Introduction
Selenium:
Selenium (Se)[1] was discovered by the Swedish chemist Jöns Jacob Berzelius in 1817 and has been recognized as an essential micronutrient element for many life forms since 1957. It is incorporated as selenocysteine (Sec) amino-acid in a wide range of proteins which are called Selenoproteins. Under physiologic conditions the Se in selenocysteine is almost fully ionised and consequently is an extremely efficient biological catalyst.
Selenocysteine:
Selenocysteine is the 21st amino-acid and is encoded by the UGA codon, whose main function is to act as a stop codon. That is the reason why the incorporation of selenium as Sec into a selenoprotein requires a specific mechanism to decode the UGA codon in mRNA, which normally operates in translation termination. Its structure differs from cysteine for presenting a selenium instead of a sulfur. Their chemical properties are similar despite selenocysteine is much more reactive due to its high nucleophilic activity. There is not any cellular selenocysteine storage for avoiding them to introduce on a cysteine concrete location. It reacts with oxygen, tioredoxine and tioredoxine reductase of mammals because it causes a fast NADPH oxidation [2].
Syntesis of selenoproteins:
The incorporation of selenium is produced in both bacteria and eukaryotic. In order to introduce Selenium it has to be Se2-. There are two different ways to incorporate Selenium, in one hand we have the inorganic way in which they go through glutathione-glutaredoxin and thioredoxin pathway and the organic way in which they are metabolized by selenocysteine liasa or by trans-selenation. That is how selenophosphate is produced in a reaction catalyzed by a key enzime: selenophosphate synthetase 2 [3].
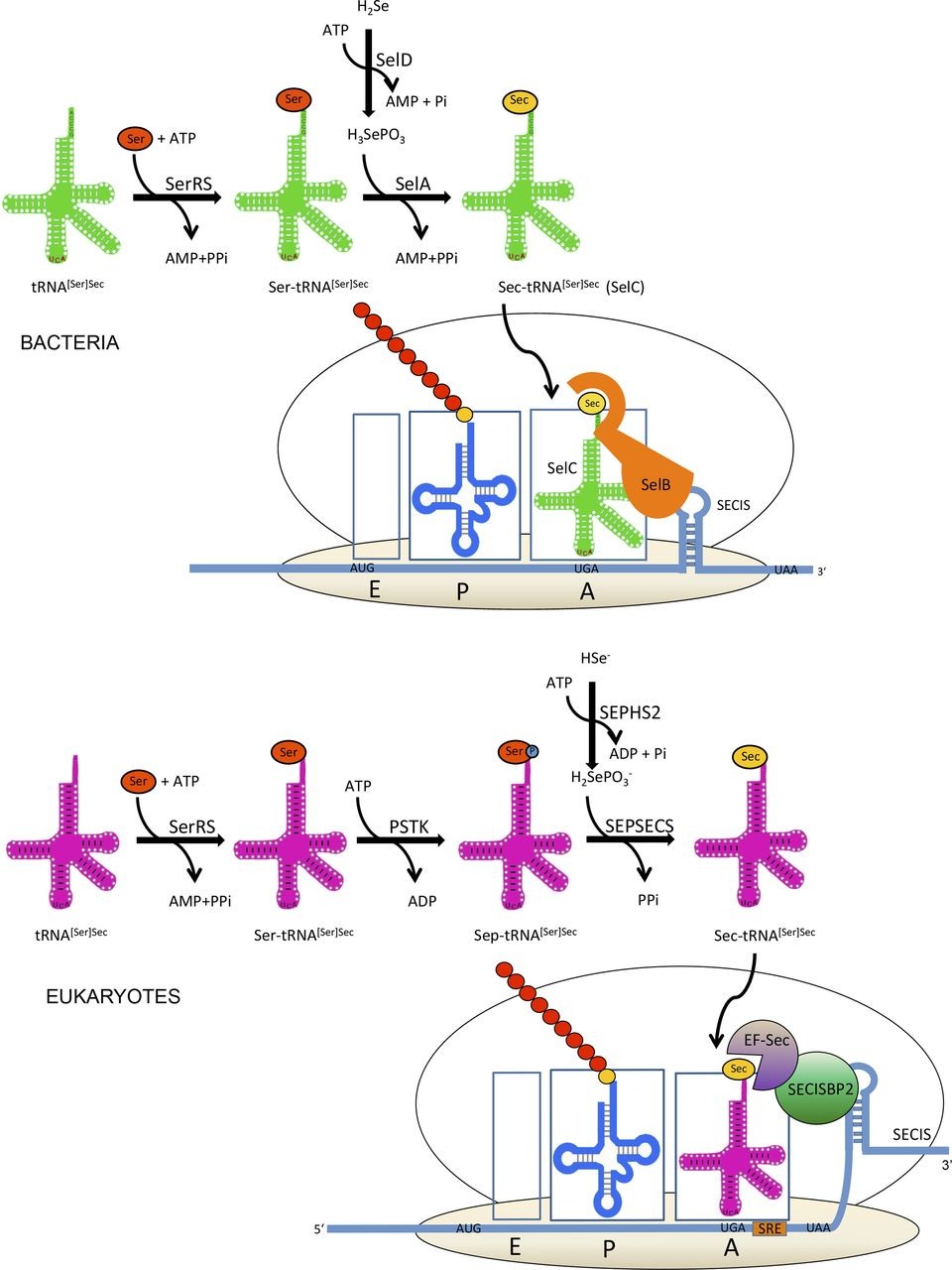
Later on, selenocysteine is incorporated to the proteins due to its tRNA which codifies for UGA codon. This codon codifies for Sec and not for a stop because it presents specific characteristics in the mRNA selenoprotein such as the insertion sequence of selenocysteine, known as SECIS, as well as a loop which has a cis function in the 3’ position of non-traduced (UTR) selenoprotein mRNA which is used as a binding site of RNA proteins.
SECIS has the ability to form complexes with two different elements: elongation factor specific for Sec (eEF-Sec) and SECIS 2 binding protein (SBP2) which let the elongation to go on. SECIS also binds other proteins such as ribosomal L30, an eukaryotic initiator factor (eIF4A3) and nucleolin [4].
eEF-Sec interacts with SBP2 and recruits tRNA[Ser]Sec in the ribosome for continuing to synthetase the protein. In some mRNA we can find what is known as redefinition selenocysteine element (SRE), near UGA codon.
Selenocysteines are synthesized directly bonded with their tRNA. This way, seryl tRNA synthetase catalyzes the gain of charges of the serine. The next step is that it is phosphorylated by PSTK, producing phosphoseryl(Sep)-tRNA[Ser]Sec and finally selenocysteine synthase catalyzes the formation of Sec-tRNA[Ser]Sec using selenophosphate as a substrate.
We can classify selenoproteins in different families[5][6][7]:
GPx:
Glutation peroxidases is composed by multiple enzimes which are involved in the H2O2 signaling pathway, hidroperoxidases detoxification and homeostatic maintainance.
GPx1: It was the first to be identified, we can find it in different parts of the body but specially on kidney and liver. It binds and reacts with H2O2 and hiperperoxidases modulating their functions. It’s also important as a protector for oxidative stress.
GPx2: It is found on the gastrointestinal epithelium and it acts as a barrier for avoiding the toxicity of hidroperoxids ingested through the diet.
GPx3: It is found on plasma and kidney. Its physiological function remains unknown but it is used as a selenium marker.
GPx4: It catalizes the reduction of lipidic peroxides.
GPx5: Located in mammals sperm.
GPx6: It is only placed on the olfactory epithelium during embrionic development.
GPx7: It is a secreted protein.
GPx8: It is located on cellular membrane.
DI
Iodotironin deiodinases (DI) are constituted by three paralogue proteins (DI1, DI2 and DI3) which are involved in regulation of thyroid hormonal activity. The three different proteins differ each other on their location: DI1 and DI3 are on plasmatic membrane, while DI2 is found on endoplasmic reticulum.
Thyroid hormone is released on its inactive form or T4. When DI1 and DI2 react, a desiodation of the external ring in T4 is produced and it changes into the active form T3. In the other hand, DI3 is responsible of the hormone inhibition. So deiodinases play a key role on thyroidic homeostasis.
TR
Tioredoxine reductases (TR) are oxidoreductases that synergically with tioredoxina (Trx) are involved on redox regulation of cellular processes. In mammals we can identify three different TR:
-TR1: Protein with 6 different isoforms formed by alternative splicing and extension of NH2 terminal extreme. Its main function is to reduce NADPH Trx1 dependent but it is also important in regulating different transcription factors, apoptosis, etc. It is located in cytoplasm and nucleus.
-TR2: Really important in the compaction on reactive oxygen.
-TR3: Protein with multiple isoforms important in the reduction of the tioredoxina mithocondrial (Trx2), that is why it is placed on the mithocondria.
Sel W, T, H and V:
They belong to the Rdz family, which is characterized for presenting a Cys-x-x-Sec motive conserved on its sequence.
-Sel W: it’s one of the most abundant selenoproteins on mammals, it is located on the cytosol and it expresses high levels in muscle and brain.
-Sel T: it’s located in the Golgi apparatus and in the endoplasmic reticulum. During the embryonic development and in adult tissue it is ubiquitinated. Its main functions are neuroendocrine and to regulate the calcium homeostasis; it also takes part in cellular adhesion reactions.
-Sel H: it presents a subcellular pattern in the nucleolus. The expression of this protein is low on adult tissues. It is able to bind DNA sequences which present heat shocks and different stress responses elements. It has glutathione peroxidase activity and it it is related with the regulation of gene transcription involved with the synthesis of glutathione.
-Sel V: it is located in the placenta of some mammals. Sel V is an homologus selenoprotein of Sel W, but it presents a terminal NH2 aditional dominium. Its function remains unknown but it is thought to be implicated in males reproduction.
Sel M and SELENO E:
M selenoprotein (Sel M) can be found in all the vertebrates, it is usually expressed on the brain and it shares a 31% of its identity with the Sel15.
In the other hand, we have SELENO E, also known as Fep15 which was generated during the genome duplication and it evolved as Sel M duplication.
Sel 15:
15 selenoprotein (Sel 15) is an ancestral selenoprotein and it can be found in all the vertebrates. It is thought to be a protection factor in front of the cancerigen effect of the Selenium taken in the diet. In the endoplasmic reticulum regulates the homeostasis and the efficiency of protein folding. It is specially expressed on the prostate, liver, kidney and testicles.
Sel R:
Mrs enzimes are responsible of repairing the oxidated residues of methionine (Met) which are susceptible to be oxidated. It lets the catalization of the sulfoxid methionine to methionine.
Sel P:
P selenoprotein (Sel P) is an ancestral protein typical from all the vertebrates. Is is released and contains 50% of the plasmic selenium. It is responsible of the maintenance of the Se storage in the periferic tissues. It is the only selenoprotein with multiple Sec residues. It is thought to have antioxidant properties.
Sel O:
O selenoprotein (Sel O) is found in all the vertebrates and expressed in different tissues.
Sel I:
I selenoprotein (Sel I) is an ancestral selenoprotein found in vertebrates. It is one of the most recent selenoproteins discoveries. It is placed in the endoplasmic reticulum as a transmembrane protein. It is thought to take part in the phospholipids biosynthesis.
Sel S and K:
They can be considered of the same family because they both have a transmembrane dominium, a segment with a big amount of glicina and aminoacids with positive charges. Despite all this they do not present any sequence similarities.
-Sel S: Ancestral selenoprotein can be found in the endoplasmic reticulum and in the cellular membrane of all the vertebrates. It has an important function in response to oxidative stress caused by missfolded proteins taking part of the degradation system related with ER (ERAD). It also prevents damage caused by ROS.
-Sel K: as well as Sel S it is an ancestral selenoprotein and it can be find in all the vertebrates. It is located in the endoplasmic reticulum and in the plasmatic membrane. It is involved in the degradation of glycosylated proteins related with the ERAD system. It also takes part in the calcium supply to immune cells.
Sel N:
It is ancestral and it can be found in the endoplasmic reticulum of all the vertebrates. It is commonly expressed in the embryonic stage as well as in the muscle, heart, lungs and placenta development. A mutation in the SEPN1 gene can cause muscular disorders known as myopathies type SEPN1.
Selenoproteins are distributed in the three domains of life: bacteria, archaea and eukaryotes; however in each lineage they perform different concrete functions. While in archaea they are involved in hydrogenotrophic methanogenesis, in bacteria their main function is to carry out redox homeostasis and compound detoxification. Finally in eukaryotic they are involved especially in redox regulation and in energy metabolism. [8]
Over the past years, selenoproteome has been widely studied, several analyses have been performed. The main conclusion is that selenoproteome analysis revealed many differences between organisms, for example aquatic organisms generally have a larger selenoproteome rather than terrestrial organisms. Mammalian selenoproteomes show a trend toward reduced use of selenoproteins. This difference may have occurred due to the evolutionary adaptation that mammals experienced to a terrestrial environment during evolution, when probably they have reduced Sec function. Across all vertebrates, individual mammalian selenoproteomes consist of 24/25 selenoproteins from a set of 28. Mammalian selenoproteome has remained relatively stable despite a number of evolutionary events changed its composition[9].
Notechis scutatus (Peters, 1861):
Notechis scutatus, also known as Tiger snake, is a highly venomous snake which inhabits in the south of Australia and in coastal islands like Tasmania.
More concretely, Tiger snakes have a non-continuous distribution within two broad areas; southeastern Australia (including the islands of Bass Strait and Tasmania), and southwestern Australia.
In the past, two species of tiger snakes were commonly recognised: the Eastern Tiger Snake Notechis scutatus (Peters, 1861), and the so-called Black Tiger Snake Notechis ater (Krefft, 1866). However, morphological differences between the two appear inconsistent, and recent molecular studies [10] have shown N. ater and N. scutatus to be genetically similar, hence it would seem that there is now just one wide-ranging species that varies greatly in size and colouration – Notechis scutatus. However, various authorities agree a revision of the genus is needed.
Taxonomy[11]
| Kingdom | Animalia |
| Phylum | Chordata |
| Class | Reptilia |
| Order | Squamata |
| Suborder | Serpentes |
| Infraorder | Alethinophidia |
| Family | Elapidae |
| Genus | Notechis (Boulenger, 1896) |
| Species | N.Scutatus (Peters, 1861) |
For more information about the Tiger snake click on the following link of the catalan Wikipedia: Notechis Scutatus
Description:
Total length varies between populations, ranging from 100cm to 240cm and the average total length for mainland tiger snakes is around 1.2m. In general, males appear to attain a larger size than females.The patterning is darker bands, strongly contrasting or indistinct, which are pale to very dark in colour. Colouration is composed of olive, yellow, orange-brown, or jet-black, and the underside of the snake is light yellow or orange.
Venom:
The venom of the tiger snake is strongly neurotoxic and coagulant, and anyone suspected of being bitten should seek medical attention immediately because it is potentially fatal to humans. When threatened, they flatten their bodies and raise their heads above the ground in a classic prestrike stance.
Diet:[12]
Tiger snakes love frogs. This is their main diet, however, given the opportunity, they will also readily take lizards, birds, small mammals and fish. In hot weather feeding often takes place at night with daytime foraging predominating at other times.
Reproduction:
As an adaptation to the temperate climate of its range, the tiger snake produces live young instead of laying eggs. The female normally produces 20-30 wriggling babies in late summer after mating in spring, although some litters as high as 70 have been recorded.
Conservation:
In the IUCN Red List of threatened spicies N. scutatus is listed as Least Concern. In most Australian states, they are protected species, and to kill or injure one incurs a fine up to $7,500, as well as a jail sentence of 18 months in some states. It is also illegal to export a native Australian snake.
Methods
lil_script.pl split_multifasta.pl
The aim of this project is to identify and annotate the selenoproteins and other related genes of Notechis scutatus, know as the Tiger Snake, using bioinformatical tools. In order to do that, the human genome was used as a reference to compare the sequences because it is the closest specie in terms of phylogenetic with well-known and detailed selenoproteome description.
Selection of species
We first wanted to compare Notechis scutatus genome with the genome of a lizard, Anolis carolinensis, because it was the most evolutionary closer specie. However the main problem it presented is that some of its proteins were really short and incomplete so it was inaccurate to make a prediction. That is why we finally decided to compare it also with Homo sapiens because despite our specie has proteins that are not included in human genome, its proteome is perfectly annotated and described. So having both species as a reference let us to reinforce our results, we could compare Notechis scutatus with the lizard and those proteins which were incompletely characterized in this specie could be compared with the Homo sapiens.
When we obtained results which make us to think that evolutionary changes have occurred on the lizard regarding the snake, we considered a third specie: the chicken, which diverged from the common ancestor of both species and is relatively closer to them. It is important to underline that when we talk about the chicken we only take into account the proteins from SelenoDB without analyzing them with our program.
Obtention of the Notechis scutatus genome
The Notechis scutatus genome was provided by the Bioinformatic teachers team in .fa format and its is available in the following directory:
/cursos/20428/BI/genomes/2018/notechis_scutatus/genome.fa
Obtention of querys
As said before, we have used Homo sapiens and Anolis carolinensis’ genomes to predict the Tiger Snake selenoproteinome. We have considered the lizard querys as our reference due to the phylogenetic distances, although in this case the protein database used was partially incomplete. That is why we also used the human proteins as a reference, for its importance, but mostly because the selenoproteins are well described and annotated. We have obtained the data from SelenoDB, a specialized selenoprotein database in multifasta format. Using the program split_multifasta.pl, we have been able to classify every protein automatically in different files in order to simplify the analysing process.
Prediction process
We have created two different Perl programmes called: split_multifasta.pl and lil_script.pl (which explained on the automatization process section), in order to generate the different files which are necessary for the protein prediction. Later on, results presentation, data analysis and final predictions were performed.
Here we present a schedule in order to make all the process more understandable:
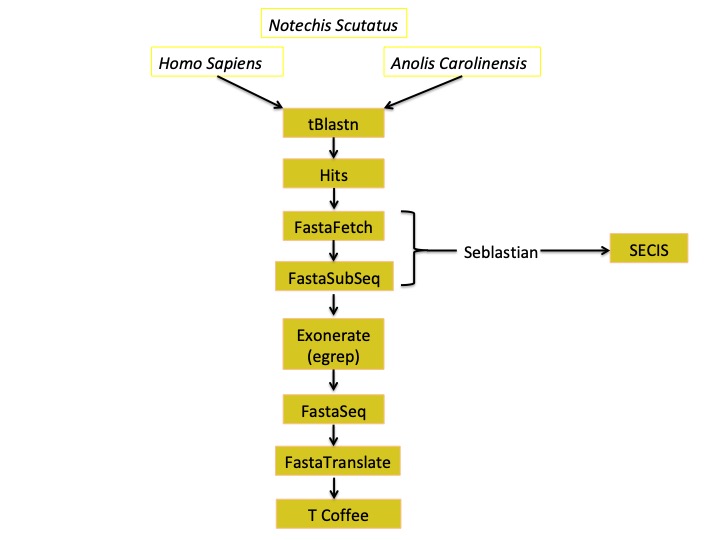
tBLASTn:
It consists on a Basic Local Alignment Search Tool, an algorithm useful for comparing DNA sequences as well as selecting significant hits for further analysis. We have performed BLAST in order to compare each query to Notechis Scutatus genome. With the BLAST an output document is generated with the following elements for each hit:
-Contig: genome fragment where the hit was found.
-Start: initial position of the hit.
-End: final position of the hit.
-E-value: measure of the significance of the hit.
To evaluate each hit significance we took E-value into account, if its result was lower than 0,0001 we took it fot further analysis. This value expresses whether is it possible to find an alignment as good as that one in your database only by chance. That is why lower E-values correspond to higher significances. It is important to underline that we only used those hits with an identity percentage above 50%.
U’s substitution for X’s
Protein sequences obtained from SelenoDB anotate selenocysteines as U’, however when running the programmes U’ are not recognised as an amino acid and an error is showed. So, we have converted all selenocysteines (U’) from tblastn output files into X.
Fasta fetch
This command lets us to extract each scaffold from the hits across the given genome, corresponding to the concrete region where the hit is contained.
Fasta Subseq
This command lets us to extract the region of interest from the output sequence of fastafetch. Fastasubseq only works on a fasta file containing one sequence. In order to obtain for sure a region of interest that contained the whole gene, the start and the end of each hit have been extended 50,000 nucleotides per site. It is important to remind that the start has to be higher than 1 and the length shorter than the contig’s length.
Exonerate
Once we have obtained our region of interest, exonerate aligned and compared the genomic region obtained by using Fastasubseq with the initial query and annotated the introns and exons of the region . Subsequently, it lets us to extract specifically the exons of the predicted gene. Later on we use egrep command in order to fuse these exons into the same file.
Fasta Seq From GFF and Fasta Translate
We use this command to generate a file containing the cDNA of the predicted protein based on the exonerate results. The next step was the Fasta Translate in which we could translate the cDNA into a protein sequence, from here we obtained an output file which corresponds to the primary structure of our predicted protein.
Substitution of *’s for X’s
When the fastatraslate finds a stop codon TGA, it writes a *, which may suppose a wrong alignment when tcoffee is executed. In order to identify an alignment between the selenocysteine of the query with a TGA or a Cysteine of the predicted protein, we changed the * for X.
T Coffee (Tree-based Consistency Objective Function for alignment Evaluation)
This programme is useful for making a global alignment between the protein sequence we have predicted and the query protein from the Homo Sapiens. Concerning to the homologous proteins, we must perform a previous alignment for identifying in which position the cysteine is located. This is important to detect if it aligns or not with our predicted sequence from the t coffee output.
SECIS and Seblastian
To corroborate the correspondance of the predictions with Selenoproteins, we used the program SECISearch3/Seblastian (seblastian.crg.es) to determine the SECIs elements. The program also predicts homologous Selenoproteins to our prediction apart from the number of exons.
If no SECIS elements were predicted, we used SecisSearch3 as a tool to predict eukaryotic SECIs elements in the genomic region of each protein.
Automatization process
The two programs created are called split_multifasta.pl and lil_script.pl. For split_multifasta.pl we copy all of the protein sequences from the reference species in an emacs automatically and then, it classifies each of them in a folder for later analysis.
For the second program, only the annotation of the protein is needed to make it start. Then it shows the blast and you choose which scaffold is the best to start the processes, by entering the name of the scaffold, and the starting and ending points. Later the program asks again for another scaffold just in case there are more duplications; you can exit the program writing "fi". Finally, the files are redirected to its matching folder for further analysis.
You can access both Perl programs following the links:
Results
The results of our study of the selenoproteasome are:
Selenoproteins machinery
Discussion
The aim of this project was to determine all selenoproteins and machinery genes present in Notechis scutatus genome. In order to determine these proteins, human and lizard genome were used to identify homologous proteins because the lizard genome is very near from the Notechis scutatus’ and the human is much better annotated. All the results obtained for every protein were analysed and discussed individually, paying special attention to the T-coffee outputs and SECIS predictions using firstly SEBLASTIAN and secondly SECISearch3.
The queries obtained from the lizard database, usually happened to be incomplete, in most cases they did not start with methionine and sometimes, he sequence consisted of no more than 20 amino acids. That is why it was convenient to compare it with human queries, because they were very complete and could be used as a reference in the case the lizard ones were not well annotated.
When we obtained some results that made us think of some evolutive changes from the lizard to the snake, we considered a third species, the chicken (Gallus gallus), which diverged from the common ancestor of these two species and is relatively near to the snake. But when we use the chicken, is only to look if the proteins are annotated in the SelenoDB, so we do not analyse them with the program. It helps us to confirm an hypothesis based on an evolutive change from the lizard to the snake and in this case the snake should be equally annotated as the chicken and differently from the lizard (it would serve as a common ancestor of the three species)
The results obtained from the T-coffee are generally better when we compare the lizard protein with its prediction rather than when comparing the human protein with its prediction. That is because our species’ proteins are more similar to the lizard ones than to the human proteins
Also, every time that we have said that the protein is a selenoprotein (due to its predictions containing a selenocysteine), SECIS elements have been found in that region. They are very important for a tRNA-selenocysteine containing amino acid recruitment. On the other hand, in most of the cases where the protein has a cysteine and thus is not a selenoprotein, SECIS elements are not predicted because that sequence is not functional anymore and can continue mutating without altering the protein translation. But in some cases we have identified some SECIS elements in a protein which did not contain any selenocysteine residues, because there have not been too many modifications and Seblastian can still predict them. This can be because it has been a short time from the change of the selenocysteine to a cysteine.
Iodothyronine deiodinase (DI)
DI1
This protein is present in humans and lizards and in both cases we obtained very similar hits from the blast when comparing them with the genome sequence of our species. Significative alignments and a good identity were obtained with the ULFQ01000014.1 scaffold between 8313119-8314103 in lizards and between 8310923-8314082 in humans, which was the region used in order to predict the protein. When comparing it with the human protein we obtained 2 exons although when comparing it with the lizard, 3 exons were obtained. Tcoffee results were very good and in both cases, the selenocysteine is conserved. Also, SECIS elements in both alignments can be seen. So DIO1 is a selenoprotein in Notechis scutatus.
DI2
Both humans and lizards have this protein and we obtained significative blasts when we compared the proteins with the Notechis scutatus genome. We saw that ULFQ01000009.1 scaffold is the one that shows better alignment with the positions 2998485-3014825 in both species. In both cases we obtained 2-exon proteins with Tcoffee results pretty good (though in lizards were better than in humans and also the conserved selenocysteine. Also, SECIS elements in both cases were obtained. So DIO2 is a selenoprotein in snake.
DI3
This deiodinase is only found in humans and not in lizards. When blasting it, we obtained the ULFQ01000129.1. scaffold hit with a very good identity leading us to think that maybe this protein is present in Notechis scutatus but not in the lizard because it may have lost it due to evolutionary divergence. To reassure our hypothesis we consulted whether the chicken (Gallus gallus) had the protein and it was present in its genome. The predicted protein contains 1 exon, the Tcoffee score is high enough, SECIS elements were found in the 3’ region of the protein and the selenocysteine is conserved. So DIO3 is a selenoprotein in snake.
Glutathione peroxidase (GPx)
GPx1
This protein is found in both human and lizard. When we ran the blast we obtained a huge amount of scaffolds where hits were found, expectable, as we knew that there are a lot of proteins in this family and they are very similar to each other. We chose the hit with the higher identity percentage from the rest in the ULFQ01000052.1 scaffold in both species at the 5007542-5010157 position in lizard’s prediction and 5007572-5010160 in human’s prediction , confirming that this protein exists in the Notechis scutatus genome. In both cases 2 exons with SECIS were predicted, conservation of the selenocysteine was observed all of this with a good Tcoffee score. So GPx1 is a selenoprotein in snake.
GPx2
The obtained results are really similar to the GPx1. We found the best hit at the ULFQ01000013.1 scaffold between the 17192429-17189038 in lizard’s prediction and between 17192429-17188694 in human’s prediction. In both exonerate predictions, the obtained proteins have 2 exons and Tcoffee scores very high. We found SECIS elements and the selenocysteine conserved. So GPx2 is a selenoprotein in snake.
GPx3
We again obtained very similar results comparing them with the other two GPx’s. The hit is found in the ULFQ01000005.1 scaffold and the prediction is located in the 3186547-3182865 nucleotides in the lizard and in the 3186544-3182671 nucleotides in human. In the human predicted protein, 5 exons were found and in the lizard one, 4. In both cases, the Tcoffee score is high, SECIS were found and the selenocysteine was conserved. So GPx3 is a selenoprotein in snake.
GPx4
Though the results found in this protein are also similar to the other ones of the family, the hit in the ULFQ01005182.1 scaffold between 7128-8512 in lizards and between 5041-7376 in humans, some differences were identified. No SECIS elements were found, neither in the human and in the lizard, although the Tcoffee score is significantly high, but with a higher value in lizards and the exonerate predicted 2 exons for the lizard and 3 for the human. The selenocysteine is conserved only in the predicted protein from the human, whereas in the predicted from the lizard is not conserved, but it does not mean that it has been substituted by a cysteine, it happens because the region where this amino acid should be located is not found in the sequence taken from the SelenoDB, so the lizard protein sequence used for the prediction was incomplete from the beginning. It is considered as a selenoprotein since the data used for the lizard prediction is incomplete, just the human ones were taken into account.
GPx5
This protein is only found in the human and not in the lizard and thus, it is not very probable that Notechis scutatus has it since, in terms of evolution, it is more near to the lizard than to the human. When comparing the human protein with the Notechis scutatus genome any blast was obtained with significant identity percentages, the hits found were in a scaffold also present in another GPx, with better identity, so we think that the results obtained were just because they are very similar instead of because the snake has this protein. The chicken also does not have GPx5 in its genome, it can be concluded that the common ancestor of the chicken, the lizard and the snake lost this duplication or it did not win it whereas the ancestor of the human did.
GPx6
In this protein, the same results as the GPx5 were obtained so Notechis scutatus does not have this protein in its genome.
GPx7
GPx7 is found in both species, so two proteins have been predicted. The scaffold used was the one with better identity, in this case ULFQ01000014.1 in the 7376806-7377072 position in lizards comparison and the 7370170-7378061 position in humans comparison. In the predicted protein from the human, 3 exons were found and in the predicted from the lizard, only 1 exon was obtained. In both cases, the Tcoffee score was very high and no SECIS elements were obtained. Finally for the selenocysteine conservation, it happens the same as in the GPx4, where the lizard protein is not well annotated and incomplete in the database (the position where the selenocysteine should be seen is not included). So only taking into account the results obtained from the human prediction, which a cysteine is found and not a selenocysteine, Notechis scutatus’ protein is not a selenoprotein.
GPx8
This case is similar to the analysed before in GPx7, but this time in a different scaffold, the ULFQ01000034.1, in the position 27920-29870 in the lizard comparison and the position 26914-30030 in the human comparison. The predicted from the human shows 3 exons and the predicted from the lizard, only 2. Also, good Tcoffee values were obtained, but not SECIS elements. As for the selenocysteine conservation, it happens exactly the same as in the GPx7 case. It is not a selenoprotein in the tiger snake, though it would be more reliable if it could be compared with the predicted protein from the lizard.
Methionine sulfoxide reductase A (MsrA I i II)
It is a protein that is found either in humans and lizards. Two significant hits were obtained in the blast with high identity in two different scaffolds leading to the thought of being a duplication of the gene. The hit from the ULFQ01000039.1 scaffold, located between 638382-518629 in the protein predicted from lizard's genome and between 638376-518471 in the protein predicted from human's genome, had better identity percentage than the hit located in the ULFQ01000081.1 scaffold at 4419105-4382144 in lizard’s predicted protein and at 4419105-43811992 in human’s predicted protein, so in the first one is where the MsrA gene and in the other one, the duplication. In all of the 4 predicted proteins, 4 exons were obtained, with the exception of the predicted from the duplication of the lizard. Tcoffee scores were in all of the cases very good and SECIS elements were not found in the duplication predictions, but were obtained in the region of the original gene (ULFQ01000039.1 scaffold). In all of the original proteins, the ones from the species and the ones predicted, a cysteine is found where a selenocysteine could be found so Notechis scutatus MsrA protein is not a selenoprotein.
Selenoprotein 15 (Sel15)
Sel15 is a protein found in humans and lizards. A very significant hit, and with a very high identity was obtained in both species in the ULFQ01000060.1 scaffold between the 5814748-5835746 positions in lizard’s prediction and between the 5814748-5827949 in humans prediction. In the protein predicted from the human, 3 exons were found and in the lizard one, 4 exons; and the Tcoffee score was good in both cases. SECIS were obtained in the 3’ region and both species conserved the selenocysteine in its original protein, as the predicted ones do too; so Sel15 is a selenoprotein in Notechis scutatus.
Selenoprotein H (SelH)
Selenoprotein H is localized in nucleus and has glutathione peroxidase activity. It is present in both species and hits were obtained from the same scaffold when (ULFQ01000205.1) comparing the human and the lizard protein against the reference genome between the positions 462096-457718 and 462084-457633 in both lizards and humans comparisons respectively. 2 exons were found in both predicted proteins and also, the Tcoffee score was good. SECIS were found in the scaffold and as for the selenocysteine, it was only observed in the predicted from the lizard but in the human one, where the selenocysteine should be found in the scaffold (Tcoffee does no align in this region since it may obtain a low score). So, selenocysteine is conserved in SelH and thus, is a selenoprotein in Notechis scutatus’ genome.
Selenoprotein I (SelI)
SelI is found in both species’ genome. When comparing the genome with the human protein, two significant hits with a high identity so the existence of a duplication in Notechis scutatus was taken into account. The original scaffold is ULFQ01000039.1 and the copy, ULFQ01000007.1 and are found between 3767438-3783556 and 156264458-156622761 in the human comparison and between 3775628-3783086 in the lizard comparison. As for the exons, 10 were predicted from the original human protein and just 1 from the duplication, and the predicted from the lizard, only 4. Tcoffee scores are significantly good in all of the cases and SECIS elements were found in every prediction. Finally, SelI is a selenoprotein because selenocysteines were found in both species in the original gene, but in the duplication, the selenocysteine residue it was aligned with a gap.
Selenoprotein K (SelK)
The protein is present in humans and lizards, and in the blast’s results hits for the same scaffold (ULFQ01000003.1) in both species were identified and the positions where the predicted protein can be found are 3675362-3672911 for he predicted protein from the lizard prediction and 3675362-3675231 for the predicted protein from the human. In both predictions, 4 exons were found and the good Tcoffee scores meant good homology with the original protein sequence of the species. SECIS elements were also found and the selenocysteine was observed at the expected position on the predicted sequences so SelK can be considered a selenoprotein in the tiger snake.
Selenoprotein M (SelM)
SelM can be found in each of the analysed species and significant hits are obtained, with a high identity at the same scaffold, ULFQ01000390.1. The prediction is found between 165099-166156 in the lizard prediction and between 165099-166174 in the human’s. Both predicted proteins have 2 exons and their Tcoffee scores were very high. On the other hand, no SECIS elements were found in this region and is impossible to determine if the predictions have a selenocysteine or not, since Tcoffee aligns these positions with gaps. Even though it can not be confirmed, it is very probable that SelM is a selenoprotein in Notechis’ genome because both the lizard and the human protein have selenocysteine.
Selenoprotein N (SelN)
SelN protein is found in both species and in the blast, the same scaffold (ULFQ01000081.1) is present showing high identity and significance in the 1162003-1178288 position in the human’s comparison and in the position 1161991-1178282 in the lizard’s comparison. The predicted protein from the human species has 10 exons, whereas the one predicted from the lizard has 11 exons. In both cases good Tcoffee values were obtained (though the lizard ones were much higher than the human ones) and SECIS elements could be predicted. Selenocysteine residue is found in both species in the correct position, so it can be concluded that is a selenoprotein in the species Notechis scutatus.
Selenoprotein O (SelO)
Found in humans and in lizards, and when comparing the human protein, we only found a hit from the blast that we could accept, in the ULFQ01000027.1 scaffold in the 1845833-1858027 position. The alignment was also found for the lizard, at the ULFQ01000043.1 scaffold, in the position between 808944-789302, meaning that it could be a duplication. In the SelenoDB, it can be seen that there are two proteins from the SelO family in lizards and also in chickens and only one in humans, but the second protein from the lizard was not well annotated in the database (only 20 amino acids). Taking this into account, the so called before as duplication seen in the blast is now considered as the second protein from the lizard, which was not used due to the results obtained from the blast. 10 exons were observed in the protein predicted from the human and only 2 from the lizard in the same scaffold as the human, but 8 exons in the other scaffold (ULFQ01000043.1), which can let to the thought of the loss of a part of the protein by the lizard, or that it is not well annotated. Tcoffee results were correct and SECIS elements were identified in the ULFQ01000027.1 scaffold, and not in the other one (ULFQ01000043.1). Finally, the predicted protein from the human has a selenocysteine whereas the predicted from the lizard has not, because where selenocysteine or cysteine should be annotated, there is no notation. So in this case, SelO is a selenoprotein in Notechis scutatus.
Selenoprotein P (SelP)
This protein is found either in humans and lizards, though in the lizard’s case, two proteins can be found in the SelenoDB, whereas in humans, only one. Two significant hits were obtained with the blast when analysing the human protein so it can be deducted from this that there is a duplication in the ULFQ01000014.1 scaffold (position 12411319-12408855) and the original gen is in the ULFQ01000016.1 scaffold 2408855-12408975). However, the blast was done with the two different proteins (the lizard ones). In the SelP1, a hit was obtained in the ULFQ01000016.1 scaffold and for the SelP2 the hit obtained was in the ULFQ01000014.1 scaffold. The chicken database also showed 2 different proteins in the same family so it can be concluded that Notechis scutatus also has the SelP1 and SelP2 proteins. In every prediction, 3 exons were found and the Tcoffee scores were significant, except in the case of the duplication (the second protein from the human), where the score was not as good as the other ones, maybe due to the fact that the human genome does not have SelP2. In both regions, SECIS elements were detected and also the conservation of the selenocysteine is seen, so it can be concluded that both SelP1 and SelP2 are selenoproteins in the Notechis scutatus genome
Methionine-R sulfoxide reductase (MSRB)
MSRB1
It is a protein present in both species. In the obtained blast, significant hits in the ULFQ01000200.1 scaffold were identified when comparing both species’ proteins, located between 1066668-167824 in the lizard predicted protein and between 1062026-1067824. 2 exons were obtained from the human prediction and 3 from the lizard prediction and also, the scores of the Tcoffee were good.SECIS elements and selenocysteine residues were identified in all of the predictions, meaning that MSRB1 is a selenoprotein in Notechis scutatus.
MSRB2
Is a protein found in both species but when he blast showed the output, the hits observed where not significant and had a low identity in both species. Moreover, this hits belonged to scaffolds from MSRB1 and 3 proteins due to its similarity. So no protein could be predicted and thus, it does not exist in Notechis scutatus.
MSRB3
MSRB3 is present in humans and lizards and the hits where from the ULFQ01000021.1 scaffold (positions 8266684-8239499 from the lizard predicted protein, and 8259503-8239338 from the human predicted protein). 6 exons were found with good Tcoffee scores, however, no SECIS elements were identified and instead of selenocysteine, a cysteine was found, so in the tiger snake, MSRB3 is not a selenoprotein.
Selenoprotein S (Sel S)
SelS is present in both species, and in the blast, significant hits were obtained in the positions 1162178-1167947 for the human and 1162184-1167986 for the lizard, in the ULFQ01000050.1 scaffold. In each prediction, 5 exons were seen and the score of the Tcoffee was better in the alignment of the lizard than in the human. This is due to he fact that the tiger snake is more similar to the lizard than to the human. Selenocysteine residues and SECIS elements were found and thus, SelS is a selenoprotein in Notechis scutatus.
Selenoprotein T (SelT)
SelT protein appears in the genome of bo the human and the lizard. ULFQ01000001.1 was the scaffold where the significant its were obtained from, precisely, between the position 34994169-34997413 in the lizard predicted protein and between 34994136-34997413. The predicted protein from the human contains 5 exons and the predicted from the lizard, 4. In both cases Tcoffee sores are good but is better he one from he lizard alignment. SECIS and selenocysteine residue were found in both predictions so it n be assured that this protein is a selenoprotein in Notechis scutatus.
Selenoprotein U (Sel U)
SelU1
SelU1 protein is present in humans and lizards. The scaffold with better hits was ULFQ01000101.1 between the positions 3646271-3655937 when comparing it with the lizard and 3646250-3655937 when comparing it with the human. In both predictions, the proteins show 3 exons and the Tcoffee scores are high and significant and SECIS elements were predicted. As for the selenocysteine residues, in the human predicted protein, a selenocysteine is aligned with a cysteine and in the lizard predicted protein a selenocysteine is lined with another selenocysteine, meaning that in Notechis scutatus SelU1 is a selenoprotein.
SelU2
Found in both species, the his detected where from the ULFQ01000002.1 scaffold between 25360140-25366186 when comparing it with the lizard protein (and 6 exons) and between 25360991-25366186 when comparing it with the human protein (and 5 exons). The Tcoffee score were good enough to consider them significant in either humans and lizards and SECIS elements were predicted in this scaffold. No selenocysteine residues were found, instead, only cysteines, meaning that SelU2 is not a selenoprotein in Notechis scutatus.
SelU3
SelU3 is only found in the human proteome. Blasting could only be performed for the humans but the hits were not significant and the identities were very low and the few hits that were significant, belonged to other SelU proteins. The final conclusion of this protein is that is no found in the snake, and it was confirmed by looking the SelenoDB of the chicken, where the protein does not appear. So, the common ancestor of the lizard, the snake and the chicken had already lost SelU3 or it did not acquire it, whereas the human ancestor did.
Selenoprotein V (SelV)
SelV is also found only in humans, and the results are exactly the same as the ones obtained with the SelU3 protein, meaning that, the protein does not appear in the chicken SelenoDB, the common ancestor deleted the protein or did not acquire it, as he human ancestor did.
Selenoprotein W (SelW)
SelW1
This protein is present in both species’ genomes. Significant hits are seen in the blast, in the ULFQ01000418.1 scaffold between the positions 243977-243045 when comparing it with the lizard protein and between the positions 243977-243903 when comparing it with the human protein. The prediction from the human protein shows 3 exons and the prediction from the lizard, 4. In both cases, Tcoffee scores were high enough to consider them significant and SECIS elements were predicted. As for the selenocysteine residue, in the lizard predicted protein, there is one of them, but is not aligned with the one from the original protein; and the human predicted protein does not contain the residue that should be corresponded with the selenocysteine and no information could be extracted. That is why SelW1 is a selenoprotein in Notechis scutatus.
SelW2
SelW2 is a protein only present in humans and not in lizards. When the blast was done with he human protein, a significant hit was obtained from the ULFQ01000121.1 scaffold between the positions 1419005-1420708. The prediction contained 3 exons and SECIS elements, and also the Tcoffee score was very high. Any selenocysteine residue was seen so Notechis scutatus has this protein but is not a selenoprotein, so the lizard ancestor may have lost tis protein during its divergence from the snakes.
Selenophpsphate synthetase (SPS or SEPHS)
SPS1
SPS1 is a protein present either in humans and lizards, and the most significant hits obtained were from the ULFQ01000484.1 scaffold, between the positions 16354-6922 when it was compared with humans and between the positions 16354-7133 when it was compared with lizards. No SECIS elements could be found, but the Tcoffee sores were very high and 8 exons in each protein were predicted. Both predictions showed a cysteine residue instead of a selenocysteine residue, the same way s the original proteins did. Through this, it was concluded that SPS1 was present in Notechis scutatus but was not a selenoprotein.
SPS2
This protein is also present in both species. Very significant hits were seen in the ULFQ01000082.1 scaffold (position 466315-472612 when the lizard was compared; and position 466294-472573 when the human was compared). The human prediction presented 7 exons and the lizard prediction, 8. Tcoffee scores were significantly high, better in the lizard than in the human alignment. Also, SECIS elements and the selenocysteine residue could be identified in both predictions so SPS2 is a selenoprotein in the Notetchis scutatus species.
Thioredoxin reductase (TR or TXNRD)
TR1
This protein is present in both species’ genomes. Positive hits were obtained from the ULFQ01000007.1 scaffold (between the positions 14742985-14707558, when comparing the lizard protein, and between the positions 14527127-14711599, when comparing the human protein). In the prediction from the human protein, 13 exons were found, and 14 exons, from the lizards. SECIS were identified and also the score of e Tcoffee was significant. Finally, selenocysteine residues were found either in the predicted proteins and the original ones, so TR1 is a selenoprotein in the Notechis scutatus’ genome.
TR2
This protein is only present in the human proteome and significant hits were obtained by analysing the ULFQ01000291.1 scaffold between the positions 568204-542100. 15 exons were predicted through the human protein, scores from the Tcoffee were very high and thus, significant, and SECIS elements were predicted, as well as the selenocysteine residue, which is present in both the original and the predicted protein. By searching in the SelenoDB for the chicken protein, it could be seen that it has the TR2 protein in its genome, so the common ancestor of the species had the protein but the lizard lost it during its evolutionary process. All in all, since the selenocysteine residue was found in both the original and the predicted protein, TR2 is present in the Notechis scutatus’ genome and as a selenoprotein.
TR3
TR3 is present in both species, and the best hit was obtained in the ULFQ01000003.1 scaffold, between the positions 10519583-10532125 when using the human protein and between the positions 10507209-10533200 when using the lizard protein. From the human predicted protein, 14 exons can be seen and from e lizard predicted protein, 16. Tcoffee scores were significant and SECIS elements were predicted. Also, selenocysteine residues could be found both in the predicted and in the original protein, so TR3 is present as a selenoprotein in the genome of Notechis scutatus.
SECIS binding protein (SBP2)
This protein is present both in humans and lizards. When running the blast, two hits from different scaffolds ULFQ01000002.1 and ULFQ01000089.1 (though the best one was the ULFQ01000002.1 scaffold (positions 22938712-22920687) showed significant identity and thus led to thought of the ULFQ01000089.1 scaffold (positions 2714063-2715870) to be a duplication in humans. But in the case of the lizard, two different proteins are annotated in the database, both of them corresponding to the same scaffolds mentioned before from the human, ULFQ01000002.1 scaffold (positions 22938712-22917132) and ULFQ01000089.1 scaffold (positions 2700345-2708415). Tcoffee scores are high enough to consider them significant, though in the case of the ULFQ01000002.1 scaffold, the scores are lower. 4 and 2 exons were identified in the case of he human protein comparison, in the original and in the duplication respectively, whereas in the lizard comparison, 11 were found in the protein with the ULFQ01000002.1 scaffold and 9 in the protein with the ULFQ01000089.1 scaffold. In every case, SECIS elements were predicted with the exception of the human duplication and any selenocysteine residues could be found, meaning that in the snake, though SBP2 exists, is not a selenoprotein. We could conclude that the common ancestor of the human, the lizard and the snake had one protein, but through evolutionary divergence, the lizard and snake ancestor acquired a second protein.
Eukaryotic elongation factor (eEFsec)
This protein is found either in humans and in lizards, though in the case of he human genome, only one protein is found, whereas in the lizard’s, three proteins can be found. When blasting the human protein, significant hit was obtained with the ULFQ01000003.1 scaffold in the positions 7848221-7676119, but when he lizard protein was blasted, only one hit was obtained that was significant, against the three expected. I corresponded to the same as in the humans, the ULFQ01000003.1 scaffold, in the positions 7757315-7757315. Chicken proteins were also consulted and only one of them was observed, same as humans. 7 exons were identified in the case of the human protein and only 1 in he lizard protein. In both cases, Tcoffee scores were very high and SECIS elements were predicted. Cysteine residues were found in every protein, the predicted and the original ones so in Notechis scutatus there is only one protein but is not a selenoprotein, so e SECIS elements are not functional in this regions. The common ancestor between the chicken, the snake and the lizard only had one eEFsec, the same as the human ancestor, but the lizard obtained the other two copies along its evolution through time
Phosphoseryl-tRNA kinase (PSTK)
This protein is not found in the human genome, only in the lizards, and when it was analysed, a significant hit was found corresponding to the ULFQ01000001.1 scaffold, at the position 3198906-3190928 of the predicted protein positions. In this case, 6 exons were found and no SECIS nor selenocysteine residue were identified, so it is not a selenoprotein in the snake.
Selenocysteine synthase (SecS)
SecS is present only in the lizard's genome and when blasting it, one significant hit was found in the ULFQ01000257.1 at the positions 571558-550117 of the predicted protein. 11 exons were identified in the protein but no SECIS elements were found and also no selenocysteine residue was detected, meaning that either in the lizard and in the snake it is not a selenoprotein, though in the chicken it can be found, possibly, due to evolutionary divergence from their ancestors.
tRNA sec 1 associated protein 1 (SECp43)
As the other two proteins, it is only present in the lizard genome, and not in the human. Only one significant hit was detected in the ULFQ01000081.1 scaffold in the 4656201-4646314 positions of the predicted protein, with 4 exons. SECIS elements were identified but no selenocysteine residue could be found. Since in the chicken database this protein can be found and is well annotated, it confirms that there have been an evolutionary divergence between the ancestor of these three species (lizard, chicken and snake), and the ancestor of the human, and also divergence between the chicken ancestor and the lizard’s and snake’s ancestor so this protein is not a selenoprotein in the Notechis scutatus’ species.
Conclusions
Selenoproteins have an important function, acting against oxidation. They are characterized because they have a selenocysteine residue (sec) in its sequence, an amino acid that carries a selenium atom. However, the selenoproteoms are no very well described in the different species because the sec residue is codified by UGA, a stop codon leading to a low recognition rate by some bioinformatic tools.
That is why, the objective of this study is to identify the selenoproteome of the Notechis scutatus specie. So, we have had to use selenoproteins from other species in order to know which proteins are susceptible to carry selenocysteine or not. The comparison of the selenoproteins with the Notechis scutatus’ genome: Anole carolinensis (a very evolutively near species to the snake) and Homo sapiens (which has a well annotated selenoproteome).
After having analysed every case separately and taking into account the comparisons with both the lizards and the humans we concluded:
- -Selenoproteins (25): DIO1, DIO2, DIO3, GPx1, GPx2, GPx3, GPx4, Sel15, SelH, SelI,SelK, SelM, SelN, SelO, SelP1, SelP2, MSRB1, SelS, SelT, SelU1, SelW1, SPS2, TR1, TR2, TR3.
- -Cysteine containing proteins (13): GPx7, GPx8, MsrA, MSRB2, MSRB3, SelU2, SelW2, SPS1, SBP2, eEFsec, SecS, SECp43, PSTK.
- -Proteins that do not exist in Notechis scutatus (4): GPx5, GPx6, SelU3, SelV.
A large amount of the predicted proteins from the lizard did not start with a methionine, mostly because the lizard proteins were not very well annotated in the database and sometimes they did not start from the beginning of the protein. In the case of the human proteins, even though they were well annotated, we still could not predict the beginning of the protein in some of the cases. This may be due to the limitations from the programmes used as blast and exonerate which may find a lower score when aligning the first amino acids and thus, not include them. A hypothesis for this problem was made which stated that it is possible that the N-terminal regions of the proteins are not well conserved and resulting in a lower alignment score so they can not be predicted, then. In order to confirm this hypothesis more exhaustive studies are needed.
One limitation of our work is that when it was time to determine the significant hits from the tblastn, only the ones with an identity of 50% or more were chosen, in order to make sure that our results were almost certainly correct. Another limitation would be that with the used program, it was impossible to predict a whole protein that was located between two scaffolds.
Another limitation was related to possible sequencing problems of the genome, we found that in some regions there were multiple N instead of the corresponding nucleotide (A, C, T or G), that probably was the reason why we got some parts that were lacking in some predicted proteins.
Overall, as multiple selenoproteins have been predicted in Notechis scutatus genome, this information could be useful for further studies about selenoproteomes in other species.
References
- 1. Trofast J. Berzelius’ Discovery of Selenium. Chem Inter. Oct 2011; 33(5).
- 2. Lu J, Holmgren A. Selenoproteins. J Biol Chem. Jan 2009; 284 (2): 723-7.
- 3. Schweizer U, Fradejas-Villar N. Why 21? The significance of selenoproteins for human health revealed by inborn errors of metabolism. FASEB J. 2016; 30(11): 3669-3681.
- 4. Cardoso BR, Roberts BR, Bush AI, Hare DJ. Selenium, selenoproteins and neurodegenerative diseases. Metallomics. 2015; 7 (8): 1213-28.
- 5. Binding of [14C]erythromycin to Escherichia coli ribosomes. Antimicrob Agents Chemother. 1974; 6 (4): 474.
- 6. Zhu, S., Li, X., Sun, X., Lin, J., Li, W., Zhang, C. and Li, J. Biochemical characterization of the selenoproteome in Gallus gallus via bioinformatics analysis: structure–function relationships and interactions of binding molecules. Metallomics. 2017; 9(2):124-131.
- 7. UniProt [Internet]. Uniprot.org (2017). Available at: http://www.uniprot.org/ [Accessed 28 Nov. 2018].
- 8. Mariotti M, Santesmasses D, Capella-Gutierrez S, Mateo A, Arnan C, Johnson R, et al. Evolution of selenophosphate synthetases: Emergence and relocation of function through independent duplications and recurrent subfunctionalization. Genome Res. 2015;25(9):1256-67.
- 9. Mariotti M et al. Composition and evolution of the vertebrate and mammalian selenoproteomes. PLoS ONE. 2012; 7: 1-18.
- 10. Browne-Cooper R, Bush B, Maryan B, Robinson D. Reptiles and Frogs in the Bush: Southwestern Australia. University of Western Australia Press. 2007: 254-255.
- 11. Boulenger (2008). ITIS Standard Report Page: Notechis. [online] archive.is. Available at: https://archive.is/20081006223342/http://www.itis.usda.gov/servlet/SingleRpt/SingleRpt?search_topic=TSN&search_value=700234 [Accessed 26 Nov. 2018].
- 12. Australian Reptile Park - Wildlife Park Sydney & Animal Encounters Australia. (2018). Mainland Tiger Snake Habitat, Diet & Reproduction - NSW. [online] Available at: https://reptilepark.com.au/animals/reptiles/snakes/venomous/mainland-tiger-snake/ [Accessed 25 Nov. 2018].
Meet the Team
Here we introduce the four members of the team:

Arnau Cañabate

Quim Fàbrega

Ireneu Folqué
