INTRODUCTION: SELENOPROTEINS1. Definition of selenoproteinSelenoproteins are Selenocysteine (Sec) containing proteins which have been identified in each of the 3 domains of life: eubacteria, archaea and eukaryotes. Sec, known as the 21st amino acid (aa), is encoded in these proteins by the codon UGA. Although it is normally a termination codon of protein synthesis, it is also used as a Sec codon [6][7].
Sec is an analog of cysteine (Cys), with the sulfur (S) atom present in the side chain of Cys being replaced by a Selenium (Se) atom. Se is an essential micronutrient which deficiency may lead to disease, otherwise in excessive amounts is recognized as toxic. Se is found in cells mostly in selenoproteins involved in redox systems and may have antioxidant protection capability. 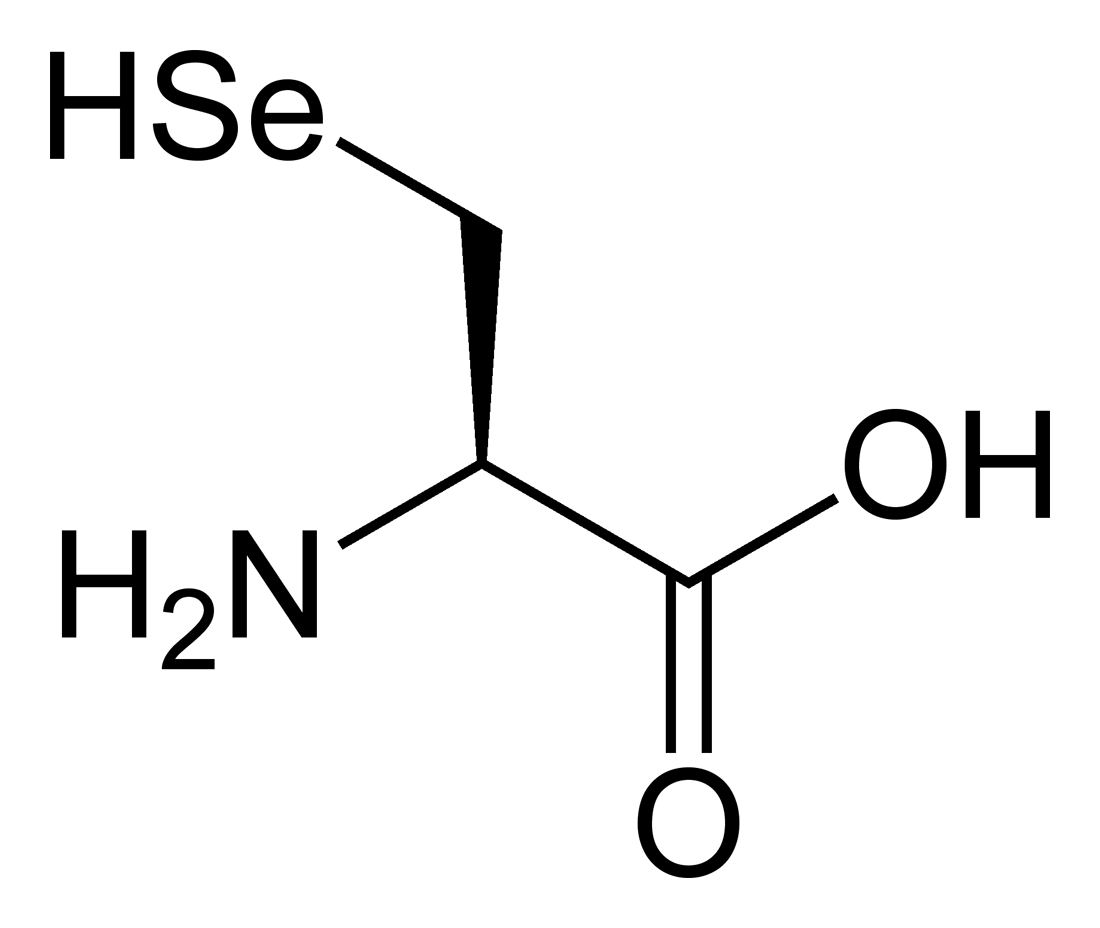 Figure 1. Structure of Selenocysteine. At the physiologic pH, its selenol is mostly deprotonated. Image extracted from Wikipedia. 2. Biosynthesis of selenoproteinsThe main form of Selenium in mammals is found as selenoproteins, that consist in the incorporation of selenocysteine (Sec or U) in their structure. The 21st amino acid is similar to cysteine only changing the Sulphur atom by Selenium, by this fact, selenocysteine could be thougth to derive from cysteine but it is really synthesised from serine [4]. Sec is codified as UGA codon in the genome, which at the same time is also a termination codon [5]. Selenocysteine has not a permanent pool in cells due to two major reasons: the first one is because of the risk of misincorporation of the amino acid in proteins replacing cysteine. This is dangerous for the cell because selenocysteine is more reactive compared to normal cysteine [6]. Second, selenite (a substrate to synthesise the amino acid) is highly reactive with oxygen and thioredoxin reductase, this leads to quick oxidation of NADPH and formation of reactive oxygen species [6]. 2.1 Synthesis and incorporation of selenocysteine in proteinsThe main source of Selenium is through Selenite and selenate, which are absorbed from aliments. Selenite, now in the body, is reduced to selenide (HSe-) by glutathione-glutaredoxin and thioredoxin systems [6].
Then, via catalysis by the selenoprotein SPS2 (selenophosphate synthetase 2), the selenide is converted to monoselenophosphate (H2SePO3-), which is the active selenium donor in the conversion from seryl-tRNAsec to Sec-tRNAsec (tRNA sepcific for selenocysteine).
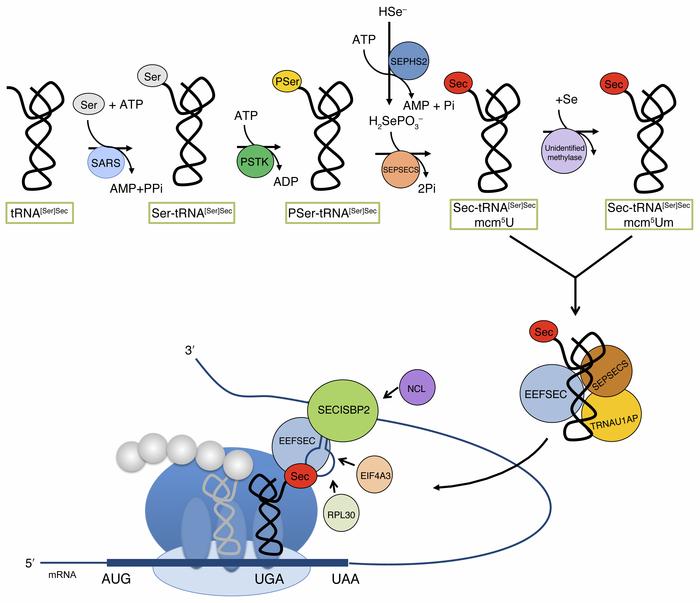 Figure 2. Selenocysteine biosynthesis pathways in eukarya [7] .3. Evolution of selenoproteins
Regarding the evolutionary history of selenoproteins, it has been found that selenoproteins are present in all live domains. Further research on model animals revealed that selenoproteins sets are wide different among them. For example, green algae shows more than 20 selenoproteins in its genome while red algae, insects and nematodes display less than five. If we look at higher plants, only on American cranberries (known as Vaccinium macrocarpon [8]), has been detected the presence of the machinery necessary for synthesise selenocysteine and there has not been reported any on fungi. 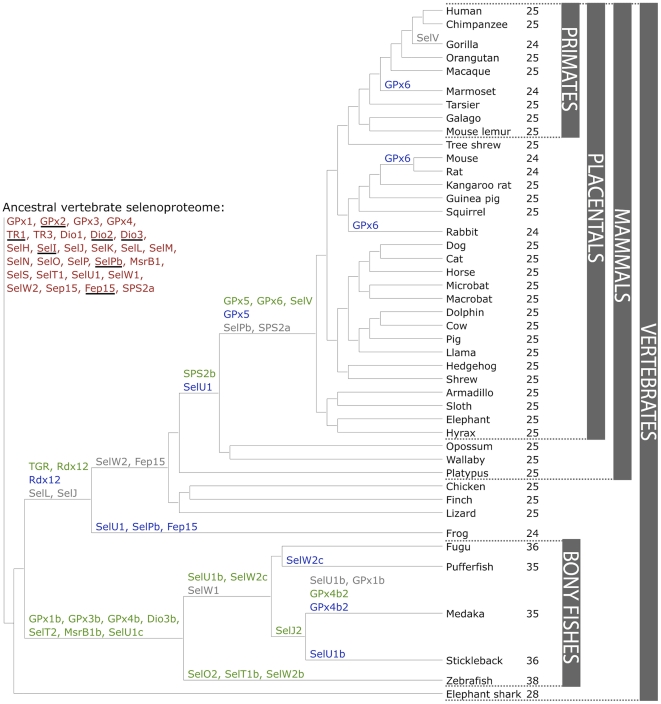 Figure 3. Composition and Evolution of the Vertebrate and Mammalian Selenoproteomes [8] . |