RESULTS
Selenoproteins machineryPhylogenetic tree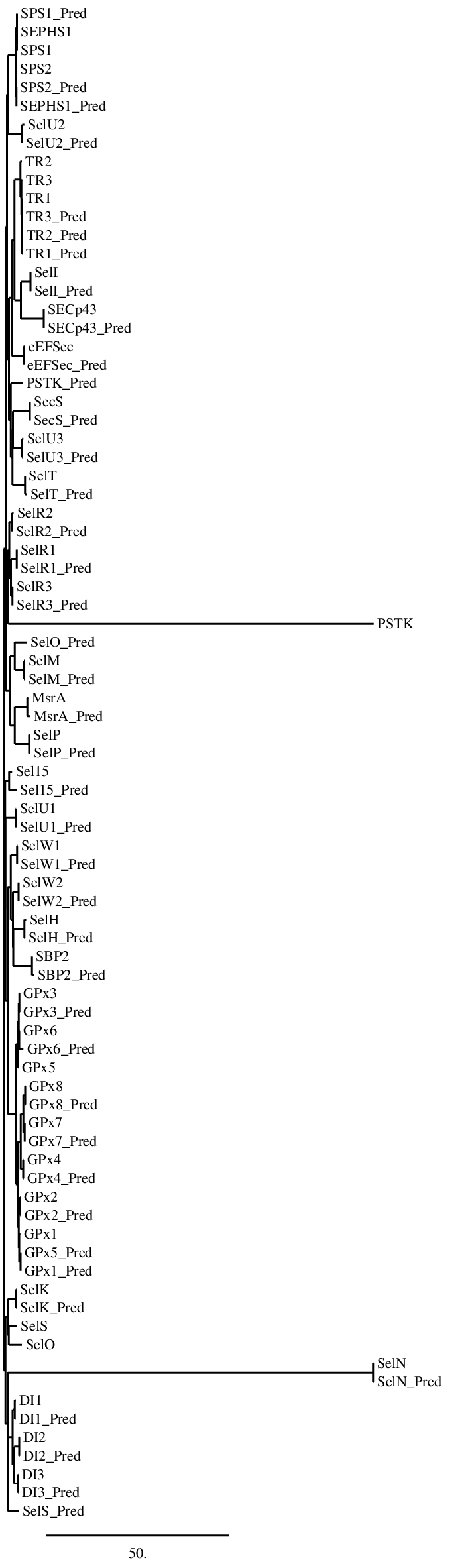 Figure 7. Phylogenetic tree comparing the selenoproteosome of Homo sapiens with the the selenoproteosome of Neophocaena asiaeorientalis This is the phylogenetic tree obtained using the program Phylogeny.fr. which compares the homology of the selenoprotein of Homo sapiens with the Neophocaena asiaeorientalis ones. In this image we can observe that the different families of selenoproteins, such as the GPxs family, are found in clusters. Considering the homology between the two species, proteins in Homo sapiens are indeed phylogenetically close with the proteins predicted. In some particular cases, for example in PSTK or SelO, the protein predicted is phylogenetically distant from the ones of Homo sapiens. |
