiodon elongatus selenoproteins and the machinery required for their synthesis. To do so, we performed an homology-based study. The reference genome we selected is Danio rerio (or zebrafish), because of its phylogenetic proximity to Ophiodon elongatus and because it has a very well annotated genome. The general scheme of the methodology we used is the following:
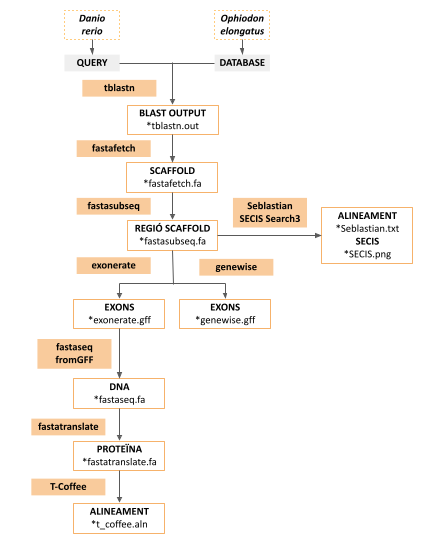
Figure 5: Methodology scheme
The O. elongatus genome was obtained from the following directory, provided by the teachers of the subject:
/mnt/NFS_UPF/soft/genomes/2021/Ophiodon_elongatus/genome.fa
We obtained the aminoacid sequences for the different selenoproteins and selenocysteines of Danio rerio from SelenoDB 2.0. We saved all the selenoprotein sequences of Danio rerio in fasta file called selenoprotDR.fa, and all the selenocysteine sequences in a fasta file called cisteinaDR.fa. These fasta files were saved in a directory called originals, inside a directory named Ophiodon_elongatus, from where we worked during all the process.
As some of the selenocysteine and selenoproteins were repeated, we manually erased the repeated protein sequences from the files.
We created a Bash program that allowed us to go through every Danio rerio query automatically and compare it (and its different scaffolds) with the genome of Ophiodon elongatus. All the results obtained are registered into a file named: results. In order to run the program, the files Ophiodon_elongatus have to be downloaded and the program has to be run in the terminal from inside this directory. You can find all these documents (Program, Results and Ophiodon_elongatus files) on the top of the web page.
The program is composed of a for loop that goes through each file of the folder iteratively and runs the different programs and commands necessary to obtain the alignment. The steps the program uses to compare the proteins are the following:
We divided the original files (selenoprotDR.fa and cisteinaDR.fa) into different files containing one protein each. Afterwards, through the loop previously mentioned, these files were saved into different folders. The folders and the files were named as the abbreviation of the protein name (and we added “.fa” for the files). To do so, we first transformed the introduction of the proteins (Ex: >SPG00000528_2.0 # Gene # 15 kDa selenoprotein (Sel15) # Zebrafish (Danio rerio) Full length # Forward) into the abbreviation of the protein (Ex: Sel15). Once that was done, we were able to create the different files and folders for each file easily.
The query sequences contained the letter “U”, which some of the programs used in later steps are unable to recognise. Some query sequences also contained other unwanted symbols at the end of the sequences, such as #, % or @. We removed the unwanted symbols and changed the letter “U” for an “X” to avoid problems.
BLAST is a program that aligns biological sequences. tBLASTn is a version of BLAST that allows the comparison of protein sequences against a nucleotide sequence. In our case, we aligned the query sequences of Danio rerio with the genome sequence of Ophiodon elongatus. We used the following command, and we saved the results in a file called: protein name + “_tblastn.out”.
tblastn -query ${foldername}/$file -db /mnt/NFS_UPF/soft/genomes/2021/Ophiodon_elongatus/genome.fa -outfmt 7 -out ${foldername}/${foldername}_tblastn.out
The files generated contained information about the alignment, such as the name of the scaffold where the hit was found, the query start and end points, the e-value (significance measure of the hit) and the query coverage (percentage of matching between the query and the hit).
The region of interest was selected by the values of coverage and e-value. The program chooses the scaffolds that have more than a 30% of identity and an e-value lower than 0.0001, in order for the alignment to be relevant to study.
For each of the scaffolds, the program runs through a while loop, and uses the command Fastafetch to extract the scaffold of interest, the region in which the tBLASTn alignment showed that we could possibly find the gene. To run the program, we used this command:
fastafetch /mnt/NFS_UPF/soft/genomes/2021/Ophiodon_elongatus/genome.fa /mnt/NFS_UPF/soft/genomes/2021/Ophiodon_elongatus/genome.index ${scaffold} > ${foldername}/${foldername}_${scaffold}_fastafetch.fa
A folder named after the name of the scaffold that is being studied is created and the output of the command, which is called “protein name”_”scaffold name”_fastafetch.fa, is saved in it. Besides, all the outputs generated for the studied scaffold will be also saved in this folder.
Then, we used the fastasubseq program to delimit the sequence of the scaffold into a much smaller sequence, in order to achieve more accurate results of the genome fragment that contains the selenoprotein. We used the start location of our hit as a reference to choose the region of the scaffold that was of our interest. We ordered the program to select regions that started 50,000bp upstream of the start location of the hit, with a length of 100,000bp - which is 50,000bp downstream of the start location of the hit.
As some scaffolds may be smaller than 100,000bp, we designed a command in order to start at position 0 of the scaffold in the case where the starting position of the hit was smaller than 50,000.
if [[ ${start} -gt 50000 ]]; then
start=$((${start}-50000))
else
start=0
fi
Another command was used in order not to over-expand the selected region upstream from the start of the hit (if the scaffold length was smaller than 100,000bp from the start point of the region we selected for the fastafetch - the start point being either 0 or 50,000bp before the start of the hit).
scaffold_length=$(grep ${scaffold} /mnt/NFS_UPF/soft/genomes/2021/Ophiodon_elongatus/ genome.lengths | cut -f 1 -d " ")
if [[ ${scaffold_length} -gt $((${start}+100000)) ]]; then
length=100000
else
length=$((${scaffold_length}-${start}))
fi
Finally, we used the following command to obtain a file which contained the region we used for the posterior analysis. The output will be named: “protein name”_”scaffold name”_”fastasubseq.fa”.
fastasubseq ${foldername}/${foldername}_${scaffold}_fastafetch.fa ${start} ${length} > ${foldername}/${foldername}_${scaffold}_fastasubseq.fa
We used exonerate to deduce the sequence of genes from our region of interest (_fastasubseq.fa files). The command we used to do so is below, where -m indicates the alignment model, which is p2g (protein to genome); -q is used to indicate where the file is found and -t indicates the sequence against which the query is compared (file generated in the previous step).
Within the region of the hit, there are both introns and exons. As we only wanted to analyze exon sequences, which are more likely to be conserved during evolution, we used a grep command. This way we generated a file with exon information only. The output of the command was saved in a file which ends with “_exonerate.gff”
exonerate -m p2g --showtargetgff -q ${foldername}/${foldername}.fa -t ${foldername}/${foldername}_${scaffold}_fastasubseq.fa | egrep -w "exon" > ${foldername}/${foldername}_${scaffold}_exonerate.gff
Then, we used the fastaseqfromGFF program, which allowed us to extract the exon, sequence in FASTA format, hence we obtained the sequence that encodes for the target protein (cDNA).
fastaseqfromGFF.pl ${foldername}/${foldername}_${scaffold}_fastasubseq.fa ${foldername}/${foldername}_${scaffold}_exonerate.gff > ${foldername}/${foldername}_${scaffold}_fastaseq.fa
Afterwards, we used fastatranslate program with the aim of translating the cDNA sequence obtained in the previous step into a protein sequence, using the following command:
fastatranslate -f ${foldername}/${foldername}_${scaffold}_fastaseq.fa -F 1 > ${foldername}/${foldername}_${scaffold}_fastatranslate.fa
where -F 1 is used to limit the selected reading frame. We obtain an output file whose name ends with “_fastatranslate.fa”.
As the generated document contained “*” where there was an UTG, we used a command to replace these “*” with “X”:
sed 's/*/X/g' -i ${foldername}/${foldername}_${scaffold}_fastatranslate.fa
Finally, we used the t-coffee program to generate a global alignment between each deduced protein sequence from the genome of Ophiodon elongatus and each initial query from Dario rerio. To obtain the results of t-coffee, we used the next commands:
t_coffee ${foldername}/${foldername}.fa ${foldername}/${foldername}_${scaffold}_fastatranslate.fa 2> /dev/null > ${foldername}/${foldername}_${scaffold}_t_coffee.aln
cat ${foldername}/${foldername}_${scaffold}_t_coffee.aln
where 2> /dev/null is used to redirect the standard error of the program to a folder that discards all data written to it. The obtained output, which will be saved into the scaffold folder, will be named: “protein name”_”scaffold name”_”t_coffee.aln”.
As previously mentioned, the program we created is a fully automatized bash program that runs through a loop for each file created from the selenoprotDR.fa and cisteinaDR.fa. It introduces the file inside a folder with the same name and the tblastn program runs.
Then a new loop, the “scaffold loop” goes through each scaffold that meets the established criteria to be analysed (identity greater than 30% and e-value lower than 0.0001). In this loop, a folder named after the scaffold is created, where all the program outputs are saved (fastafetch, fastasubseq, exonerate, genewise, fastaseqfromGFF, fastatranslate and t-coffee).
To predict the SECIS elements present in our predicted protein, we will use the Seblastian server. This server is a web page which uses SECISearch3 to identify possible SECIS elements in the analysed genome (from the output of the fastasubseq program) and then predicts selenoprotein sequences encoded upstream of these elements.
The output generated from this program will be an image (.png) of the predicted SECIS elements, downstream of our potential selenoproteins and a text file (.txt) that gives information about the predicted selenoprotein (alignment, starting and ending positions, strand…) as well as information about the SECIS elements (starting and ending positions, strand…).
In order to analyse the phylogenetic relationships between different proteins of the main families of selenoproteins studied in this project (GPx, DI, Sel, TR, Sec machinery), we perform a phylogenetic study through the Phylogeny.fr server. The main objective of this process is to confirm if the results from the previous steps were coherent.
We input a multifasta file containing the query of Danio rerio and the predicted proteins from Ophiodon elongatus (which is the output of the fastatranslate program). The output of the program is a phylogenetic tree that illustrates the distance between the proteins.
The results of the analysis of Ophiodon elongatus are sumarized in the following table:
| Legend | |
 |
information available |
 |
information not available |
As explained in the introduction, Idothreonine deiodinase family is a commonly present protein family in mammals. This family is composed of three different proteins but only DI and DI1 are present in both Danio rerio and Ophiodon elongatus.
After running the tblastn, seven hits on six different scaffolds were identified but only four of the scaffolds had an e-value (smaller than 0.0001) and identity (bigger than 30%) good enough to be studied. However, results obtained from the complete analysis allowed us to reduce the possible scaffolds to one (JACSYO010000262.1), which had the best T-coffee alignment
As we can observe with Exonerate, the gene is located in the Ophiodon elongatus genome between positions 917287 and 918078, comprising 1 exon in the reverse strand (-). The T-coffee alignment between the predicted protein in O.elongatus and the protein of reference (from D.rerio) had high similarity (with a score of 985) so we can conclude that a coding sequence for this protein is found in the studied organism. Moreover, the analysed protein contains Sec in the same position as the reference protein.
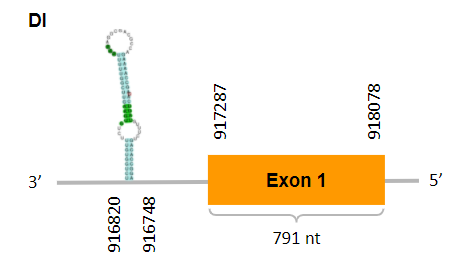
The analysis conducted in Seblastian produced a better prediction, as the sequence obtained contains 1 exon in the reverse strand (-) and includes a Met at the beginning of the protein (which did not happen with the Exonerate prediction). SECIS Search3 identified a SECIS element for this sequence with good infernal and covels score which is found in the 3’-UTR segment of the reverse strand, specifically between positions 916820 and 916748 of the JACSYO010000262.1 scaffold.
In conclusion, we can say that the gene DI is a selenoprotein found in Ophiodon elongatus, due to the presence of a Sec residu and a SECIS element. Moreover, from the information displayed in the phylogenetic tree we can also conclude that it is an homologous protein of the zebrafish DI.
After running the tblastn, six hits on four different scaffolds were identified and all of them had an e-value (smaller than 0.0001) and identity (bigger than 30%) good enough to be studied. However, results obtained from the complete analysis allowed us to reduce the possible scaffolds to one (JACSYO010002145.1), which had the best T-coffee alignment.
As we can observe with Exonerate, the gene is located in the Ophiodon elongatus genome between positions 2097519 and 2098905, comprising 3 exons in the forward strand (+). The T-coffee alignment between the predicted protein in O.elongatus and the protein of reference (from D.rerio) had high similarity (with a score of 998) so we can conclude that a coding sequence for this protein is found in the studied organism. Moreover, the analysed protein contains Sec in the same position as the reference protein.
The analysis conducted in Seblastian produced a better prediction, as the sequence obtained contains 4 exons in the forward strand (+) and includes a Met at the beginning of the protein (which did not happen with the Exonerate prediction). SECIS Search3 identified two SECIS elements for this sequence with good infernal and covels score which is found in the 3’-UTR segment of the reverse strand, specifically between positions 2099993-2100065 and 2099729-2099795 of the JACSYO010002145.1 scaffold. Despite the fact that both of the SECIS elements could be correct, the representation will only show one of them.
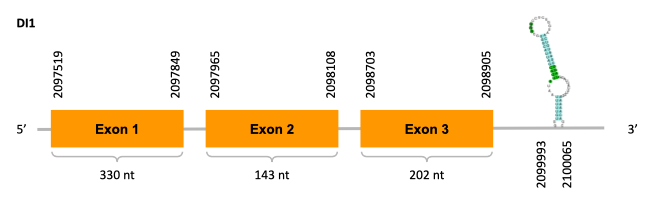
In conclusion, we can say that the gene DI1 is a selenoprotein found in Ophiodon elongatus, due to the presence of a Sec residu and a SECIS element. Moreover, from the information displayed in the phylogenetic tree we can also conclude that it is an homologous protein of the zebrafish DI1.
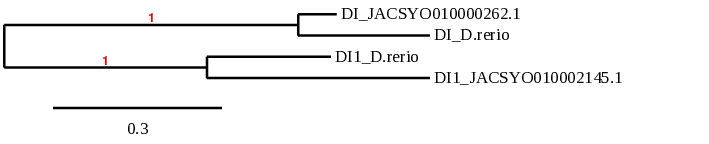
The results observed in this phylogenetic tree are coherent with the ones we extracted from our data. The proteins of the Ophiodon elongatus are closely related to their homologues in Danio rerio and afterwards they link the whole DI family.
After running the tblastn, twenty hits on nine different scaffolds were identified and all of them had an e-value (smaller than 0.0001) and identity (bigger than 30%) good enough to be studied. However, results obtained from the complete analysis allowed us to reduce the possible scaffolds to two (JACSYO010000406.1 and JACSYO010000471.1), which both had good T-coffee alignments.
As we can observe with Exonerate, the gene of the scaffold JACSYO010000406.1 is located in the Ophiodon elongatus genome between positions 810181 and 811527, comprising 2 exons in the forward strand (+). The T-coffee alignment between the predicted protein in O.elongatus and the protein of reference (from D.rerio) had high similarity (with a score of 998) so we can conclude that a coding sequence for this protein is found in the studied organism. Moreover, the analysed protein contains a Sec in the same position as the reference protein.
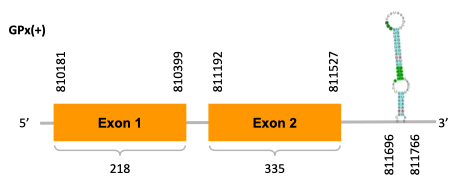
The gene of the scaffold JACSYO010000471.1 is located in the Ophiodon elongatus genome between positions 2900773 and 2902385, comprising 2 exons in the reverse strand (-). The T-coffee alignment between the predicted protein in O.elongatus and the protein of reference (from D.rerio) had high similarity (with a score of 999) so we can conclude that a coding sequence for this protein is found in the studied organism. Moreover, the analysed protein contains a Sec in the same position as the reference protein.
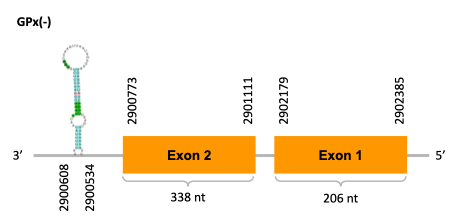
Because the two scaffolds of Ophiodon elongatus contain the GPx protein, we think that it might have duplicated.
The analysis conducted in Seblastian also predicted a sequence that contains 2 exons for each protein of each of the scaffolds, in the reverse and forward strand (as we had found with exonerate). SECIS Search3 identified a SECIS element for this sequence in each of the scaffolds. For the scaffold JACSYO010000406.1 the SECI element is found in the 3’-UTR segment of the forward strand, between positions 811696 and 811766. For the scaffold JACSYO010000471.1 the SECIS element is found in the 3’-UTR segment of the reverse strand, between positions 2900534 and 2900608.
In conclusion, we can say that the gene GPx is a selenoprotein found in Ophiodon elongatus, due to the presence of a Sec residu and a SECIS element. In addition, we can say that this selenoprotein has been duplicated in the Ophiodon elongatus genome.
After running the tblastn, twenty-one hits on eleven different scaffolds were identified and nine scaffolds had an e-value (smaller than 0.0001) and identity (bigger than 30%) good enough to be studied. However, results obtained from the complete analysis allowed us to reduce the possible scaffolds to one (JACSYO010002173.1), which had the best T-coffee alignment.
As we can observe with Exonerate, the gene is located in the Ophiodon elongatus genome between positions 858742 and 813855, comprising 2 exons in the forward strand (+). The T-coffee alignment between the predicted protein in O.elongatus and the protein of reference (from D.rerio) had very high similarity (with a score of 1000) so we can conclude that a coding sequence for this protein is found in the studied organism. Moreover, the region containing the selenocystein aligns with the protein of Danio rerio perfectly.
The analysis conducted in Seblastian also predicted a sequence that contains 4 exons in the forward strand (+) and includes a Met at the beginning of the protein (which also happened with the Exonerate prediction). SECIS Search3 identified a SECIS element for this sequence with good infernal and covels score which is found in the 3’-UTR segment of the forward strand, specifically between positions 859962 and 860029 of the JACSYO010002173.1 scaffold.
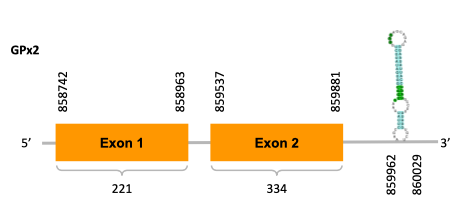
In conclusion, we can say that the gene GPx2 is a selenoprotein found in Ophiodon elongatus, due to the presence of a Sec residu and a SECIS element.
After running the tblastn, eighteen hits on nine different scaffolds were identified and all of them had an e-value (smaller than 0.0001) and identity (bigger than 30%) good enough to be studied. However, results obtained from the complete analysis allowed us to reduce the possible scaffolds to one (JACSYO010000535.1), which had the best T-coffee alignment.
As we can observe with Exonerate, the gene is located in the Ophiodon elongatus genome between positions 68906 and 70533, comprising 4 exons in the forward strand (+). The T-coffee alignment between the predicted protein in O.elongatus and the protein of reference (from D.rerio) had high similarity (with a score of 985) so we can conclude that a coding sequence for this protein is found in the studied organism. Moreover, the Selenocysteine of protein sequence of Danio rerio aligns with the one from Ophiodon elongatus.
The analysis conducted in Seblastian was not able to predict any sequence, but the SECIS Search3 identified two SECIS elements for this sequence. We selected only one of the SECIS predicted, because it had a grade A (the other one had a grade B), and it is near the exonic sequence of the gene. It is found in the 3’-UTR segment of the forward strand, specifically between positions 70629 and 70706 of the JACSYO010000535.1 scaffold.
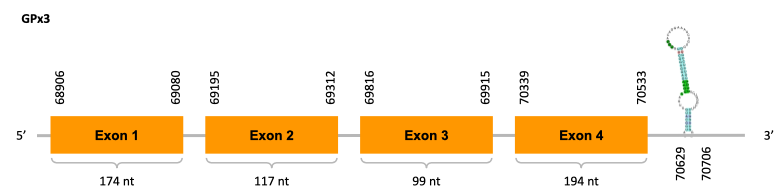
In conclusion, we can say that the gene GPx3 is a selenoprotein found in Ophiodon elongatus, due to the presence of a Sec residu and a SECIS element.
After running the tblastn, eighteen hits on eleven different scaffolds were identified but only eight of the scaffolds had an e-value (smaller than 0.0001) and identity (bigger than 30%) good enough to be studied. However, results obtained from the complete analysis allowed us to reduce the possible scaffolds to one (JACSYO010001924.1), which had the best T-coffee alignment.
As we can observe with Exonerate, the gene is located in the Ophiodon elongatus genome between positions 1623795 and 1625702, comprising 3 exons in the reverse strand (-). The T-coffee alignment between the predicted protein in O.elongatus and the protein of reference (from D.rerio) had very high similarity (with a score of 1000) so we can conclude that a coding sequence for this protein is found in the studied organism. Moreover, the analysed protein contains Sec in the same position as the reference protein.
The analysis conducted in Seblastian produced a quite different prediction, as the sequence obtained contains 2 exons in the reverse strand (-), instead of 3. SECIS Search3 identified a SECIS element for this sequence with good infernal and covels score which is found in the 3’-UTR segment of the reverse strand, specifically between positions 1622955 and 1623034 of the JACSYO010001924.1 scaffold.
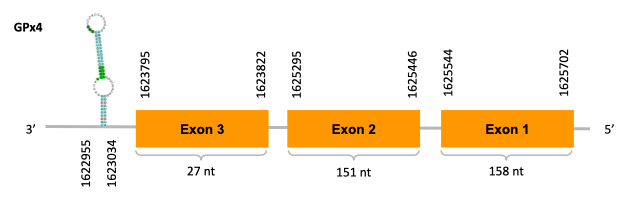
In conclusion, we can say that the gene GPx4 is a selenoprotein found in Ophiodon elongatus, due to the presence of a Sec residu and a SECIS element.
After running the tblastn, seventeen hits on twelve different scaffolds were identified but only six of the scaffolds had an e-value (smaller than 0.0001) and identity (bigger than 30%) good enough to be studied. However, results obtained from the complete analysis allowed us to reduce the possible scaffolds to one (JACSYO010002134.1), which had the best T-coffee alignment.
As we can observe with Exonerate, the gene is located in the Ophiodon elongatus genome between positions 801284 and 802063, comprising 2 exons in the reverse strand (-). The T-coffee alignment between the predicted protein in O.elongatus and the protein of reference (from D.rerio) had high similarity (with a score of 994) so we can conclude that a coding sequence for this protein is found in the studied organism. The analysed protein does not contain any Selenocysteine, but it has two cysteines, indicating that it probably is a cystein-homologous protein.
The analysis with Seblastian was not able to predict any protein alignment. SECIS Search3 identified 12 SECIS element for this sequence. As all of them had a grade B and had similar Infernal score values, we were not able to select the one(s) that fits best the Ophiodon elongatus genome. An example of one of the SECIS found is the one represented in the image, which is found in the 3’-UTR segment of the reverse strand, between positions 778084 and 778151 of the JACSYO010002134.1 scaffold.
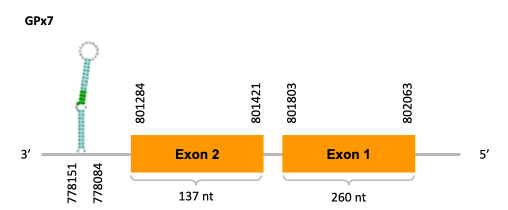
In conclusion, we can say that the gene GPx7 is a cysteine-homolgue protein found in Ophiodon elongatus, due to the presence of a Cys residue and even though a SECIS element was identified through SECIS Search3 (this indicates that the protein could have been a selenoprotein on the past but lost the Sec residue).
After running the tblastn, twenty-one hits on fourteen different scaffolds were identified but only seven of the scaffolds had an e-value (smaller than 0.0001) and identity (bigger than 30%) good enough to be studied. However, results obtained from the complete analysis allowed us to reduce the possible scaffolds to two (JACSYO010000919.1 and JACSYO010002134.1), which had the best T-coffee alignments.
As we can observe with Exonerate, the gene, for the scaffold JACSYO010000919.1 is located in the Ophiodon elongatus genome between positions 388811 and 390588, comprising 3 exons in the reverse strand (-). The T-coffee alignment between the predicted protein in O.elongatus and the protein of reference (from D.rerio) had very high similarity (with a score of 1000) so we can conclude that a coding sequence for this protein is found in the studied organism.
For the scaffold JACSYO010002134.1, the gene is located in the Ophiodon elongatus genome between positions 801287 and 805048, comprising 3 exons in the reverse strand (-). The T-coffee alignment between the predicted protein in O.elongatus and the protein of reference (from D.rerio) had high similarity (with a score of 996) so we can conclude that a coding sequence for this protein is found in the studied organism.
The analysed protein does not contain Sec, but it has a common Cys in both sequences aligned, making GPx8 a possible Cys-containing homologue protein.
The analysis conducted in Seblastian for the first scaffold (JACSYO010000919.1) was not able to make any prodictions. No SECIS element was predicted for this scaffold either. For the scaffold JACSYO010002134.1, Seblastian was not able to make any predictions, but SECIS Search3 identified twelve different SECIS, all of them in the reverse strand, with similar positions and infernal score, so we were not able to elucidate which one(s) are present in the genome of Ophiodon elongatus.
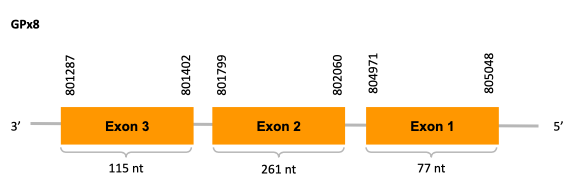
In conclusion, we can say that the gene GPx8 is a cysteine-containing homologue found in Ophiodon elongatus, due to the presence of a Cys residue, despite the fact that a SECIS element was identified through SECIS Search3 (this indicates that the protein could have been a selenoprotein on the past but lost the Sec residue).
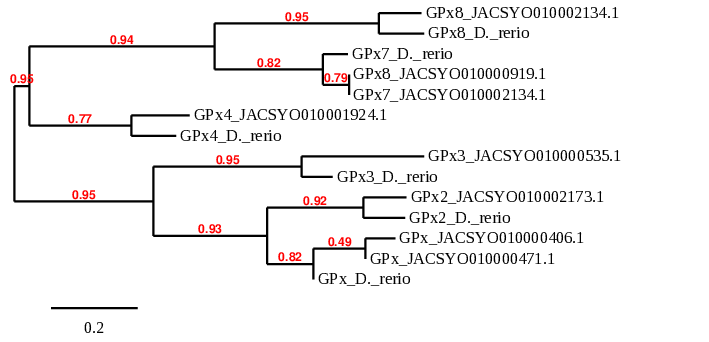
Regarding the phylogenetic tree of the GPx protein family, we can see that GPx2, GPx3 and GPx4 proteins predicted from Ophiodon elongatus’ genome pair specifically with their homologues in Danio rerio, confirming that these proteins are Selenoproteins present in Ophiodon elongatus.
However, GPx has similar alignments with proteins predicted from two different scaffolds of Ophiodon elongatus, which affirms our hypothesis that this protein might be duplicated in the Ophiodon elongatus’ genome.
Considering the GPx8 protein, in which we could not decide between two scaffolds of Ophiodon elongatus, we can see that the scaffold which pairs better with Danio rerio’s GPx8 protein is JACSYO010002134.1, which is the only of the two scaffolds analysed for which SECIS3 Search found possible SECIS sequences. This confirms that GPx8 protein of Danio rerio has an homologous protein in Ophiodon elongatus, whose sequence is found in the scaffold JACSYO010002134.1.
When observing protein GPx7, even though the protein predicted from Ophiodon elongatus pairs better with a GPx8 protein also predicted from the analysed organism genome, we found that this GPx8 predicted protein is probably not a good prediction.
In addition, proteins GPx, GPx2 and GPx3 are related, as seen in the phylogenetic tree, confirming relationship between them. GPx7 and GPx8 are related, and they are also related with GPx4, indicating that they evolved from GPx4.
In conclusion, the phylogenetic tree obtained confirms that our predicted selenoproteins and cysteine-containing proteins are homologous of the human ones, and they evolved in the same way.
After running the tblastn, twelve hits on five different scaffolds were identified. Since the hits from only two of the scaffolds (JACSYO010002166.1 and JACSYO010001682.1) had an e-value (smaller than 0.0001) and identity (bigger than 30%) good enough to be studied, the other three scaffolds were not further studied. We completed the analysis with both of the scaffolds, and both showed a good T-coffee alignment.
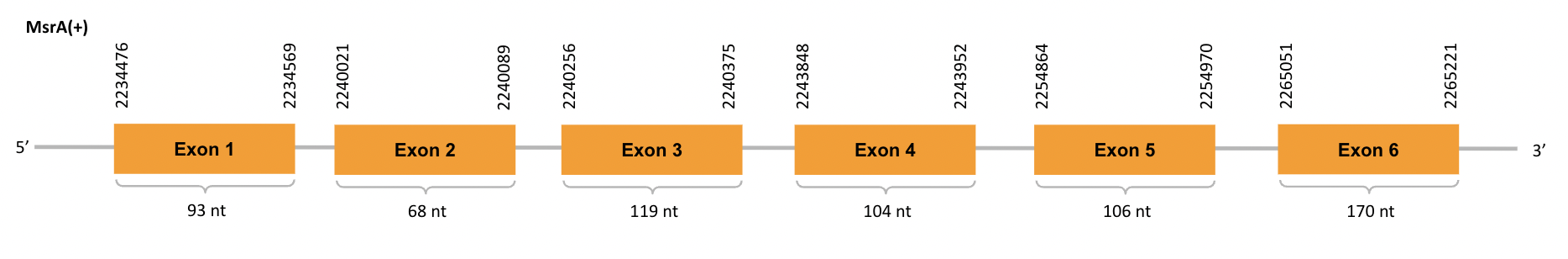
For the scaffold JACSYO010002166.1, as we can observe with Exonerate, the gene is located in the Ophiodon elongatus genome between positions 2194476 and 2265221, comprising 6 exons in the forward strand (+). The T-coffee alignment between the predicted protein in O.elongatus and the protein of reference (from D.rerio) had high similarity (with a score of 993).
For the scaffold JACSYO010001682.1, as we can observe with Exonerate, the gene is located in the Ophiodon elongatus genome between positions 1851193 and 1867206, comprising 4 exons in the forward strand (+). The T-coffee alignment between the predicted protein in O.elongatus and the protein of reference (from D.rerio) had high similarity (with a score of 999)
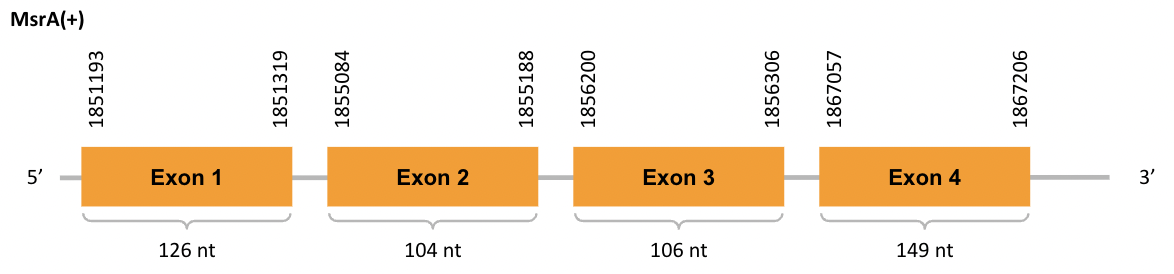
We can conclude that a coding sequence for this protein is found in the studied organism. Moreover, the analysed protein contains Cys in the same position as the reference protein.
The analysis by Seblastian didn’t find any results for none of the two scaffolds. SECIS Search3 only identified a SECIS element for the scaffold JACSYO010001682.1 of this sequence between positions 1812445 and 1812512. Because this SECIS element found in the scaffold are previous than the exons of the scaffold, we can not conclude that this SECIS element corresponds to the protein analysed and if the protein is indeed a cystein-containing protein in Ophiodon elongatus
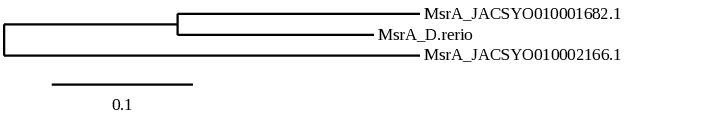
When doing the phylogeny, more proximity is observed between the scaffold JACSYO010002166.1 of Ophiodon elongatus MsrA protein and the MsrA protein of Danio rerio, meaning that the protein found in the JACSYO010002166.1 scaffold is probably the cystein-containing homologous protein of Ophiodon elongatus.
After running the tblastn, four hits on two different scaffolds were identified and both had an e-value (smaller than 0.0001) and identity (bigger than 30%) good enough to be studied. However, results obtained from the complete analysis allowed us to reduce the possible scaffolds to one (JACSYO010002146.1), which had the best T-coffee alignment.
As we can observe with Exonerate, the gene is located in the Ophiodon elongatus genome between positions 1123108 and 1125051, comprising 4 exons in the reverse strand (-). The T-coffee alignment between the predicted protein in O.elongatus and the protein of reference (from D.rerio) had high similarity (with a score of 978) so we can conclude that a coding sequence for this protein is found in the studied organism. Moreover, the analysed protein contains a Sec in the same position as the reference protein.
The analysis conducted in Seblastian also predicted a sequence that contains 4 exons in the reverse strand (-) and includes a Met at the beginning of the protein (which did not happen with the Exonerate prediction). SECIS Search3 identified a SECIS element for this sequence with good infernal and covels score which is found in the 3’-UTR segment of the reverse strand, specifically between positions 1122987 and 1123079 of the JACSYO010002146.1 scaffold.
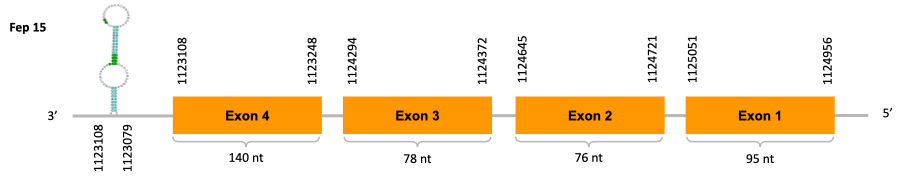
In conclusion, we can say that the gene Fep15 is a selenoprotein found in Ophiodon elongatus, due to the presence of a Sec residu and a SECIS element. Moreover, from the information displayed in the phylogenetic tree we can also conclude that it is an homologous protein of the zebrafish Fep15.
After running the tblastn, four hits on three different scaffolds were identified but only one scaffold had an e-value (smaller than 0.0001) and identity (bigger than 30%) good enough to be studied. Therfore, the study was only performed on the scaffold JACSYO010001861.1.
As we can observe with Exonerate, the gene is located in the Ophiodon elongatus genome between positions 2709194 and 2713255, comprising 4 exons in the reverse strand (-). The T-coffee alignment between the predicted protein in O.elongatus and the protein of reference (from D.rerio) had high similarity (with a score of 1000) so we can conclude that a coding sequence for this protein is found in the studied organism. Moreover, the analysed protein contains Sec in the same position as the reference protein.
The analysis conducted in Seblastian produced a better prediction, as the sequence obtained contains 5 exons in the reverse strand (-) and includes a Met at the beginning of the protein (which did not happen with the Exonerate prediction). SECIS Search3 identified a SECIS element for this sequence with good infernal and covels score which is found in the 3’-UTR segment of the reverse strand, specifically between positions 2708249 and 2708322 of the JACSYO010001861.1 scaffold.
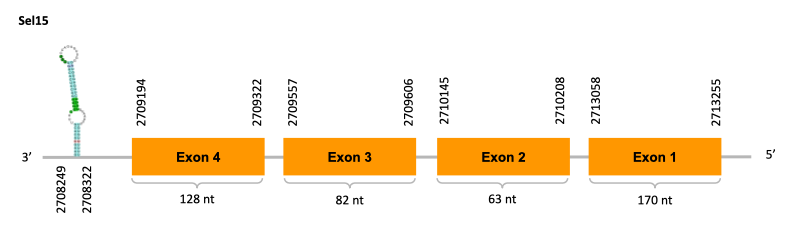
In conclusion, we can say that the gene Sel15 is a selenoprotein found in Ophiodon elongatus, due to the presence of a Sec residu and a SECIS element. Moreover, from the information displayed in the phylogenetic tree we can also conclude that it is an homologous protein of the zebrafish Sel15.
After running the tblastn, three hits on two different scaffolds were identified but only one scaffold had an e-value (smaller than 0.0001) and identity (bigger than 30%) good enough to be studied. Therfore, the study was only performed on the scaffold JACSYO010000469.1.
As we can observe with Exonerate, the gene is located in the Ophiodon elongatus genome between positions 3211813 and 3213920, comprising 3 exons in the reverse strand (-). The T-coffee alignment between the predicted protein in O.elongatus and the protein of reference (from D.rerio) had high similarity (with a score of 922) so we can conclude that a coding sequence for this protein is found in the studied organism. Moreover, the analysed protein contains Sec in the same position as the reference protein.
The analysis conducted in Seblastian produced a similar prediction, because despite the sequence obtained contains 2 exons in the reverse strand (-), it does not include a Met at the beginning of the protein (the same as in the Exonerate prediction). SECIS Search3 identified a SECIS element for this sequence with good infernal and covels score which is found in the 3’-UTR segment of the reverse strand, specifically between positions 3210334 and 3210249 of the JACSYO010000469.1 scaffold.
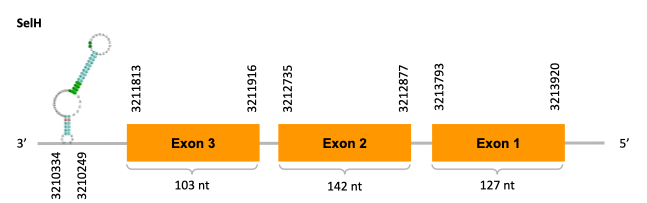
In conclusion, we can say that the gene SelH is a selenoprotein found in Ophiodon elongatus, due to the presence of a Sec residu and a SECIS element. Moreover, from the information displayed in the phylogenetic tree we can also conclude that it is an homologous protein of the zebrafish SelH.
After running the tblastn, thirteen hits on four different scaffolds were identified but only three of the scaffolds had an e-value (smaller than 0.0001) and identity (bigger than 30%) good enough to be studied. However, results obtained from the complete analysis allowed us to reduce the possible scaffolds to one (JACSYO010001293.1), which had the best T-coffee alignment
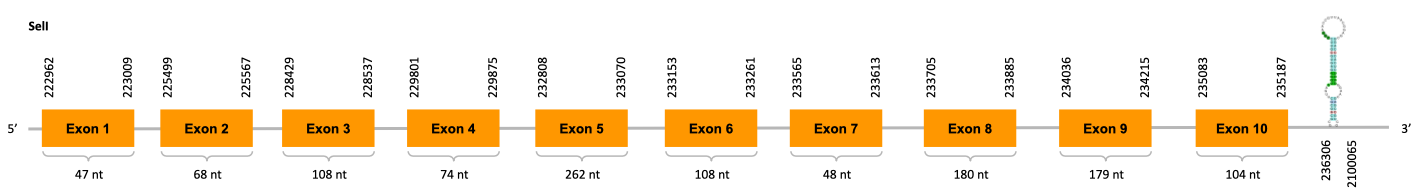
As we can observe with Exonerate, the gene is located in the Ophiodon elongatus genome between positions 222962 and 235187, comprising 10 exons in the forward strand (+). The T-coffee alignment between the predicted protein in O.elongatus and the protein of reference (from D.rerio) had high similarity (with a score of 922) so we can conclude that a coding sequence for this protein is found in the studied organism. Moreover, the analysed protein contains Sec in the same position as the reference protein.
The analysis conducted in Seblastian produced a better prediction, as the sequence obtained contains 10 exons in the forward strand (+) and includes a Met at the beginning of the protein (which did not happen with the Exonerate prediction). SECIS Search3 identified a SECIS element for this sequence with good infernal and covels score which is found in the 3’-UTR segment of the reverse strand, specifically between positions 236306 and 236379 of the JACSYO010001293.1 scaffold.
In conclusion, we can say that the gene SelI is a selenoprotein found in Ophiodon elongatus, due to the presence of a Sec residu and a SECIS element. Moreover, from the information displayed in the phylogenetic tree we can also conclude that it is an homologous protein of the zebrafish SelI.
After running the tblastn, fifteen hits on two different scaffolds were identified and both of the scaffolds had an e-value (smaller than 0.0001) and identity (bigger than 30%) good enough to be studied. However, results obtained from the complete analysis allowed us to reduce the possible scaffolds to one (JACSYO010000511.1), as it had a better T-Coffee alignment around de Sec position.
As we can observe with Exonerate, the gene is located in the Ophiodon elongatus genome between positions 798545 and 800865, comprising 9 exons in the reverse strand (-). The T-coffee alignment between the predicted protein in O.elongatus and the protein of reference (from D.rerio) had high similarity (with a score of 999) so we can conclude that a coding sequence for this protein is found in the studied organism. Moreover, the analysed protein contains Sec in the same position as the reference protein.
The analysis conducted in Seblastian produced a similar prediction, as the sequence obtained contains 9 exons in the reverse strand (-) and does not include a Met at the beginning of the protein (the same as in the Exonerate prediction). SECIS Search3 identified a SECIS element for this sequence with good infernal and covels score which is found in the 3’-UTR segment of the reverse strand, specifically between positions 798405 and 798478 of the JACSYO010000511.1 scaffold.
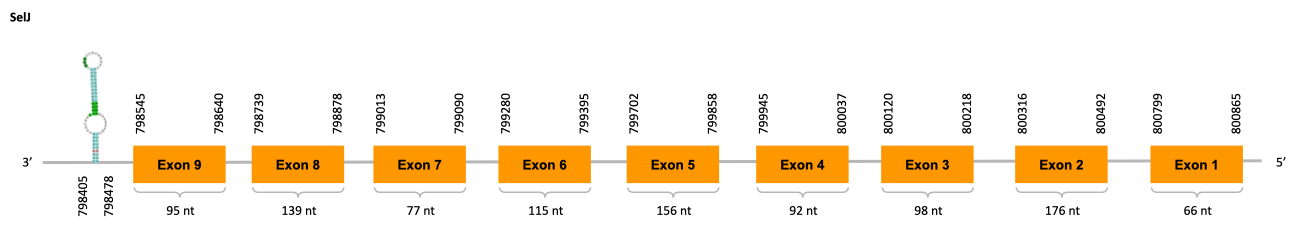
In conclusion, we can say that the gene SelJ is a selenoprotein found in Ophiodon elongatus, due to the presence of a Sec residu and a SECIS element. Moreover, from the information displayed in the phylogenetic tree we can also conclude that it is an homologous protein of the zebrafish SelJ.
After running the tblastn, two hits on two different scaffolds were identified but only one of the scaffolds (JACSYO010000981.1) had an e-value (smaller than 0.0001) and identity (bigger than 30%) good enough to be studied.
As we can observe with Exonerate, the gene is located in the Ophiodon elongatus genome between positions 267080 and 268351, comprising 4 exons in the forward strand (+). The T-coffee alignment between the predicted protein in O.elongatus and the protein of reference (from D.rerio) had high similarity (with a score of 983) so we can conclude that a coding sequence for this protein is found in the studied organism. Moreover, the analysed protein contains Sec in the same position as the reference protein.
The analysis conducted in Seblastian could not predict any selenoprotein but SECIS Search3 identified two SECIS elements for this sequence. However, one of them was found on the reverse strand (-), so it is rejected because it is not found on the same strand as the predicted gene. The other one has a good infernal and covels score and is found in the 3’-UTR segment of the forward strand (+), specifically between positions 268623 and 268712 of the JACSYO010000981.1 scaffold.
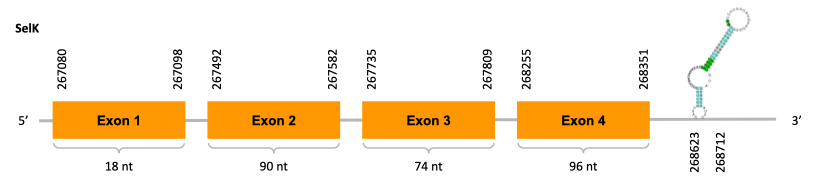
In conclusion, we can say that the gene SelK is a selenoprotein found in Ophiodon elongatus, due to the presence of a Sec residu and a SECIS element. Moreover, from the information displayed in the phylogenetic tree we can also conclude that it is an homologous protein of the zebrafish SelK.
After running the tblastn, eight hits on two different scaffolds were identified and both of the scaffolds had an e-value (smaller than 0.0001) and identity (bigger than 30%) good enough to be studied. However, results obtained from the complete analysis allowed us to reduce the possible scaffolds to one (JACSYO010001682.1), as it was the only one which presented a Sec residue in the same position as the reference protein.
As we can observe with Exonerate, the gene is located in the Ophiodon elongatus genome between positions 2876969 and 2879747, comprising 4 exons in the reverse strand (-). The T-coffee alignment between the predicted protein in O.elongatus and the protein of reference (from D.rerio) had high similarity (with a score of 997) so we can conclude that a coding sequence for this protein is found in the studied organism. Moreover, the analysed protein contains Sec in the same position as the reference protein.
The analysis conducted in Seblastian produced a similar prediction, because despite the sequence obtained contains 8 exons in the reverse strand (-), it does not include a Met at the beginning of the protein (the same as in the Exonerate prediction). SECIS Search3 identified a SECIS element for this sequence with good infernal and covels score which is found in the 3’-UTR segment of the reverse strand, specifically between positions 2876719 and 2876796 of the JACSYO010001682.1 scaffold.
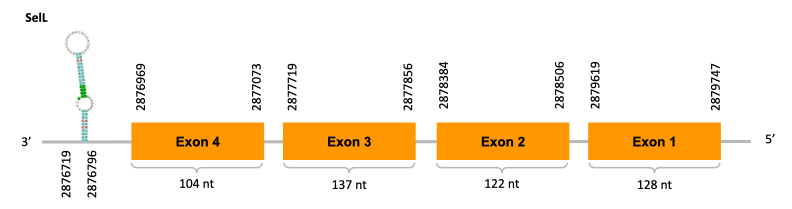
In conclusion, we can say that the gene SelL is a selenoprotein found in Ophiodon elongatus, due to the presence of a Sec residu and a SECIS element. Moreover, from the information displayed in the phylogenetic tree we can also conclude that it is an homologous protein of the zebrafish SelL.
After running the tblastn, five hits on four different scaffolds were identified and two of the scaffolds had an e-value (smaller than 0.0001) and identity (bigger than 30%) good enough to be studied. However, results obtained from the complete analysis allowed us to reduce the possible scaffolds to one (JACSYO010000247.1), which had the best T-coffee alignment.
As we can observe with Exonerate, the gene is located in the Ophiodon elongatus genome between positions 719097 and 721152, comprising 5 exons in the forward strand (+). The T-coffee alignment between the predicted protein in O.elongatus and the protein of reference (from D.rerio) had high similarity (with a score of 999) so we can conclude that a coding sequence for this protein is found in the studied organism. Moreover, the analysed protein contains Sec in the same position as the reference protein.
The analysis conducted in Seblastian produced a better prediction, as the sequence obtained contains 5 exons in the forward strand (+) and includes a Met at the beginning of the protein (which did not happen with the Exonerate prediction). SECIS Search3 identified a SECIS element for this sequence with good infernal and covels score which is found in the 3’-UTR segment of the forward strand (+), specifically between positions 721635 and 721716 of the JACSYO010000247.1 scaffold.
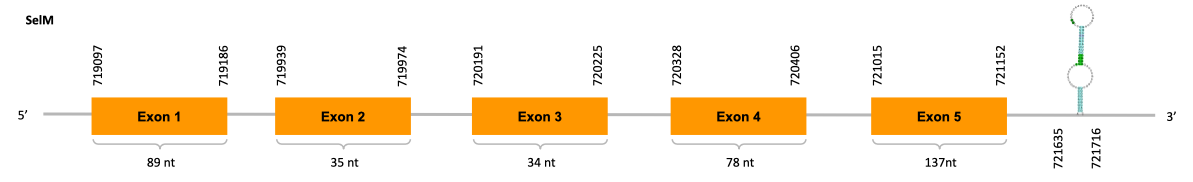
In conclusion, we can say that the gene SelM is a selenoprotein found in Ophiodon elongatus, due to the presence of a Sec residu and a SECIS element. Moreover, from the information displayed in the phylogenetic tree we can also conclude that it is an homologous protein of the zebrafish SelM.
After running the tblastn, eleven hits on three different scaffolds were identified and only one of the scaffolds (JACSYO010002166.1) had an e-value (smaller than 0.0001) and identity (bigger than 30%) good enough to be studied.
As we can observe with Exonerate, the gene is located in the Ophiodon elongatus genome between positions 2926593 and 2931928, comprising 9 exons in the forward strand (+). The T-coffee alignment between the predicted protein in O.elongatus and the protein of reference (from D.rerio) had high similarity (with a score of 994) so we can conclude that a coding sequence for this protein is found in the studied organism. Moreover, the analysed protein contains Sec in the same position as the reference protein.
The analysis conducted in Seblastian produced a better prediction, as the sequence obtained contains 12 exons in the forward strand (+) and includes a Met at the beginning of the protein (which did not happen with the Exonerate prediction). SECIS Search3 identified a SECIS element for this sequence with good infernal and covels score which is found in the 3’-UTR segment of the forward strand (+), specifically between positions 2932459 and 2932532 of the JACSYO010002166.1 scaffold.
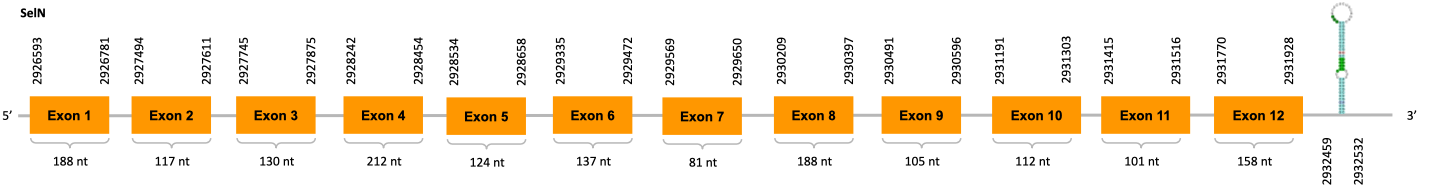
In conclusion, we can say that the gene SelN is a selenoprotein found in Ophiodon elongatus, due to the presence of a Sec residu and a SECIS element. Moreover, from the information displayed in the phylogenetic tree we can also conclude that it is an homologous protein of the zebrafish SelN.
After running the tblastn, twenty-one hits on three different scaffolds were identified and all the scaffolds had an e-value (smaller than 0.0001) and identity (bigger than 30%) good enough to be studied. However, results obtained from the complete analysis allowed us to reduce the possible scaffolds to one (JACSYO010000740.1), which had the best T-coffee alignment.
As we can observe with Exonerate, the gene is located in the Ophiodon elongatus genome between positions 133486 and 138062, comprising 9 exons in the forward strand (+). The T-coffee alignment between the predicted protein in O.elongatus and the protein of reference (from D.rerio) had high similarity (with a score of 997) so we can conclude that a coding sequence for this protein is found in the studied organism. Moreover, the analysed protein contains Sec in the same position as the reference protein.
The analysis conducted in Seblastian produced a better prediction, as the sequence obtained contains 9 exons in the forward strand (+) and includes a Met at the beginning of the protein (the same as in the Exonerate prediction). SECIS Search3 identified a SECIS element for this sequence with good infernal and covels score which is found in the 3’-UTR segment of the forward strand (+), specifically between positions 138546 and 138624 of the JACSYO010000740.1 scaffold.
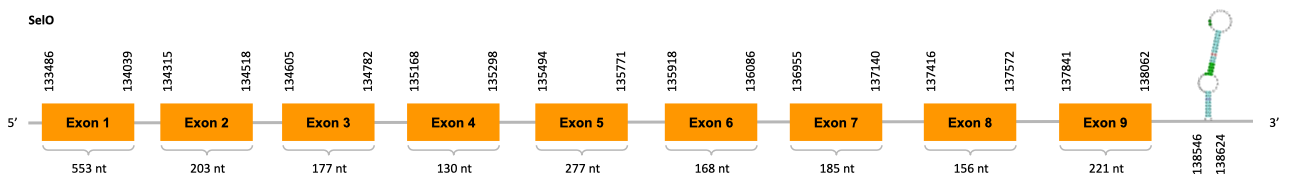
In conclusion, we can say that the gene SelO is a selenoprotein found in Ophiodon elongatus, due to the presence of a Sec residu and a SECIS element. Moreover, from the information displayed in the phylogenetic tree we can also conclude that it is an homologous protein of the zebrafish SelO.
After running the tblastn, eight hits on four different scaffolds were identified and two of the scaffolds had an e-value (smaller than 0.0001) and identity (bigger than 30%) good enough to be studied. However, results obtained from the complete analysis allowed us to reduce the possible scaffolds to one (JACSYO010000335.1), which had the best T-coffee alignment.
As we can observe with Exonerate, the gene is located in the Ophiodon elongatus genome between positions 348133 and 348881, comprising 2 exons in the forward strand (+). The T-coffee alignment between the predicted protein in O.elongatus and the protein of reference (from D.rerio) had high similarity (with a score of 969) so we can conclude that a coding sequence for this protein is found in the studied organism. Moreover, the analysed protein contains Sec in the same position as the reference protein.
The analysis conducted in Seblastian produced a better prediction, as the sequence obtained contains 4 exons in the forward strand (+) and includes a Met at the beginning of the protein (which did not happen with the Exonerate prediction). SECIS Search3 identified two SECIS elements for this sequence but we selected the one with better infernal and covels score which is found in the 3’-UTR segment of the forward strand (+), specifically between positions 350194 and 350194 of the JACSYO010000335.1 scaffold.
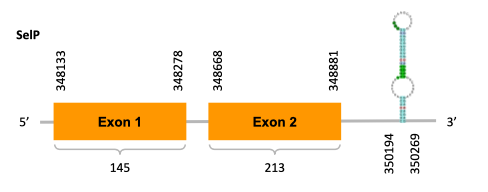
In conclusion, we can say that the gene SelP is a selenoprotein found in Ophiodon elongatus, due to the presence of a Sec residu and a SECIS element. Moreover, from the information displayed in the phylogenetic tree we can also conclude that it is an homologous protein of the zebrafish SelP.
After running the tblastn, ten hits on three different scaffolds were identified and two of the scaffolds had an e-value (smaller than 0.0001) and identity (bigger than 30%) good enough to be studied. Results obtained from the complete analysis could not allow us to reduce the possible scaffolds to one due to the fact that the T-Coffee results were good for both indicating that there could be a duplication. Therefore, we will analyse both scaffolds.
As we can observe with Exonerate, the gene is located in the JACSYO010001932.1 of the Ophiodon elongatus genome between positions 1586927 and 1588059, comprising 3 exons in the forward strand (+). The T-coffee alignment between the predicted protein in O.elongatus and the protein of reference (from D.rerio) had high similarity (with a score of 1000) so we can conclude that a coding sequence for this protein is found in the studied organism. Moreover, the analysed protein contains Sec in the same position as the reference protein.
The analysis conducted in Seblastian produced a similar prediction, as the sequence obtained contains 3 exons in the forward strand (+) and includes a Met at the beginning of the protein (the same as in the Exonerate prediction). SECIS Search3 identified a SECIS elements for this sequence with good infernal and covels score which is found in the 3’-UTR segment of the forward strand (+), specifically between positions 1589047 and 1589121 of the JACSYO010001932.1 scaffold.
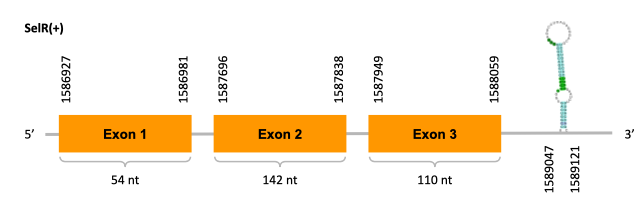
On the other scaffold (JACSYO010002129.1), the gene is located between positions 3429717 and 3433976, comprising 3 exons in the reverse strand (-). The T-coffee alignment between the predicted protein in O.elongatus and the protein of reference (from D.rerio) had high similarity (with a score of 1000) so we can conclude that a coding sequence for this protein is found in the studied organism. Moreover, the analysed protein contains Sec in the same position as the reference protein.
The analysis conducted in Seblastian produced a similar prediction, because despite the sequence obtained contains 4 exons in the reverse strand (-), it includes a Met at the beginning of the protein (the same as in the Exonerate prediction). SECIS Search3 identified two SECIS elements for this sequence but one of them was discarded because it is predicted before the end of our gene. Therfore, we selected the one which is found in the 3’-UTR segment of the reverse strand, specifically between positions 3429498 and 3429571 of the JACSYO010002129.1 scaffold.
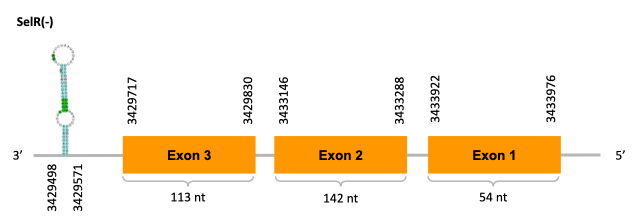
In conclusion, we can say that the gene SelR is a selenoprotein found in Ophiodon elongatus, due to the presence of a Sec residu and a SECIS element. Moreover, from the information displayed in the phylogenetic tree we can also conclude that it is an homologous protein of the zebrafish SelR and SelR1.
After running the tblastn, ten hits on three different scaffolds were identified and two of the scaffolds had an e-value (smaller than 0.0001) and identity (bigger than 30%) good enough to be studied. Results obtained from the complete analysis could not allowed us to reduce the possible scaffolds to one due to the fact that the T-Coffee results were good for both indicating that there could be a duplication. Therefore, we will analyse both scaffolds.
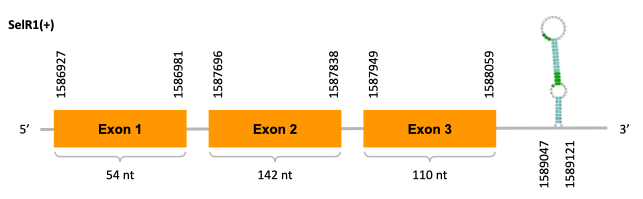
As we can observe with Exonerate, the gene is located in the JACSYO010001932.1 of the Ophiodon elongatus genome between positions 1586927 and 1588059, comprising 3 exons in the forward strand (+). The T-coffee alignment between the predicted protein in O.elongatus and the protein of reference (from D.rerio) had high similarity (with a score of 1000) so we can conclude that a coding sequence for this protein is found in the studied organism. Moreover, the analysed protein contains Sec in the same position as the reference protein. In fact, this prediction is exactly the same as the one on SelR, which we will discuss later on in the discussion of the phylogenetic tree.
The analysis conducted in Seblastian produced a similar prediction, as the sequence obtained contains 3 exons in the forward strand (+) and includes a Met at the beginning of the protein (the same as in the Exonerate prediction). SECIS Search3 identified a SECIS element for this sequence with good infernal and covels score which is found in the 3’-UTR segment of the forward strand (+), specifically between positions 1589047 and 1589121 of the JACSYO010001932.1 scaffold.
On the other scaffold (JACSYO010002129.1), the gene is located between positions 3432772 and 3433976, comprising 3 exons in the reverse strand (-). The T-coffee alignment between the predicted protein in O.elongatus and the protein of reference (from D.rerio) had high similarity (with a score of 1000) so we can conclude that a coding sequence for this protein is found in the studied organism. Moreover, the analysed protein contains Sec in the same position as the reference protein. In fact, the prediction of the protein is exactly the same as the one on SelR, it only shows a difference in the position of the third exon. This will discuss later on in the discussion of the phylogenetic tree.
However, Seblastian does not predict any selenoprotein nor SECIS element inside the sequence studied.
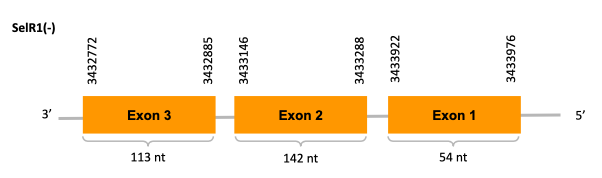
In conclusion, we cannot affirm that the gene SelR1 is a selenoprotein found in Ophiodon elongatus, because even though it presents a Sec residue on the same position as the reference protein, no SECIS element was found. Moreover, from the information displayed in the phylogenetic tree we can also conclude that it is an homologous protein of the zebrafish SelR and SelR1.
After running the tblastn, thirteen hits on seven different scaffolds were identified and two of the scaffolds had an e-value (smaller than 0.0001) and identity (bigger than 30%) good enough to be studied. However, results obtained from the complete analysis allowed us to reduce the possible scaffolds to one (JACSYO010000605.1), which had the best T-coffee alignment.
As we can observe with Exonerate, the gene is located in the Ophiodon elongatus genome between positions 1743106 and 1744400, comprising 4 exons in the forward strand (+). The T-coffee alignment between the predicted protein in O.elongatus and the protein of reference (from D.rerio) had high similarity (with a score of 999) so we can conclude that a coding sequence for this protein is found in the studied organism.
The analysis conducted in Seblastian could not produce a prediction but SECIS Search3 identified a SECIS element for this sequence with good infernal and covels score which is found in the 3’-UTR segment of the forward strand (+), specifically between positions 1705384 and 1705384 of the JACSYO010000605.1 scaffold.
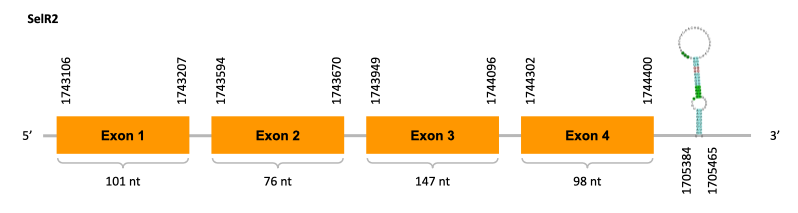
In conclusion, we can say that the gene SelR2 is on the Ophiodon elongatus genome but is not a selenoprotein (because it has not a Sec residue nor SECIS elements). In fact, it is a cysteine-containing homolog found in Ophiodon elongatus, due to the presence of a Cys residue, even though a SECIS element was identified through SECIS Search3 (this indicates that the protein could have been a selenoprotein on the past but lost the Sec residue). Moreover, from the information displayed in the phylogenetic tree we can also conclude that it is an homologous protein of the zebrafish SelR2.
After running the tblastn, fifteen hits on six different scaffolds were identified and three of the scaffolds had an e-value (smaller than 0.0001) and identity (bigger than 30%) good enough to be studied. However, results obtained from the complete analysis allowed us to reduce the possible scaffolds to one (JACSYO010001636.1), which had the best T-coffee alignment.
As we can observe with Exonerate, the gene is located in the Ophiodon elongatus genome between positions 1500866 and 1509463, comprising 6 exons in the forward strand (+). The T-coffee alignment between the predicted protein in O.elongatus and the protein of reference (from D.rerio) had high similarity (with a score of 980) so we can conclude that a coding sequence for this protein is found in the studied organism. Moreover, the analysed protein contains Sec in the same position as the reference protein.
Seblastian does not predict any selenoprotein nor SECIS element inside the sequence studied, which is expected as this protein doesn’t have a Sec residue which indicates it is a Cys-containing homolog.
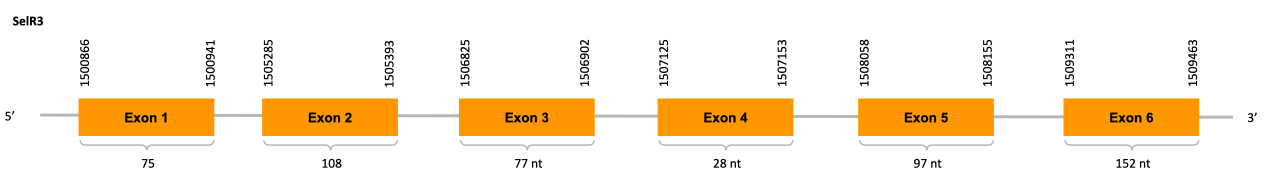
In conclusion, we can say that the gene SelR3 is on the Ophiodon elongatus genome but is not a selenoprotein (because it has not a Sec residue nor SECIS elements). In fact, it is a Cys-containing homolog found in Ophiodon elongatus, due to the presence of a Cys residue and the absence of SECIS elements and Seblastian prediction. Moreover, from the information displayed in the phylogenetic tree we can also conclude that it is an homologous protein of the zebrafish SelR3.
After running the tblastn, five hits on three different scaffolds were identified but only one scaffold (JACSYO010001703.1) had an e-value (smaller than 0.0001) and identity (bigger than 30%) good enough to be studied.
As we can observe with Exonerate, the gene is located in the Ophiodon elongatus genome between positions 922421 and 926886, comprising 5 exons in the forward strand (+). The T-coffee alignment between the predicted protein in O.elongatus and the protein of reference (from D.rerio) had high similarity (with a score of 946) so we can conclude that a coding sequence for this protein is found in the studied organism. Moreover, the analysed protein contains Sec in the same position as the reference protein.
The analysis conducted in Seblastian could not produce a prediction, but SECIS Search3 identified two SECIS elements for this sequence from where we selected the one with better infernal and covels score which is found in the 3’-UTR segment of the forward strand (+), specifically between positions 927177 and 927253 of the JACSYO010001703.1 scaffold.
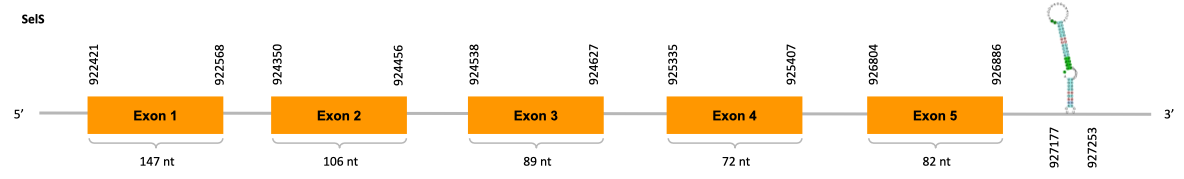
In conclusion, we can say that the gene SelS is a selenoprotein found in Ophiodon elongatus, due to the presence of a Sec residu and a SECIS element. Moreover, from the information displayed in the phylogenetic tree we can also conclude that it is an homologous protein of the zebrafish SelS.
After running the tblastn, nine hits on three different scaffolds were identified and two of the scaffolds had an e-value (smaller than 0.0001) and identity (bigger than 30%) good enough to be studied. However, results obtained from the complete analysis allowed us to reduce the possible scaffolds to one (JACSYO010001924.1), which had the best T-coffee alignment.
As we can observe with Exonerate, the gene is located in the Ophiodon elongatus genome between positions 3746598 and 3750901, comprising 5 exons in the reverse strand (-). The T-coffee alignment between the predicted protein in O.elongatus and the protein of reference (from D.rerio) had high similarity (with a score of 970) so we can conclude that a coding sequence for this protein is found in the studied organism. Moreover, the analysed protein contains Sec in the same position as the reference protein.
The analysis conducted in Seblastian produced a similar prediction, as the sequence obtained contains 5 exons in the reverse strand (-) and includes a Met at the beginning of the protein (the same as in the Exonerate prediction). SECIS Search3 identified a SECIS element for this sequence with good infernal and covels score which is found in the 3’-UTR segment of the revserse strand (-), specifically between positions 3746256 and 3746329 of the JACSYO010001924.1 scaffold.
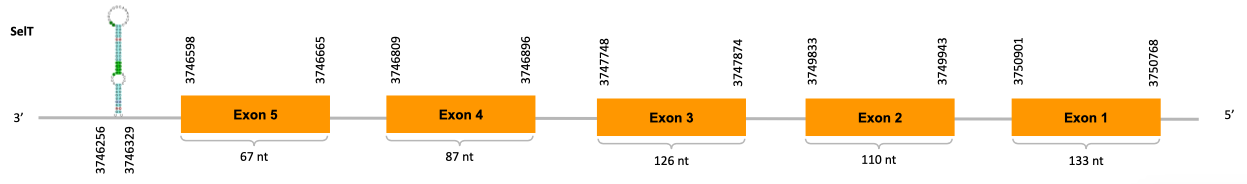
In conclusion, we can say that the gene SelT is a selenoprotein found in Ophiodon elongatus, due to the presence of a Sec residu and a SECIS element. Moreover, from the information displayed in the phylogenetic tree we can also conclude that it is an homologous protein of the zebrafish SelT.
After running the tblastn, twelve hits on three different scaffolds were identified and all the scaffolds had an e-value (smaller than 0.0001) and identity (bigger than 30%) good enough to be studied. However, we could only obtain Exonerate results for two of them and results obtained from the complete analysis allowed us to reduce the possible scaffolds to one (JACSYO010001669.1), which had the best T-coffee alignment.
As we can observe with Exonerate, the gene is located in the Ophiodon elongatus genome between positions 53588 and 54961, comprising 4 exons in the forward strand (+). The T-coffee alignment between the predicted protein in O.elongatus and the protein of reference (from D.rerio) had high similarity (with a score of 982) so we can conclude that a coding sequence for this protein is found in the studied organism. Moreover, the analysed protein contains Sec in the same position as the reference protein.
The analysis conducted in Seblastian produced a better prediction, because the sequence obtained contains 5 exons in the forward strand (+) and includes a Met at the beginning of the protein (which did not happen with the Exonerate prediction). SECIS Search3 identified a SECIS element for this sequence with good infernal and covels score which is found in the 3’-UTR segment of the forward strand (+), specifically between positions 55090 and 55160 of the JACSYO010001669.1 scaffold.
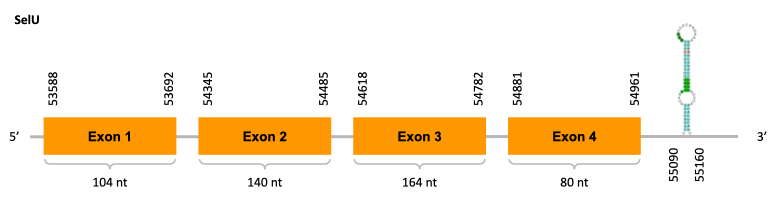
In conclusion, we can say that the gene SelU is a selenoprotein found in Ophiodon elongatus, due to the presence of a Sec residu and a SECIS element. Moreover, from the information displayed in the phylogenetic tree we can also conclude that it is an homologous protein of the zebrafish SelU.
After running the tblastn, eight hits on three different scaffolds were identified and only one of the scaffolds (JACSYO010002009.1) had an e-value (smaller than 0.0001) and identity (bigger than 30%) good enough to be studied.
As we can observe with Exonerate, the gene is located in the Ophiodon elongatus genome between positions 707478 and 709150, comprising 6 exons in the reverse strand (-). The T-coffee alignment between the predicted protein in O.elongatus and the protein of reference (from D.rerio) had high similarity (with a score of 994) so we can conclude that a coding sequence for this protein is found in the studied organism.
The analysis conducted in Seblastian could not produce a prediction but SECIS Search3 identified three SECIS elements for this sequence. All have good infernal and covels score, but two of them are is discarded because they were predicted before the end of our gene. The other one is found in the 3’-UTR segment of the revserse strand (-), specifically between positions 682679 and 682763 of the JACSYO010002009.1 scaffold.
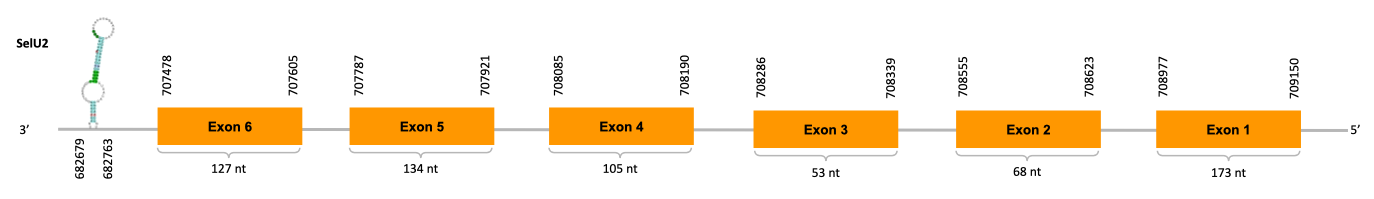
In conclusion, we can say that the gene SelU2 is on the Ophiodon elongatus genome but is not a selenoprotein (because it has not a Sec residue nor SECIS elements). In fact, it is a Cys-containing homolog found in Ophiodon elongatus, due to the presence of a Cys residue, even though a SECIS element was identified through SECIS Search3 (this indicates that the protein could have been a selenoprotein on the past but lost the Sec residue). Moreover, from the information displayed in the phylogenetic tree we can also conclude that it is an homologous protein of the zebrafish SelU2.
After running the tblastn, eight hits on four different scaffolds were identified and only one of the scaffolds (JACSYO010001618.1) had an e-value (smaller than 0.0001) and identity (bigger than 30%) good enough to be studied.
As we can observe with Exonerate, the gene is located in the Ophiodon elongatus genome between positions 840617 and 845246, comprising 6 exons in the reverse strand (-). The T-coffee alignment between the predicted protein in O.elongatus and the protein of reference (from D.rerio) had high similarity (with a score of 1000) so we can conclude that a coding sequence for this protein is found in the studied organism.
Seblastian does not predict any selenoprotein nor SECIS element inside the sequence studied, which is expected as this protein doesn’t have a Sec residue which indicates it is a Cys-containing homolog.
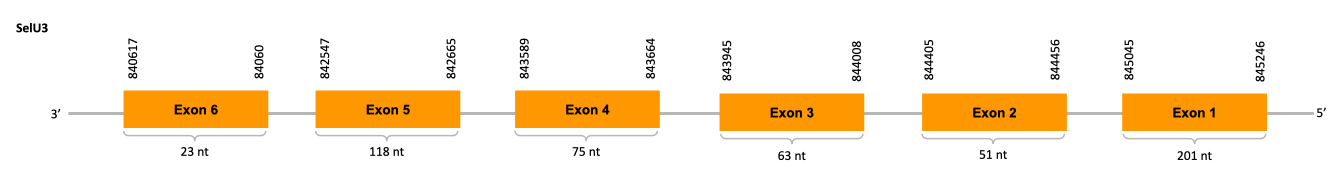
In conclusion, we can say that the gene SelU3 is on the Ophiodon elongatus genome but is not a selenoprotein (because it has not a Sec residue nor SECIS elements). In fact, it is a Cys-containing homolog found in Ophiodon elongatus, due to the presence of a Cys residue and the absence of SECIS elements and Seblastian prediction. Moreover, from the information displayed in the phylogenetic tree we can also conclude that it is an homologous protein of the zebrafish SelU3.
After running the tblastn, two hits on one scaffold (JACSYO010000671.1) were identified and they had an e-value (smaller than 0.0001) and identity (bigger than 30%) good enough to be studied.
As we can observe with Exonerate, the gene is located in the Ophiodon elongatus genome between positions 1718117 and 1719393, comprising 4 exons in the reverse strand (-). The T-coffee alignment between the predicted protein in O.elongatus and the protein of reference (from D.rerio) had high similarity (with a score of 1000) so we can conclude that a coding sequence for this protein is found in the studied organism. Moreover, the analysed protein contains Sec in the same position as the reference protein.
The analysis conducted in Seblastian produced a similar prediction, as the sequence obtained contains 4 exons in the forward strand (+) and includes a Met at the beginning of the protein (the same as in the Exonerate prediction). SECIS Search3 identified a SECIS element for this sequence with good infernal and covels score which is found in the 3’-UTR segment of the reverse strand (-), specifically between positions 1717844 and 1717917 of the JACSYO010000671.1 scaffold.
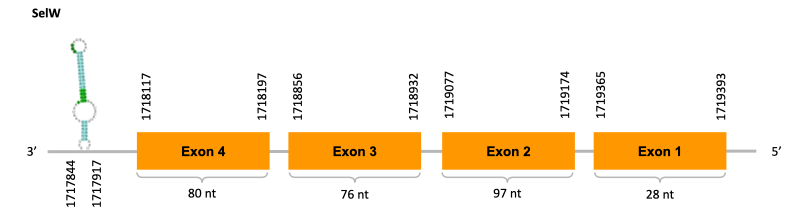
In conclusion, we can say that the gene SelW is a selenoprotein found in Ophiodon elongatus, due to the presence of a Sec residu and a SECIS element. Moreover, from the information displayed in the phylogenetic tree we can also conclude that it is an homologous protein of the zebrafish SelW.
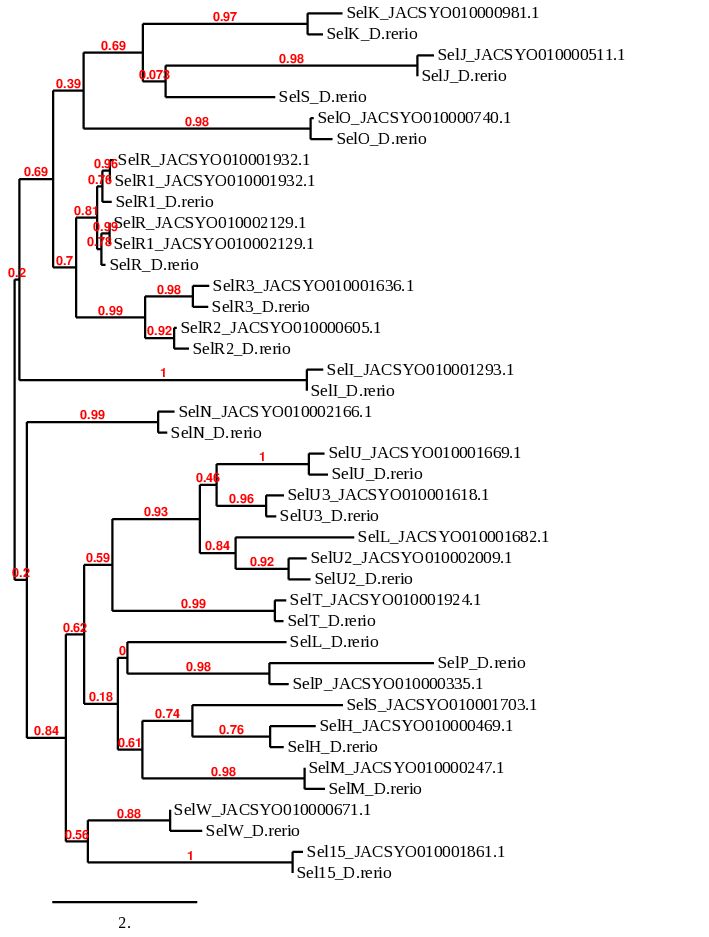
In this phylogenetic tree, most of the predicted proteins of Ophiodon elongatus are closely related to their homologues in Danio rerio, so the results observed are coherent with the ones we extracted from our data.
However, some inconsistencies can be observed on the phylogenetic tree. By one hand, SelL and SelS are not as closely related to their homologues as the other Selenoproteins. To elucidate the problem, we performed another phylogenetic study on the SelL using all of the studied scaffolds of Ophiodon elongatus. The resulting phylogenetic tree corroborates our results because the scaffold that showed more similarity with the SelL from Danio rerio was the one we selected during our analysis (JACSYO0100011682.1). Therefore, we can conclude that, even though these proteins are not as closely related to their homologues in Danio rerio, they are still homologues.
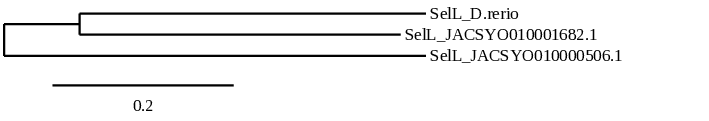
By the other hand, as we previously mentioned, we can see that SelR and SelR1 present some inconsistencies in the phylogenetic relation. Despite the fact that they are both closely related to their homologues in each species, if we look at the scaffolds from which the proteins were extracted, we can see that the scaffold JACSYO010001932.1 from both proteins is closely related to SelR1 of Danio rerio whereas the scaffold JACSYO010002129.1 from both proteins is closely related to SelR. Even though we hypothesized that the second scaffold of SelR1 could not be a selenoprotein because it lacked a SECIS element, we think that it possibly had one and the SECIS Search3 could not find it due to a server’s intrinsic error. All this, together with the fact that the proteins of the same scaffold have an almost identical structure, suggests that these selenoprotein genes are extremely similar.
In addition, we can see two big groups of related proteins. Proteins SelK, SelJ and SelO are related, confirming relationship between them, and they are also related to the SelR subfamily (containing SelR, SelR1, SelR2, SelR3). This group also contains the SelI protein. On the other grup, we can find the protein SelT and the subfamily SelU (containing SelU, SelU2 and SelU3), which are also related to SelP, SelH and SelM. Finally, we can also relate them with SelW and Sel15
In conclusion, the phylogenetic tree obtained confirms that our predicted selenoproteins and cysteine-containing proteins are homologous of the zebrafish ones, and they evolved in the same way.
After running the tblastn, thirty-two hits on five different scaffolds were identified but four of the scaffolds had an e-value (smaller than 0.0001) and identity (bigger than 30%) good enough to be studied. However, results obtained from the complete analysis allowed us to reduce the possible scaffolds to one (JACSYO010001694.1), which had the best T-coffee alignment.
As we can observe with Exonerate, the gene is located in the Ophiodon elongatus genome between positions 1737072 and 1756998, comprising 15 exons in the forward strand (+). The T-coffee alignment between the predicted protein in O.elongatus and the protein of reference (from D.rerio) had high similarity (with a score of 999) so we can conclude that a coding sequence for this protein is found in the studied organism. Moreover, the analysed protein contains a Sec in the same position as the reference protein.
The analysis conducted in Seblastian does a quite different prediction, as the sequence obtained contains 12 exons instead of 15. SECIS Search3 identified a SECIS element for this sequence with good infernal and covels score which is found in the 3’-UTR segment of the forward strand, specifically between positions 1759150 and 1759221 of the JACSYO010000262.1 scaffold.
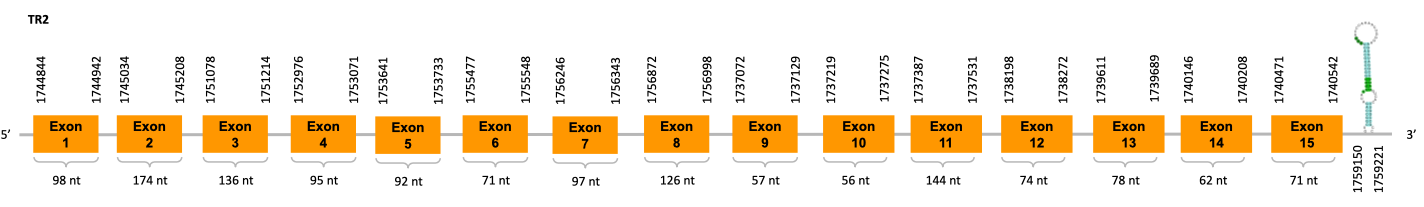
In conclusion, we can say that the gene TR2 is a Selenoprotein found in Ophiodon elongatus, due to the presence of a Sec residu and a SECIS element.
After running the tblastn, thirty-three hits on seven different scaffolds were identified but five of the scaffolds had an e-value (smaller than 0.0001) and identity (bigger than 30%) good enough to be studied. However, results obtained from the complete analysis allowed us to reduce the possible scaffolds to one (JACSYO010000324.1), which had the best T-coffee alignment.
As we can observe with Exonerate, the gene is located in the Ophiodon elongatus genome between positions 14528 and 25421, comprising 16 exons in the forward strand (+). The T-coffee alignment between the predicted protein in O.elongatus and the protein of reference (from D.rerio) had high similarity (with a score of 998) so we can conclude that a coding sequence for this protein is found in the studied organism. Moreover, the analysed protein contains a Sec in the same position as the reference protein.
The analysis conducted in Seblastian also predicted 16 exons, and the SECIS Search3 identified a SECIS element for this sequence with good infernal and covels score which is found in the 3’-UTR segment of the forward strand, specifically between positions 25737 and 25817 of the JACSYO010000324.1 scaffold.
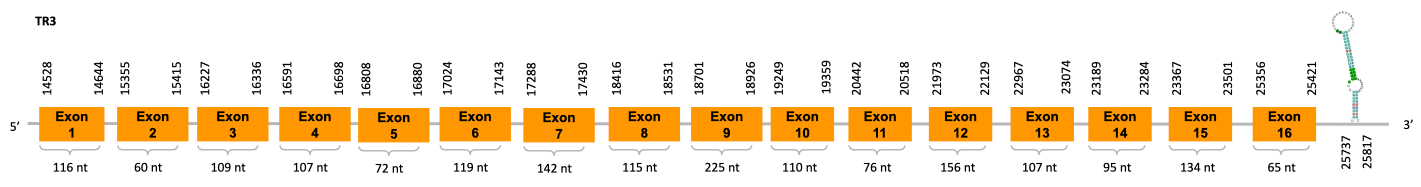
In conclusion, we can say that the gene TR2 is a Selenoprotein found in Ophiodon elongatus, due to the presence of a Sec residu and a SECIS element.
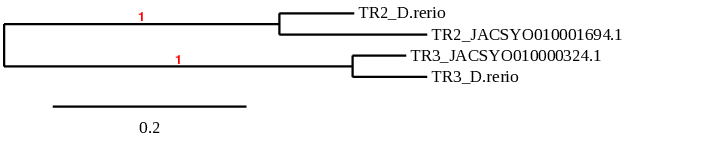
The phylogeny of TR family proteins confirms our predictions that Ophiodon elongatus has both TR2 and TR3 proteins in its genome, and they both pair specifically with these selenoproteins homologous of Danio rerio.
After running the tblastn, eleven hits on five different scaffolds were identified and two scaffolds had an e-value (smaller than 0.0001) and identity (bigger than 30%) good enough to be studied. However, results obtained from the complete analysis allowed us to reduce the possible scaffolds to one (JACSYO010002072.1), which had the best T-coffee alignment.
As we can observe with Exonerate, the gene is located in the Ophiodon elongatus genome between positions 69993 and 72296, comprising 7 exons in the reverse strand (-). The T-coffee alignment between the predicted protein in O.elongatus and the protein of reference (from D.rerio) had high similarity (with a score of 987) so we can conclude that a coding sequence for this protein is found in the studied organism.
The analysis conducted in Seblastian could not produce a prediction and, even though SECIS Search3 identified a SECIS element for this sequence, it was not on the same strand as the analysed gene so it was discarded.
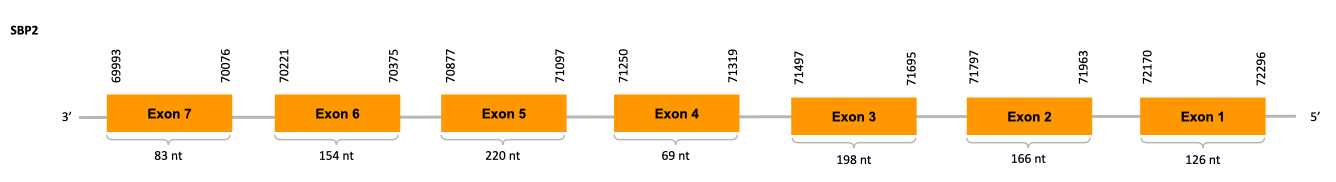
In conclusion, we can say that the gene SBP2 is found on the Ophiodon elongatus genome but is not a selenoprotein (because it has neither a Sec residue nor SECIS elements). Moreover, from the information displayed in the phylogenetic tree we can also conclude that it is an homologous protein of the zebrafish SBP2.
After running the tblastn, eighteen hits on four different scaffolds were identified and two scaffolds had an e-value (smaller than 0.0001) and identity (bigger than 30%) good enough to be studied. However, results obtained from the complete analysis allowed us to reduce the possible scaffolds to one (JACSYO010000597.1), which had the best T-coffee alignment.
As we can observe with Exonerate, the gene is located in the Ophiodon elongatus genome between positions 274032 and 278999, comprising 8 exons in the reverse strand (-). The T-coffee alignment between the predicted protein in O.elongatus and the protein of reference (from D.rerio) had high similarity (with a score of 995) so we can conclude that a coding sequence for this protein is found in the studied organism.
The analysis conducted in Seblastian could not produce a prediction but SECIS Search3 identified a SECIS element for this sequence with good infernal and covels score, but it was discarded because it was predicted before the end of our gene.
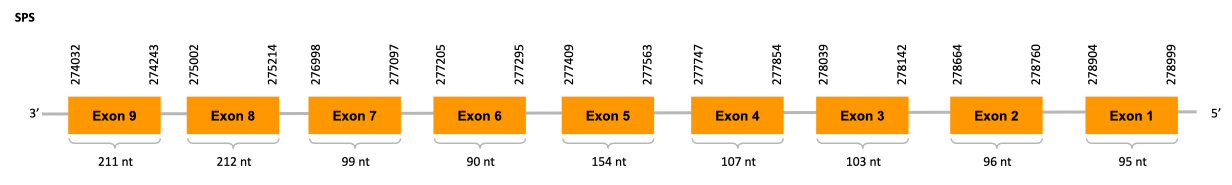
In conclusion, we can say that the gene SPS is on the Ophiodon elongatus genome but is not a selenoprotein (because it has neither a Sec residue nor SECIS elements). IMoreover, from the information displayed in the phylogenetic tree we can also conclude that it is an homologous protein of the zebrafish SPS.
After running the tblastn, eighteen hits on four different scaffolds were identified and two scaffolds had an e-value (smaller than 0.0001) and identity (bigger than 30%) good enough to be studied. However, results obtained from the complete analysis allowed us to reduce the possible scaffolds to one (JACSYO010000005.1), which had the best T-coffee alignment.
As we can observe with Exonerate, the gene is located in the Ophiodon elongatus genome between positions 138644 and 144509, comprising 9 exons in the reverse strand (-). The T-coffee alignment between the predicted protein in O.elongatus and the protein of reference (from D.rerio) had high similarity (with a score of 987) so we can conclude that a coding sequence for this protein is found in the studied organism. Moreover, the analysed protein contains a Sec in the same position as the reference protein.
The analysis conducted in Seblastian produced a better prediction, as the sequence obtained contains 8 exons in the reverse strand (-) and includes a Met at the beginning of the protein (which did not happen with the Exonerate prediction). SECIS Search3 identified a SECIS element for this sequence with good infernal and covels score which is found in the 3’-UTR segment of the reverse strand, specifically between positions 137835 and 137921 of the JACSYO010000005.1 scaffold.
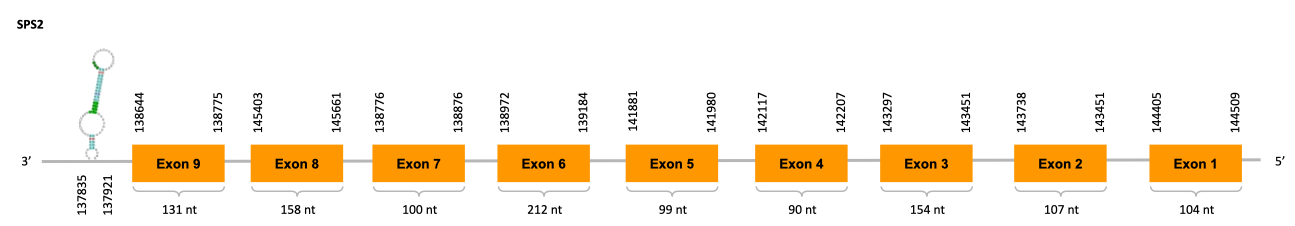
In conclusion, we can say that the gene SPS2 is on the Ophiodon elongatus genome and it is a selenoprotein (because it has a Sec residue and a SECIS element). Moreover, from the information displayed in the phylogenetic tree we can also conclude that it is an homologous protein of the zebrafish SPS2.
After running the tblastn, thirteen hits on three different scaffolds were identified and only one scaffold (JACSYO010001725.1) had an e-value (smaller than 0.0001) and identity (bigger than 30%) good enough to be studied.
As we can observe with Exonerate, the gene is located in the Ophiodon elongatus genome between positions 887062 and 895283, comprising 9 exons in the forward strand (+). The T-coffee alignment between the predicted protein in O.elongatus and the protein of reference (from D.rerio) had high similarity (with a score of 1000) so we can conclude that a coding sequence for this protein is found in the studied organism.
The analysis conducted in Seblastian could not produce a prediction and SECIS Search3 could not identify any SECIS element for this sequence.In conclusion, we can say that the gene SecS is found on the Ophiodon elongatus genome but is not a selenoprotein (because it has not a Sec residue nor SECIS elements). Moreover, from the information displayed in the phylogenetic tree we can also conclude that it is an homologous protein of the zebrafish SecS.
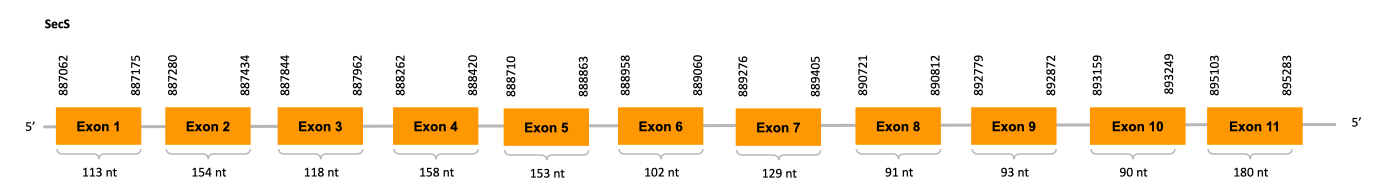
In conclusion, we can say that the gene SecS is found on the Ophiodon elongatus genome but is not a selenoprotein (because it has not a Sec residue nor SECIS elements). Moreover, from the information displayed in the phylogenetic tree we can also conclude that it is an homologous protein of the zebrafish SecS.
After running the tblastn, 44 hits on 26 different scaffolds were identified and seven scaffolds had an e-value (smaller than 0.0001) and identity (bigger than 30%) good enough to be studied. However, we could only obtain Exonerate results for two of them and results obtained from the complete analysis allowed us to reduce the possible scaffolds to one (JACSYO010000096.1), which had the best T-coffee alignment.
As we can observe with Exonerate, the gene is located in the Ophiodon elongatus genome between positions 210120 and 213656, comprising 4 exons in the reverse strand (-). The T-coffee alignment between the predicted protein in O.elongatus and the protein of reference (from D.rerio) had high similarity (with a score of 1000) so we can conclude that a coding sequence for this protein is found in the studied organism.
The analysis conducted in Seblastian could not produce a prediction and, even though SECIS Search3 identified a SECIS element for this sequence, it was not on the same strand as the analysed gene so it was discarded.
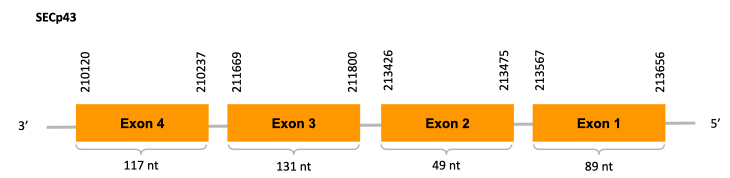
In conclusion, we can say that the gene SECp43 is found on the Ophiodon elongatus genome but is not a selenoprotein (because it has neither a Sec residue nor SECIS elements). Moreover, from the information displayed in the phylogenetic tree we can also conclude that it is an homologous protein of the zebrafish SECp43.
After running the tblastn, six hits on four different scaffolds were identified and only one scaffold (JACSYO010001669.1) had an e-value (smaller than 0.0001) and identity (bigger than 30%) good enough to be studied.
As we can observe with Exonerate, the gene is located in the Ophiodon elongatus genome between positions 3430986 and 3432365, comprising 3 exons in the reverse strand (-). The T-coffee alignment between the predicted protein in O.elongatus and the protein of reference (from D.rerio) had high similarity (with a score of 958) so we can conclude that a coding sequence for this protein is found in the studied organism.
The analysis conducted in Seblastian could not produce a prediction and SECIS Search3 could not identify any SECIS element for this sequence.
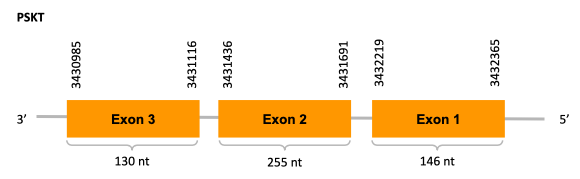
In conclusion, we can say that the gene PSTK is found on the Ophiodon elongatus genome but is not a selenoprotein (because it has neither a Sec residue nor SECIS elements). Moreover, from the information displayed in the phylogenetic tree we can also conclude that it is an homologous protein of the zebrafish PSTK.
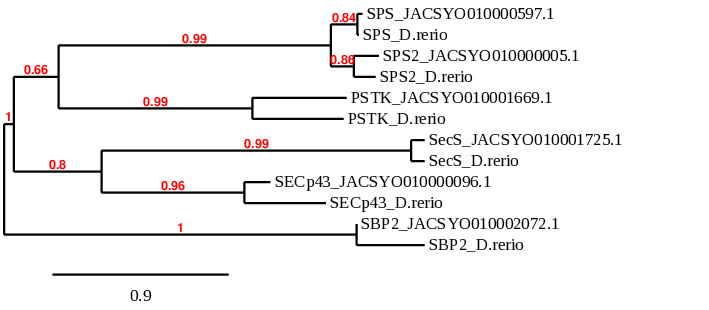
As we can see in the phylogeny, all machinery proteins predicted from Ophiodon elongatus’ genome pair specifically with their homologues in zebrafish. So the results observed are coherent with the ones we extracted from our data. In the phylogeny we can also see that SPS and SPS2 are related proteins, as well as SecS and SECp43, indicating that they could have been originated from the same protein, respectively.
Selenoproteins are proteins which are not well described among species. Accordingly, the aim of this project is to analyse and identify the selenoproteins present in Ophiodon elongatus through the comparison with the characterized selenoproteins in Danio rerio. The project includes the analysis of 25 selenoproteins, 7 cysteine-containing homologues of existing selenoproteins and 6 machinery proteins that participate in the synthesis of selenoproteins.
After analyzing all the proteins, we found that Ophiodon elongatus’ genome has all of the selenoproteins of zebrafish, which are: DI, DI1, Sel15, SelH, SelI, SelJ, SelK, SelL, SelM, SelN, SelO, SelP, SelR, SelR1, SelS, SelT, SelU, SelW, GPx, GPx2, GPx3, GPx4, TR2, TR3, Fep15. In addition, we hypothesize that GPx may be duplicated in the genome of Ophiodon elongatus. Moreover, regarding the SelR and SelR1 issue further research is needed to elucidate whether the observation is due to an internal bug of the analysis or a special phylogenetic relation.
Considering cysteine-containing homologues, we predicted all of these proteins in O. elongatus’ genome, some of them had SECIS elements (SelR2, SelU2, GPx7, GPx8), while others did not (MsrA, SelR3, SelU3).
Finally, regarding the selenoprotein machinery, we could also predict machinery proteins in Ophiodon elongatus, homologous from the ones found in zebrafish. These predicted proteins are SBP2, SPS, SPS2, SecS, SECp43, PSTK and SPS2. In this case, all of the predicted proteins are cysteine-containing homologues except from SPS2, which is a selenoprotein due to the fact that it contains a Sec residue and a SECIS element.
Regarding the predictions of SECIS elements using Seblastian, it detected SECIS for almost all of the Selenoproteins we studied. For the cysteine-containing proteins, SECIS elements were predicted for some of the proteins, whereas not for others. The absence of SECIS elements in cysteine-containing proteins may be due to the loss of these elements during evolution, as there is no need to maintain them when there is no selenoprotein to be translated. Some of the cysteine-homologous proteins did contain these SECIS elements, indicating that they probably evolved from Selenoproteins and still have not lost these SECIS elements.
In addition, SECIS Search3 was able to identify SECIS elements for SBP2, SPS and SECp43. Nevertheless, they were discarded because they were predicted before the end of our gene or were not located on the same strand as the analysed gene.
During this study, we found some limitations. First of all, we were not able to obtain results from the Genewise program, that is why we did not compare our results with the ones we would have obtained from the t-coffee after using Genewise.
An other limitation was that the annotation of the genome analysed is fractioned, and some of the proteins were found in different scaffolds of Ophiodon elongatus’ genome. In addition, for some proteins there were large number of aminoacids missing, complicating our predictions. Many of the predicted selenoproteins and Zebrafish proteins didn’t start with a Methionine. This could mean that we are missing parts of the beginning of the gene so the predictions could only be a fragment of the whole protein. By using Seblastian, we partially corrected this problem because in this program almost all proteins started with a methionine.
To sum up, this project allowed us to computationally characterize for the first time the selenoproteome of Ophiodon elongatus. However, further research is needed to elucidate if the inconsistencies in the predicted proteins are due to limitations of the methods used or real evolutionary changes. A possible future research line to solve this problem could be aligning the genome of Ophiodon elongatus with other organisms more closely related phylogenetically.
[1] Labunskyy, V. M., Hatfield, D. L., & Gladyshev, V. N. (2014). Selenoproteins: molecular pathways and physiological roles. Physiol rev. 94(3): 739-777.
[2] Xu, X. J., Zhang, D. G., Zhao, T., Xu, Y. H., & Luo, Z. (2020). Characterization and expression analysis of seven selenoprotein genes in yellow catfish Pelteobagrus fulvidraco to dietary selenium levels. J Trace Elem Med Biol. 62: 126600.
[3] Papp, L. V., Lu, J., Holmgren, A., & Khanna, K. K. (2007). From selenium to selenoproteins: synthesis, identity, and their role in human health. Antioxid Redox Signal. 9(7): 775-806.
[4] Lu, Jun, & Arne Holmgren (2009). Selenoproteins. Journal of Biological Chemistry. 284 (2): 723-727.
[5] Girard, C.F. (1854). Descriptions of new fishes, collected by Dr. A. L. Heermann, naturalist attached to the survey of the Pacific railroad route, under Lieut. R. S. Williamson, U. S. A. Proc Acad Nat Sci Phila. 7: 129-140.
[6] Starr, R. M., O'Connell, V., & Ralston, S. (2004). Movements of lingcod (Ophiodon elongatus) in southeast Alaska: potential for increased conservation and yield from marine reserves. Can J Fish Aquat Sci. 61(7): 1083-1094..
[7] Lingcod (c2021). EDF Seafood Selector. https://seafood.edf.org/lingcod?tagID=15732
[8] Martell, S. J., Walters, C. J., & Wallace, S. S. (2000). The use of marine protected areas for conservation of lingcod (Ophiodon elongatus). Bull Mar Sci. 66(3): 729-743.
