Selenoproteins (Sec) are a peculiar type of proteins that include at least one amino acid of selenocysteine in their structure. Selenium plays an indispensable role as a micronutrient for all animals, consequently, selenoproteins exist in the three major domains of life, eukaryotes, bacteria and archaea. The distinctiveness of these proteins reside in the presence of selenium in the 21st amino acid, in which instead of a cysteine atom, it has a sulfur one. This 21st amino acid is encoded by an UGA codon, which normally has a stop function except in the case of selenoproteins in which it serves to insert a selenocysteine amino acid. Selenoproteins have a wide variety of functions. Commonly, they function as enzymes that catalyze oxidation-reduction reactions.
The aim of this project is to study the selenoproteome and the machinery that enables its production in the species Otocyon megalotis, also known as the bat-eared fox.
Otocyon megalotis is a canidae that populates eastern and southern Africa. For the purpose of analyzing the selenoproteome of the bat-eared fox a comparison between the chosen species and Mus musculus as an homologous species was performed via the design of an automatic bash program. With the results obtained from the tBLAStn we applied Exonarate and T-Coffee. These programs facilitate the results of the alignment between the genomic sequences of the homologous species, Mus musculus and the predicted sequence of Otocyon megalotis’ selenoproteins. The Seblastian webserver was used to find the SECIs elements in the 3-UTR regions in order to locate the selenoproteins’ genes. Our results show that...
Otocyon megalotis, also known as Bat-eared fox, is a species of fox that inhabits the African savanna. It is part of the canidae family and is considered the only extant species of the genus Otocyon. The archaeological records indicate this canid first appeared during the middle Pleistocene. [1] [11]
| Kingdom | Animalia |
| Phylum | Chordata |
| Class | Mammalia |
| Order | Carnivora |
| Family | Canidea |
| Genus | Otocyon |
| Species | Otocyon megalotis |
The Bat-eared fox is named after its large ears that can reach a length of 1.14 to 1.35 cm and play a crucial role in thermoregulation. These specimens are considered as relatively small canids, ranging in weight from 3 kg to 5.3 kg. Head and body length is between 46–66 cm, tail length reaches 23–34 cm and shoulder height is 30–40 cm, its legs are relatively short compared with other species of foxes. The coloring of his body is generally a yellow-brown tone, the fur throat and underparts are pale. The lower legs, feet, tail tip, the outsides of the ears and the face-mask fur which reminds a raccoon are black. Apart from its large ears, the bat-eared fox is differentiated from other foxes by its unique dentition. It has more teeth than any other heterodont placental mammal with a total between 46 and 50 dental pieces. Whereas in all other canids there are no more than two upper and three lower molars, the bat-eared fox has at least three upper and four lower molars. Another peculiar feature is a large step-like protrusion on the lower jaw that anchors the large digastric muscle used for rapid chewing of insects. [1] [2] [11]

The bat-eared fox inhabits the arid and semi-arid regions of Eastern and Southern Africa. Two allopatric populations are known representing each one a subspecies. Otocyon megalotis megalotis inhabits southern Africa, spanning from Angola through Namibia and Botswana to South Africa, and extends as far east as Mozambique and Zimbabwe.In the case of Otocyon megalotis virgatus is found in eastern Africa, extending from southern Sudan, Ethiopia and Somalia, through Uganda and Kenya to southwestern Tanzania. The closest territories of the different subspecies are separated by approximately 1,000 km. [1] [11]

The Otocyon megalotis are gregarious animals, they usually don’t distance more than 200 metres apart from the herd and they practice the allogroom or social grooming commonly performed between mammals. They tend to be monogamous, although there are some exceptions like polygyny or females can raise pups that are not theirs. The south african bat eared foxes are mainly diurnal during winter and nocturnal during summer while the ones found in the Serengeti have 85% of their activity during the night. In South Africa, home ranges usually overlap. Population density may reach 10 individuals per km2. [11]
Bat-eared foxes are carnivores, specifically insectivores. Their food habits basically consist of insects like termites and other arthropods, and occasionally small rodents, lizards and eggs and chicks of birds and a variety of plants. Even so, 80% of their diet is filled by the Harvester termite and the dung beetle (Macdonald, 1984). Curiously, bat-eared foxes obtain most of their water supply from the intake of these insects that often feed on grass. [11]
Bat-eared foxes breed annually, they are monogamous with some exceptions of males with two female partners. The breeding season varies according to the habitat zone. Normally it coincides with the rainy season which brings an increase in insect populations. Females give birth in a den, 17 days after the pups come out of the burrow. Until the 4th - 5th week of life, their fur won’t change to the adult colors, remaining gray. By the 5th or 6th month, pups are fully grown. Some females will remain with their natal herd and breed but most of them, as the males, will spread searching for other groups to breed. [11]
There are no major threats for this species although it is are subject to subsistence hunting for skins. Commercial use is very limited, but winter pelts are valued and sold as blankets. They are also sold as hunting trophies in South Africa. Populations usually fluctuate due to diseases, especially rabies and canine distemper, which can cause short-term drastic declines in the number of individuals or drought, which depresses insect populations. [2]
For further information about Otocyon megalotis click on the following link corresponding to both our Viquipedia and Wikipedia, respectively (also available in other languages): Viquipèdia Wikipedia
Selenium is a chemical element in the periodic table whose symbol is Se and which belongs to the nonmetal family. Despite being an element present in low quantities in nature, it is one of the nine essential trace elements for humans. This element is also an essential micronutrient for a great diversity of organisms. Selenium plays an important role in many vital functions such as improving cognition, immune system function and fertility. Selenium homeostasis is crucial for a proper regulation of both cellular and organismal functions. An excess is toxic for the organism, while a selenium deficiency causes severe and diverse pathophysiological conditions such as Keshan‘s disease, cancer, neuromuscular disorders and male infertility. [4] [6] [12]
Selenocysteine, abbreviated as Sec or U, was discovered in 1986 by Thressa Stadman and is considered the amino acid number 21. The chemical structure of cysteine is significantly similar to that of selenocysteine with the exception of a sulphur atom that is replaced by a selenium one in the selenocysteines residues.
The selenocysteine has the peculiarity that in the mRNA is encoded from a UGA triplet. The UGA codon is one of the 3 STOP human codons that codes for the termination of a protein’s translation. In 99.9% of cases, the UGA codon is responsible for finishing the translation, while the remaining 0.1% is involved in the generation of selenocysteines. When in a polypeptide sequence there is a UGA codon followed by a SecIS sequence -Selenocysteine insertion sequence- in the 3 'UTR region, it encodes for the incorporation of a selenocysteine amino acid. [6] [7]
The biological effects of the selenium are mediated by selenium-containing proteins known as selenoproteins. In this way, selenoproteins are those which contain one or more selenocysteine residues within its amino acid chain. Selenoproteins play a vital role as catalytic enzymes, concretely intervening in the degradation of the organism’s compounds for the subsequent obtaining of energy. These proteins commonly act as oxidoreductases in biological reduction-oxidation reactions. Selenoproteins also play an important role in antioxidant defense, as well as in immune responses and the thyroid hormone’s metabolism.
Selenoproteins are present in all 3 domains of life - eukarya, archaea and eubacteria - and play important roles in all of them. A total of 25 genes encoding for selenoproteins have been identified (GPx1, GPx2, GPx3, GPx4, GPx6, DI1, DI2, DI3, TR1, TR2, TR3, Sel15, SelH, SelI, SelK, SelM, SelN, SelO, SelP, SelR1, SelS, SelT, SelV, SelW, SPS2). Thus, the human selenoproteome is encoded by a total of 25 selenoproteins, although this number varies according to the different taxonomic species.
The study of the evolution of selenoproteins has made possible to establish that in orthologous species the selenocysteine residues have been replaced by cysteine ones within several selenoproteins. Although the basic pathways underneath of synthesis of selenocysteine have been studied in great detail, more research is needed on both the function and evolution of selenoproteins, as they still remain mostly unknown. [3] [4] [7]
The first step in the synthesis of selenoproteins is carried out by the tRNA-synthetase -SerRS-, which ligates serine to tRNA-Sec. Later on, the enzyme Ser-tRNA kinase -pstk- is going to use ATP to phosphorylate the hydroxyl groups present in Ser-tRNASec. The next reaction is leaded by the selenocysteine synthase -SecS- which uses ATP to make a conversion that results in Sec-tRNA[Ser]Sec.
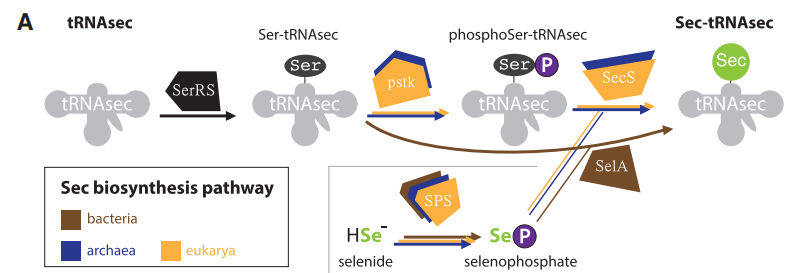
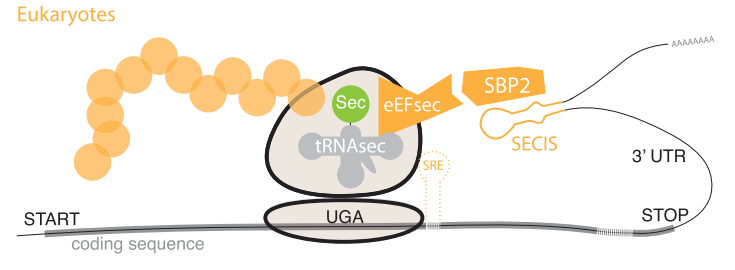
To synthesize selenoproteins in Eukaryotes, it is necessary to incorporate selenocysteine residues present in Sec-tRNA[Ser]Sec into the amino acid sequence that is being processed by a ribosome. To do so, the presence of a Selenocysteine insertion sequence -SECIS- region is needed. The SECIS element can be simply described as a mRNA sequence composed by approximately 60 nucleotides and that are commonly located downstream of a UGA codon. The particularity of this element is that it adopts a stem-loop secondary structure that allows it to bind with the SECIS Binding Protein 2 -SBP2-. Additionally, Sec-specific translation elongation factor -eEFSec- binds to SBP2 and facilitates the final incorporation of selenocysteine residues within the amino acid sequence by recluting these residues from Sec-tRNA[Ser]Sec. [8] [10] [13]
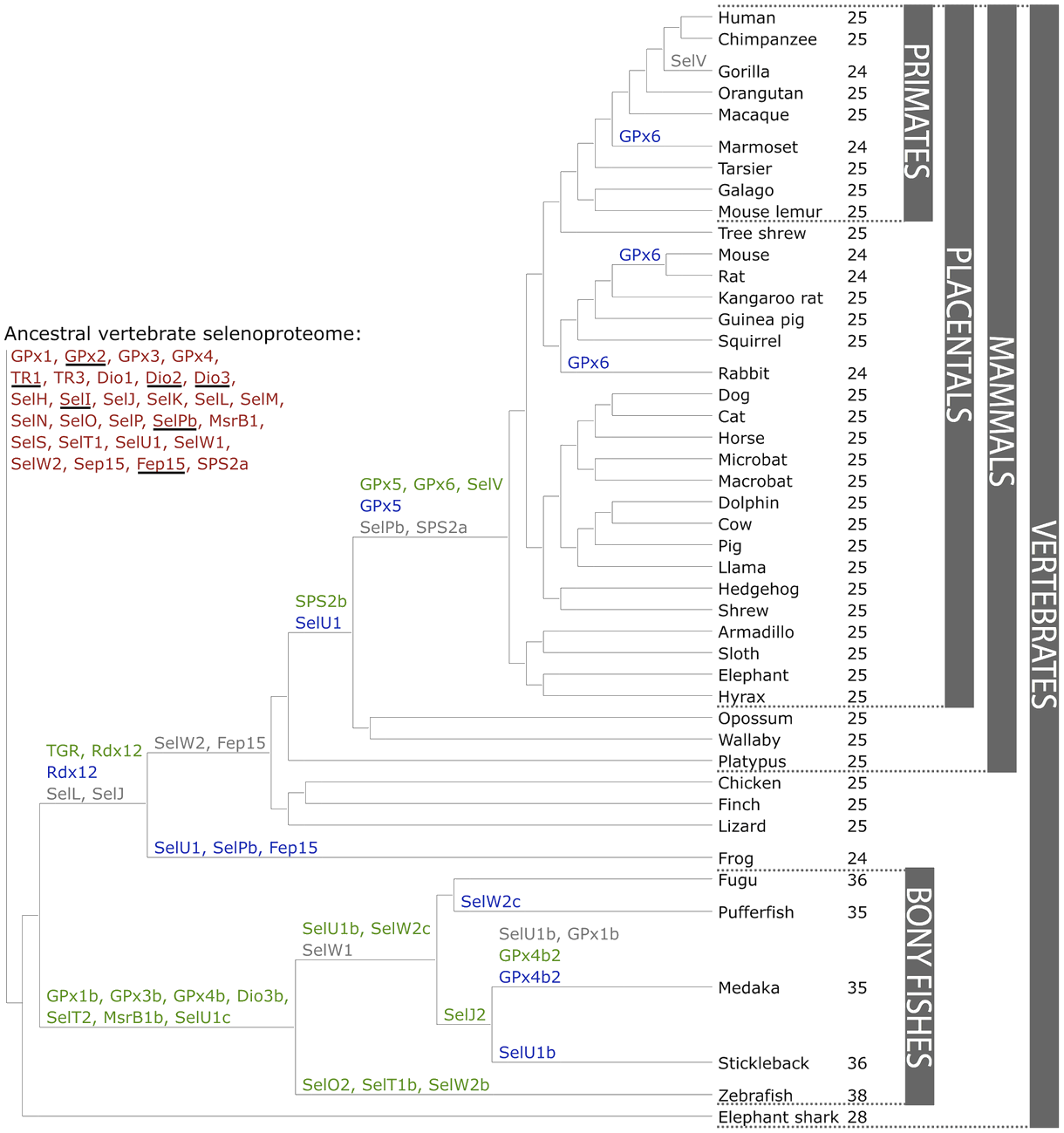
There are currently 45 known subfamilies of selenoproteins, some of which can be found in all vertebrates such as the 21 selenoproteins: Px1-4, TR1, TR3, Dio1, Dio2, Dio3, SelH, SelI, SelK, SelM, SelN, SelO , SelP, MsrB1 -methionine-R-sulfoxide reductase 1-, SelS, SelT1, SelW1, Sep15 and others that are lineage specific or that have been generated from duplications or changes between Se and Cys or vice versa during evolution. In vertebrates, 28 families of selenoproteins have been found, and mammals specifically have 25.
Some duplicate selenoprotein genes have been found in bony fish, which is why it is believed that the entire genome may have duplicated during the early evolution of ray-finned fishes generating selenoproteins: GPx1b, GPx3b, GPx4b, Dio3b, SelT2, MsrB1b and SelU1c. Mammalian-specific selenoproteins were generated after the split with fishes.
Thioredoxin/glutathione reductase (TGR) appeared after the separation of tetrapods from the duplication of an ancestor of TR1 and SelV and GPx6 appeared in placental mammals from duplication of SelW and GPx3 respectively. When vertebrates colonized the terrestrial environment several selenoproteins were lost and mammals reduced the use of Sec compared to fish. Among other existing ones, one of the reasons why the selenoproteome was reduced was because the specific codons of Sec UGA mutated into Cys codons and where there had been Sec there became cysteines. Although Sec and cysteines did not have an identical operation and the concessions are not neutral it is not clear why they happened. [7] [8]
The selenoproteins GPx family are in all three domains of life. Mammals have 8 Px paralogs five of which (GPx1, GPx2, GPx3, GPx4 and GPx6) have a selenocysteine residue at the active site. The other three have a cysteine residue instead of a selenocysteine one.
GPx family has various functions in the body, but is involved in hydrogen peroxide -H2O2- signaling, detoxification of hydroperoxides, and maintaining cellular redox homeostasis. The active site residues of GPx selenoproteins are highly conserved and their catalytic mechanisms are identical.
The DIO family is present in mammals and consists of three parallel proteins whose function is to regulate the activity of thyroid hormones from reductive deodination. Other homologues of DIOs can also be found in eukaryotes and bacteria, but their function is unknown.
DIO 1 and DIO2 are involved in the inactive activation of thyroxine (T4) to which they make an outer ring deiodination. In contrast, DIO3 and sometimes DIO1 can inactivate T3 and T4 from the removal of the iodine inner ring. Therefore, the DIO family can be said to play an important role in maintaining stable thyroid hormone levels.
The TXNRDS are a family of oxidoreductases that, together with thioredoxins, form the most important sulfide reduction system in the cell.There are three types of thioredoxin reductases in mammalian cells and they all have a Sec residue at COOH-terminal penultimate position.
These TXNRDs are flavoenzymes that contain selenocysteine and have a major role in reducing thioredoxins, as well as in redox homeostasis. The three isoenzymes differ, among others, in the cell localization and its function:
It is a family of selenoproteins that is characterized by containing zinc. Its function is to catalyze repair of the R enantiomer of oxidized methionine residues in proteins.
The most common selenoprotein of MsrB in mammals is MsrB1 and is located in the nucleus and cytosol. Of this family, however, we can also find two other homologues, although they are expressed in different locations: MsrB2 in the mitochondria and MsrB3 in the endoplasmic reticulum, which are less expressed in mammals, but have a catalytic efficiency similar to the first homologue.
The MsrA family is a highly conserved family of selenoproteins, has a similar and complementary function to MsrB1, but they do not share structure or sequence. This selenoprotein repairs the L enantiomer of oxidized methionine-S-sulfide.
Participates in the biosynthesis of Sec catalyzing the synthesis of the active Se donor selenophosphate. In vertebrates, SPS2 has a Sec residue at the active site, but in lower eukaryotes it has a Cys residue.
It is a family of selenoproteins that share a thioredoxin-like fold and a conserved motif: Cys-x-x-Sec. They are thought to be thiol-based oxidoreductases, but their exact function is unknown.
SelI is found only in vertebrates, which is why it is believed to be a protein that has evolved recently. It is a transmembrane protein that has two highly conserved motifs. Although the function of the motifs is known, further research is needed to determine the function of selenoprotein.
It is a thioredoxin-like fold ER-resident protein whose expression is regulated by the amount of Se ingested in the diet. It is involved in the quality control of proteins in the endoplasmic reticulum as there are studies that suggest that Sep15 expression is induced when misfolded proteins accumulate in the ER.
Selenoprotein M is a distant homologous of Sep15 and, like SelM, is also a thioredoxin-like fold ER-resident protein, but in this case where there is more expression of the protein is in the brain. The function of SelM is believed to be neuroprotection as overexpression in neurons protects them from oxidative damage by H2O2.
These two selenoproteins do not have significant sequence similarity, but constitute a family because they share topology. It is the family that can be found in most eukaryotic organisms. Both SelK and SelS are transmembrane proteins that are in the endoplasmic reticulum and have a Sec in the second or third position of the terminal COOH. They are related to the degradation in the ER of the misfolded proteins, but the exact function is unknown.
The two selenoproteins are differentiated by a coiled-coil domain: SelK has this domain in the cytosolic part of the protein that mediates the interaction with other proteins.
It is a human selenoprotein that is poorly characterized because it has no structural or biochemical information. Homologues of this protein have been found in other species of animals, plants, bacteria, etc. SelO has a Sec residue at the penultimate position of the COOH-terminal end, but only in the case of vertebrates, in the case of other homologous in this position do they have a cysteine.
It was the first selenoprotein family to be identified from bioinformatics. It is an ER-resident transmembrane glycoprotein that is expressed during embryonic development.
It is expressed in large quantities, 50% of the Se in the blood plasma corresponds to the SelP and it is one of the selenoproteins that has more than one selenocysteine residue. Depending on the species of vertebrate, selenoproteins can be found that have from 7 to 17 selenocysteines as found in zebrafish.
The SelP family has recently evolved, and for this reason their homologs are found mainly in vertebrates.
Most SelP is synthesized in the liver, but expresses its mRNA in all tissues. Its function is thought to be that of Se supplier to peripheral tissues, but especially in the brain and testicles as has been observed with studies evaluating the relationship between ApoER2 and megalin receptors with SelPs.
It is a family of selenoproteins found in fish. They have redox activity and belong to the thioredoxin-like superfamily.
It is a family of selenoproteins found in only a few vertebrates. Its role is unknown, but it is believed to have a structural function.
SelL belongs to the thioredoxin superfamily. It has a restricted phylogenetic distribution to aquatic organisms and it is not found in mammals.
It is a protein that is expressed in the endoplasmic reticulum of fish, Fep15 was a part of the ancestral vertebrate selenoproteome and was lost prior to the split of reptiles. The function of this family of selenoproteins is unknown.
[3] [4] [7]
The aim of this study is, therefore, to predict and annotate the full selenoproteome of Otocyon megalotis, inculing both the selenoprotein machinery and Cys-containing proteins homologs.
We characterised the full selenoproteome and selenoproteine machinery present in Otocyon megalotis by applying an homology-based approach. For the genome sequencing and proper protein annotation, we selected Mus musculus as the referential organism. Mus musculus was chosen due to its phylogenetic proximity to Otocyon megalotis and also because its selenoproteins are well-annoted and described.
The summary protocol of the performing analysis is shown in the scheme below:

The Otocyon megalotis genome was obtained from a mounted directory provided by the Bioinformatics teachers of Universitat Pompeu Fabra. All the necessary files containing our species genome, including both the genome in fasta format and genome index, could be found in the following sources, respectively:
To obtain the total set of selenoprotein sequences present in Otocyon megalotis we used a database specific for selenoproteins, known as SelenoDB 2.0. All the selenoprotein sequences were collected and saved in independent emacs files. These emacs files were stored in a shared folder within a shared directory provided by our university for posterior analysis. Such directory is the following one:
An important step to emphasize is that the different softwares needed for the analysis did not recognise the amino acid number 21 -Sec-, abbreviated as “U”, as an amino acid. To solve this inconvenient, all the “U” within the selenoproteine sequences were replaced by an “X”, which is an abbreviation for any kind amino acid. These modified sequences were settled as the protein queries and saved in a fasta format for posterior analysis. The command to change the "U" by "X" is the following one:
The final selection is made up of a total of 35 queries, which are the following:
Sel15, eEFsec, GPx1, GPx2, GPx3, GPx4, GPx5, GPx6, GPx7, DI1, DI2, DI3, MsrA, PSTK, SBP2, SecS, SPS, SelH, SelI, SelK, SelM, SelN, SelO, SelP, SelR1, SelR2, SelR3, SelS, SelT, SelU, SelW, TR1, TR2, TR3, SECp43.
BLAST (Basic Local Alignment Search Tool) is a program that uses different algorithms to perform local alignments between a selected query and a genome. As our queries are protein sequences, we used the tBlastn’s option within the BLAST program.
tBlastn compares our selected protein queries from Mus musculus to the Otocyon megalotis genome in order to find contigs with significant hits. In order to discard non-significant hits, we only considered those whose e-value was equal to or less than 0.01.
The final outputs for each hit contained the following information:
The fasta fetch command is used to extract the contigs containing the significant hits found by tBlasnt. For the execution of this command, the indexed genome provided by our university was applied.
The fasta subseq command is used to extract the genomic sequence of the hits contained in the different contigs. For a more accurate analysis, it is necessary to take into account that the command tBlastn provides hits located exclusively in exons. To avoid loss of information due to the presence of introns within the genomic sequence, we extended 50.000 nucleotides upstream the final positions and subtract 50.000 nucleotides downstream the start positions given by the tblastn output. In this way, we make sure that the fasta subseq execution will not result in the loss of coding sequences.
However, there may be cases where this nucleotide extension might lead to errors in the program’s execution, as it may overcome the contig’s length either in 3’ or 5’. In those specific cases, two possible situations could appear:
To overcome this drawbacks, we stablished some commands in the bash program to make sure that the start position was not less than zero not the final position greater than the contig sequence itself.
Once we had the subsequences given by the fasta subseq execution, we ran an exonerate command. The combination of the command exonerate together with egrep one, allow us to select the exons and separate them from introns. Thus, after carrying out this step, we obtained the exonic sequence -cDNA- of the region of interest.
Once we have predicted the multiple exon coordinates, we applied a fasta seq from GFF to extract the cDNA that encodes for the predicted selenoprotein.
The fastatranslate command was required to convert the fastaseqfromGFF output to the translation of the cDNA sequence, in order to obtain the amino acid sequence coding for our predicted protein.
Lastly, we run a t-coffee command to perform a global alignment between the Mus musculus initial query and the predicted selenoprotein sequence of Otocyon megalotis.
In order to automatize all this process, we created a semiautomatic program that analysed and aligned the sequences of selenoproteins from the Otcyon megalotis genome with the ones in the Mus musculus genome. Bash programming language was used. This program includes all the steps explained previously in this section and some checkpoints so the person running the program knows that it is working correctly. To do this automatization, we used different loops. The first for loop is used for the program to read all the protein files and analyse them individually. Another for loop is used for the program to calculate the minimum and maximum positions of each contig. Next, we used an iterative command -if- to calculate the starting and finishing positions of each contig. We also used an if loop to detect inverted sequences and be able to analyse them. This program also provided the file ready to use in both, SECISearch3 and Seblastian analysis. The program can be downloaded clicking here.
As SECIS elements are indicative of a selenoprotein sequence, since they participate in the incorporation of Sec amino acids within the coding mRNA, we performed both a SECISearch3 and Seblastian server. This analysis allowed us to identify such SECIS elements in our predicted sequences, as well as if there was any sequence that could match with a selenoprotein sequence. In this way, we could confirm if our predicted selenoprotein sequences coincided with selenoprotein sequences.
To provide a clear representation of the results obtained during the analysis of Otocyon megalotis’ selenoproteome, we created a table of the results. The following table contains clear information of all the files that have been obtained during this project.
| Family | Protein | ID | tBLASTn | Contig | Exonerate | T-coffee | SECIS | Seblastian |
|---|---|---|---|---|---|---|---|---|
| Sel15 | Sel15 | SPP00001563_2.0 | ✔ | JAEUCK010002617.1 | ✔ | ✔ | ✔ | ✖ | JAEUCK010006653.1 | ✔ | ✔ | ✖ | ✖ |
| eEFsec | eEFsec | SPP00001563_2.0 | ✔ | JAEUCK010003379.1 | ✔ | ✔ | ✔ | ✖ |
| JAEUCK010005760.1 | ✔ | ✔ | ✖ | ✖ | ||||
| JAEUCK010010429.1 | ✔ | ✔ | ✖ | ✖ | ||||
| GPx | GPx1 | SPP00001546_2.0 | ✖ | ✖ | ✖ | ✖ | ✖ | ✖ |
| GPx2 | SPP00001547_2.0 | ✖ | ✖ | ✖ | ✖ | ✖ | ✖ | |
| GPx3 | SPP00001548_2.0 | ✖ | ✖ | ✖ | ✖ | ✖ | ✖ | |
| GPx5 | SPP00001549_2.0 | ✔ | JAEUCK010000467.1 | ✔ | ✔ | ✖ | ✖ | |
| JAEUCK010002264.1 | ✔ | ✔ | ✔ | ✔ | ||||
| JAEUCK010003412.1 | ✔ | ✔ | ✔ | ✔ | ||||
| GPx6 | SPP00001550_2.0 | ✔ | JAEUCK010007861.1 | ✔ | ✔ | ✖ | ✖ | |
| JAEUCK010007880.1 | ✔ | ✔ | ✔ | ✖ | ||||
| JAEUCK010008055.1 | ✔ | ✔ | ✖ | ✖ | ||||
| JAEUCK010010914.1 | ✔ | ✔ | ✔ | ✔ | ||||
| GPx7 | SPP00001551_2.0 | ✔ | JAEUCK010000467.1 | ✔ | ✔ | ✖ | ✖ | |
| JAEUCK010002137.1 | ✔ | ✔ | ✖ | ✖ | ||||
| JAEUCK010003412.1 | ✔ | ✔ | ✔ | ✔ | ||||
| JAEUCK010007861.1 | ✔ | ✔ | ✖ | ✖ | ||||
| GPxd | SPP00001555_2.0 | ✔ | JAEUCK010007861.1 | ✔ | ✔ | ✖ | ✖ | JAEUCK010009202.1 | ✔ | ✖ | ✖ | ✖ |
| DI | DI1 | SPP00001543_2.0 | ✔ | JAEUCK010006202.1 | ✔ | ✔ | ✖ | ✖ |
| JAEUCK010003376.1 | ✔ | ✔ | ✖ | ✖ | ||||
| JAEUCK010003714.1 | ✔ | ✔ | ✖ | ✖ | ||||
| DI2 | SPP00001544_2.0 | ✔ | JAEUCK010003714.1 | ✔ | ✔ | ✖ | ✖ | |
| JAEUCK010006202.1 | ✔ | ✔ | ✖ | ✖ | ||||
| JAEUCK010003376.1 | ✔ | ✔ | ✖ | ✖ | ||||
| DI3 | SPP00001545_2.0 | ✔ | JAEUCK010006202.1 | ✔ | ✔ | ✔ | ✖ | |
| JAEUCK010003714.1 | ✔ | ✔ | ✖ | ✖ | ||||
| MsrA | MsrA | SPP00001556_2.0 | ✔ | JAEUCK010000992.1 | ✔ | ✔ | ✔ | ✖ |
| PSTK | PSTK | SPP00001599_2.0 | ✔ | JAEUCK010007423.1 | ✔ | ✔ | ✖ | ✖ |
| JAEUCK010008664.1 | ✔ | ✔ | ✖ | ✖ | ||||
| SBP2 | SBP2 | SPP00001557_2.0 | ✔ | JAEUCK010005358.1 | ✔ | ✔ | ✖ | ✖ |
| JAEUCK010010458.1 | ✔ | ✔ | ✖ | ✖ | ||||
| SecS | SecS | SPP00001562_2.0 | ✔ | JAEUCK010009065.1 | ✔ | ✔ | ✔ | ✖ |
| SPS | SPS2 | SPP00001561_2.0 | ✖ | ✖ | ✖ | ✖ | ✖ | ✖ |
| SPSa | SPP00001560_2.0 | ✖ | ✖ | ✖ | ✖ | ✖ | ✖ | |
| SelH | SelH | SPP00001576_2.0 | ✔ | JAEUCK010003649.1 | ✔ | ✔ | ✔ | ✔ |
| SelI | SelI | SPP00001577_2.0 | ✔ | JAEUCK010008070.1 | ✔ | ✔ | ✔ | ✖ |
| JAEUCK010009040.1. | ✔ | ✔ | ✖ | ✖ | ||||
| JAEUCK010009754.1 | ✔ | ✔ | ✖ | ✖ | ||||
| SelK | SelKa | SPP00001564_2.0 | ✔ | JAEUCK010006495.1 | ✔ | ✔ | ✔ | ✔ |
| JAEUCK010008780.1 | ✔ | ✔ | ✖ | ✖ | ||||
| JAEUCK010010833.1 | ✔ | ✔ | ✔ | ✔ | ||||
| SelM | SelM | SPP00001578_2.0 | ✔ | JAEUCK010003309.1 | ✔ | ✔ | ✔ | ✔ |
| SelN | SelN | SPP00001579_2.0 | ✖ | ✖ | ✖ | ✖ | ✖ | ✖ |
| SelO | SelO | SPP00001580_2.0 | ✔ | JAEUCK010006337.1 | ✔ | ✔ | ✔ | ✖ |
| JAEUCK010004982.1 | ✔ | ✔ | ✔ | ✖ | ||||
| SelP | SelP | SPP00001581_2.0 | ✔ | JAEUCK010004288.1 | ✔ | ✔ | ✔ | ✔ |
| SelR | SelR1 | SPP00001582_2.0 | ✔ | JAEUCK010007266.1 | ✔ | ✔ | ✔ | ✔ |
| JAEUCK010002698.1 | ✔ | ✔ | ✔ | ✖ | ||||
| SelR2 | SPP00001583_2.0 | ✔ | JAEUCK010009932.1 | ✔ | ✔ | ✖ | ✖ | |
| JAEUCK010002140.1 | ✔ | ✔ | ✖ | ✖ | ||||
| JAEUCK010001505.1 | ✔ | ✔ | ✖ | ✖ | ||||
| SelR3 | SPP00001584_2.0 | ✔ | JAEUCK010002140.1 | ✔ | ✔ | ✔ | ✖ | |
| JAEUCK010009932.1 | ✔ | ✔ | ✖ | ✖ | ||||
| SelS | SelS | SPP00001585_2.0 | ✔ | JAEUCK010008033.1 | ✔ | ✔ | ✔ | ✖ |
| JAEUCK010000639.1 | ✔ | ✔ | ✔ | ✔ | ||||
| SelT | SelT | SPP00001586_2.0 | ✔ | JAEUCK010000881.1 | ✔ | ✔ | ✔ | ✖ |
| SelU | SelU1 | SPP00001587_2.0 | ✔ | JAEUCK010005122.1 | ✔ | ✔ | ✔ | ✖ |
| SelU2 | SPP00001588_2.0 | ✔ | JAEUCK010010378.1 | ✔ | ✔ | ✖ | ✖ | |
| SelU3 | SPP00001589_2.0 | ✔ | JAEUCK010004025.1 | ✔ | ✔ | ✖ | ✖ | |
| SelW | SelW | SPP00001592_2.0 | ✔ | JAEUCK010010366.1 | ✔ | ✔ | ✖ | ✖ |
| JAEUCK010003396.1 | ✔ | ✔ | ✔ | ✔ | ||||
| TR | TR1 | SPP00001595_2.0 | ✔ | JAEUCK010009245.1 | ✔ | ✔ | ✖ | ✖ |
| JAEUCK010006363.1 | ✔ | ✔ | ✔ | ✔ | ||||
| JAEUCK010004863.1 | ✔ | ✔ | ✖ | ✖ | ||||
| JAEUCK010001568.1 | ✔ | ✔ | ✔ | ✔ | ||||
| TR2 | SPP00001596_2.0 | ✔ | JAEUCK010009245.1 | ✔ | ✔ | ✖ | ✖ | |
| JAEUCK010006363.1 | ✔ | ✔ | ✔ | ✔ | ||||
| JAEUCK010004863.1 | ✔ | ✔ | ✖ | ✖ | ||||
| JAEUCK010001568.1 | ✔ | ✔ | ✔ | ✔ | ||||
| TR3 | SPP00001597_2.0 | ✔ | JAEUCK010009245.1 | ✔ | ✔ | ✖ | ✖ | JAEUCK010006363.1 | ✔ | ✔ | ✔ | ✔ |
| JAEUCK010004863.1 | ✔ | ✔ | ✖ | ✖ | ||||
| JAEUCK010001568.1 | ✔ | ✔ | ✔ | ✔ | ||||
| SECp43 | SECp43 | SPP00001600_2.0 | ✖ | ✖ | ✖ | ✖ | ✖ | ✖ |
In order to collect all the information obtained during the study of the Otocyon megalotis proteome a compilatory table has been attached in the beginning of this section. All the files with the information obtained from the different programs used are stored in it, allowing its download by clicking on the icons.
In addition, an explanation of the results obtained from the selenoproteins is detailed below.
Sel 15 was located in two contigs. The first one, JAEUCK010002617.1, is located in the + strand between 613963 and 648736 positions. One gene with five exons was found between positions 42041 and 84773 in the + strand.
The second contig was JAEUCK010006653.1, located between positions 38167 and 38427. e value 3.00e-29. One gene with one exon was found starting at position 38160 and finishing at position 38426 in the + strand. No SECIS elements nor selenoproteins were predicted for this contig.
eEFsec was located in three different contigs. The first one, JAEUCK010003379.1 is located between positions 7411 and 101126 (-) strand. One gene with four exons was found starting from the beginning of the contig and finishing at position 101125 in the forward strand. One SECIS element was predicted, specifically between positions 46850 - 46929 (+) strand. No protein was predicted for this gene. e value 2.39e-119. Deletion 1.
The second contig was JAEUCK010005760.1 is located between positions 70682 70936 (-) strand. e value 2.60e-18. One gene with two exons was found starting from 50000 position and finishing at 50254. Neither SECIS elements nor proteins were predicted per this gene.
The third contig was JAEUCK010010429.1 located between positions 5664 and 51385 (+) strand.e value 3.02e-21. One gene with two exons was found between positions 5663 and 51396 (+) strand. Neither SECIS elements nor proteins were predicted per this gene.
No data regarding selenoprotein Gpx1 could be found in Otocyon megalotis genome when compared to Mus musculus’ genome using the homology-based methods mentioned.
No data regarding selenoprotein Gpx2 could be found in Otocyon megalotis genome when compared to Mus musculus’ genome using the homology-based methods mentioned.
No data regarding selenoprotein Gpx3 could be found in Otocyon megalotis genome when compared to Mus musculus’ genome using the homology-based methods mentioned.
GPx5 was located in three contigs. The first one, JAEUCK010000467.1, is located in the - strand between 369569 and 369835 positions. One gene with one exon was found between positions 50111 and 50266. 3 insertions. Neither SECIS elements nor proteins were predicted per this gene. e value 5.23e-10.
The second contig was JAEUCK010002264.1, located between positions 203799 and 244302. e value 9.16e-29. Two different genes were found on this contig. The first one comprises five exons, between positions 81851 and 90554 (+) strand. The second one, comprising four exons, between positions 49794 and 54382 (-). Two SECIS elements were predicted for this gene in positions 91074 - 91146 (+) and 58785 - 58702 (-). The Protein predicted was glutathione peroxidase 6 precursor (GPx6).
The third contig was JAEUCK010003412.1, located between positions 163454 and 164279 (+). e value 6.40e-27. One gene comprising two exons was found between positions 50000 and 50822 (+). 15 insertions. One SECIS element was found in this gene between positions 50888 - 50962 (+). The protein predicted was glutathione peroxidase 1 (GPx1).
GPx6 was located in four different contigs. The first one, JAEUCK010007861.1 was located between positions 14379-13948 (-).e value 2.56e-15. One gene with one exon was found between positions 14178 and 14378 (-). 3 insertions. Neither SECIS elements nor proteins were predicted per this gene.
The second contig JAEUCK010007880.1 was located between positions 39039 and 41680 (+) e value 3.10e-27 One gene comprising four exons was found between positions 39038 and 41673 (+). Two SECIS candidates were found in positions 42323 - 42402 (+) and 18643 - 18560 (-).No selenoprotein was predicted for this gene.
The third contig JAEUCK010008055.1 was located between positions 218023 and 217587 (-). e value 2.98e-11 One gene comprising one exon was found between positions 50045 - 50203 (-). Neither SECIS elements nor proteins were predicted per this gene.
The fourth contig JAEUCK010010914.1 was located between positions 128540 and 131729 (+). e value 1.23e-22 One gene comprising two exons was found between positions 50000 and 53189 (+). 15 insertions. One SECIS element was predicted for this gen at positions 53411 - 53474 (+). The protein predicted for this gene was glutathione peroxidase 2 (GPx2).
GPx7 was found in four different contigs. The first one JAEUCK010000467.1 is located in positions 369844 - 367856 (-). e value 8.12e-49 One gene comprising two exons was found in positions 49872-51860 (-). 6 insertions Neither SECIS elements nor proteins were predicted per this gene.
The second contig JAEUCK010002137.1 was found between positions 29984 - 25980 (-) e value 2.11e-29 One gene comprising two exons was found between positions 29983- 25979 (-). 1 deletion. Neither SECIS elements nor proteins were predicted per this gene.
The third contig JAEUCK010003412.1 was located in positions 163502 - 163663 (-). e value 3.01e-07. One gene comprising one exon was found between positions 49997 - 50161 (+). One SECIS element was predicted for this gene at positions 50840 - 50914 (+). The selenoprotein predicted for this gene was glutathione peroxidase 1 (GPx1).
The fourth contig JAEUCK010007861.1 was located between positions 14367-13963 (-) e value 9.10e-42. One gene comprising one exon was found between positions 14372- 13965(-). 6 insertions Neither SECIS elements nor proteins were predicted per this gene.
GPx was found in two different contigs. The fist one JAEUCK010007861.1 was located at positions 14352 - 14086 (-). e value 8.31e-13. One gene comprising one exone was found between positions 14351 - 14085 (-). 3 insertions. Neither SECIS elements nor proteins were predicted per this gene.
The second contig JAEUCK010009202.1 was located between positions 79219 - 79484 (+). e value 1.60e-13. One gene comprising one exon was found between positions 50000 - 50265 (+). Deletions 1 ; frameshifts 2. No SECIS elements nor selenoproteins were predicted for this gene.
DI1 was found in three different contigs. The first contig JAEUCK010006202.1 one was located between positions 657946 - 658506 (+). e value 4.80e-43. One gene comprising one exon was found between positions 50069 - 50560 (+). 6 insertions. Neither SECIS elements nor proteins were predicted per this gene.
The second contig JAEUCK010003376.1 was located in positions 1066794 - 1053594 (-). e value 1.44e-42. One gene comprising four exons was found between positions 62982 - 47188 (-). 5 deletions. Neither SECIS elements nor proteins were predicted per this gene.
The third contig JAEUCK010003714.1 was located between positions 10358 - 10864 (+) e value 1.64e-33 One gene comprising one exon was found between positions 10393 - 10863 (+). 21 insertions. Neither SECIS elements nor proteins were predicted per this gene.
DI2 was found in three different contigs. The first one, JAEUCK010003714.1, was located between positions 2114 - 10879 (+). e value 4.76e-109. One gene comprising two exons was found between positions 49964 - 50788 (+). Neither SECIS elements nor proteins were predicted per this gene.
The second contig JAEUCK010006202.1 was located between positions 657769 - 658506 (+). e value 5.08e-42. One gene comprising two exons was found between positions 10420 - 10863 (+). 18 insertions. Neither SECIS elements nor proteins were predicted per this gene.
The third contig JAEUCK010003376.1 was located between positions 1058928 - 1058764 (-). e value 2.11e-08 One gene comprising two exons was found between positions 55125 - 54976 (-). Neither SECIS elements nor proteins were predicted per this gene.
DI3 was found in two different contigs. The first contig JAEUCK010006202.1 was located between positions 657769 - 658506(+) .1.30e-159. One SECIS element was found, while no proteins were predicted for this gene.
The second one, JAEUCK010003714.1, was located between positions 10304 - 10864 (+). e value 2.44e-34. One gene comprising one exone was found between positions 10420 - 10863 (+). 18 insertions. Neither SECIS elements nor proteins were predicted per this gene.
MsrA was found in one contig JAEUCK010000992.1 located between the positions 607153 - 986481 (+). e value 4.18e-24 Two genes were found. One gene comprising five exons between the positions 175845 - 429328 (-) and the other one located at 50000- 50149 (+) 3 insertions. Two SECIS elements were predicted for this gene, located at positions 409943 - 409867 (-) and 104202 - 104276 (+). No selenoprotein was predicted for this gene.
PSTK selenoprotein was found in two contigs. The first one JAEUCK010007423.1 located between the positions 58323 - 69062 (+) e value 6.25e-34. One gene comprising six exones between the positions 50000 - 60739 (+). Neither SECIS elements nor proteins were predicted per this gene.
The second contig JAEUCK010008664.1 was located in positions 204306 - 203686 (-) e value 1.86e-82. One gene comprising one exone between the position 50000 - 50563 (-). Neither SECIS elements nor proteins were predicted per this gene.
SBP2 was found in two different contigs. The first one JAEUCK010005358 was located between positions 249015 - 297610 (-) e value 7.97e-59. One gene comprising sixteen exons was found between the positions 98116 - 49521(-). 18 insertions Neither SECIS elements nor proteins were predicted per this gene.
The second contig JAEUCK010010458.1 was located between positions 286458 - 279797 (-). e value 2.91e-22. One gene comprising one exon was found between the positions 50563 - 50000 (-). Neither SECIS elements nor proteins were predicted per this gene.
SecS was found in one contig JAEUCK010009065.1 located between the positions 204629- 172385 (-). e value 3.98e-38. One gene comprising eleven exons was found between positions 49722 - 81966 (+). 3 insertions. One SECIS element was predicted for this gene at positions 48935 - 49003 (+). No selenoprotein was predicted for this gene.
No data regarding selenoprotein SPS2 could be found in Otocyon megalotis genome when compared to Mus musculus’ genome using the homology-based methods mentioned.
No data regarding selenoprotein SPSa could be found in Otocyon megalotis genome when compared to Mus musculus’ genome using the homology-based methods mentioned.
SelH was found in one contig JAEUCK010003649.1 located between positions 240560 - 241123 (+). e value 3.52e-22. One gene was found comprising one exon between positions 50000 - 50563 (+). Three candidate SECIS were located at positions (51276 - 51343) (+), (52294 - 52220) (-), (17869 - 17949) (+). Protein prediction matched the protein analysed: SelH.
SelS was found in three different contigs. The first contig JAEUCK010008070.1 was found between the positions 15673- 33956 (+). e value 2.03e-35. One gene comprising nine exons was found between positions 15157 - 33955 (+) 1 deletion Two candidate SECIS were found at positions 35295 - 35374 (+), 37985 - 38075 (+). The protein predicted for this gene was ethanolaminephosphotransferase 1 .
he second contig JAEUCK010009040.1. was found between position 1803 - 1946 (+) e value 3.26e-07. One gene comprising one exon was found between positions 1802 - 1945 (+). Neither SECIS elements nor proteins were predicted per this gene.
The third contig JAEUCK010009754.1 was located between positions 195831 - 195980 (+) e value 6.66e-09. One gene comprising two exons was found between positions 44338 - 50149(+). 1 deletion. Neither SECIS elements nor proteins were predicted per this gene.
SelI was found in three different contigs. The first contig JAEUCK010006495.1 was located between positions 13322 - 13420 (). e value 6.09e-11. One gene comprising three exons was founded between positions 13321 - 15655 (+).
The second contig JAEUCK010008780.1 was located between positions 7002 - 7142 (+). e value 5.61e-11. One gene comprising two exons was founded between positions 7001 - 7262 (+). 1 deletion, 2 frameshifts.
The third contig JAEUCK010010833.1 was located between positions 1241368 - 1241466 (+). e value 6.13e-11. One gene comprising four exons was found between positions 46607 - 52314 (+).
SelM was found in one contig JAEUCK010003309.1 located between positions 43608 - 43925 (+). e value 1.70e-25. One gene comprising five exons was found between positions 41530 - 43924 (+). 3 deletions.
No data regarding selenoprotein SelN could be found in Otocyon megalotis genome when compared to Mus musculus’ genome using the homology-based methods mentioned.
SelO was found in two contigs. The first contig JAEUCK010006337.1 was located between positions 320296 - 327257 (+). e value 4.00e-53. One gene comprising ten exones was found between positions 49941 - 62961 (+). 15 insertions, 2 deletions and 2 frameshifts.
The second contig JAEUCK010004982.1 was located between positions 1023005 - 1023398 (). e value 3.11e-08. One gene comprising three exons was found between positions 50141 - 57787 (+). 2 deletions.
SelP was found in one contig JAEUCK010004288.1located between positions 132498 - 139507 (+). e value 9.51e-23. One gene comprising four exons was founded between positions 50000 - 57069 (+). 27 insertions.
SelR1 was found in two contigs. The first contig JAEUCK010007266.1 was located between positions 160615 - 161201 (+). e value 4.70e-21. One gene comprising three exons was found between positions 48242 - 50586 (+).
SelR2 was found in three contigs. The first contig JAEUCK010009932.1 was located between positions 2361941 - 2372811 (+). e value 1.79e-20. One gene comprising one exon was found between positions 49988 - 50104 (+).
The second contig JAEUCK010002140.1 was located between positions 206000 - 206095 (+). e value 6.29e-08. One gene comprising one exon was founded between positions 50000 - 50095 (+).
The third contig JAEUCK010001505.1 was located between positions 341260 - 341670 (+). e value 2.32e-16. One gene comprising one exon was founded between positions 50000 - 50293 (+). 3 deletions, 3 frameshifts.
SelR3 was found in two different contigs. The first one JAEUCK010002140.1 was located between positions 82362 -206095 (+). e value 4.26e-15. One gene comprising five exons was found between positions 164973 - 85946 (+). 3 deletions.
The second contig JAEUCK010009932.1 was located between positions 2372716 - 2372811 (+). e value 7.75e-08. One gene comprising two exons was founded between positions 48521 - 50095 (+). 2 deletions.
SelS was found in two different contigs. The first one JAEUCK010008033.1 was located between positions 712521 - 713010 (+). e value 4.06e-17. One gene comprising one exon was found between positions 50183 - 50489 (+). frameshifts 1. One SECIS element was predicted at psotions 50879 - 50953(+). No selenoprotein was predicted for this gene.
The second contig JAEUCK010000639.1 was found between positions 713963 - 710814 (-). e value 3.05e-15. One gene comprising five exons was found between positions 55922- 47766 (-). frameshifts 1. Two SECIS were predicted for this gene at positions 47421 - 47342(-)and 20695 - 20780 (+).Protein prediction matched the protein analysed: SelS.
SelU1 was found in one contig. JAEUCK010000881.1 is located in positions 1314184-1314315(+). e value 1.42e-20. One gene comprising five exons was found in positions 49952-70035 (+). 0 insertions, 0 deletions. Proteins were not predicted per this gene, but SECIS elements were.
SelU1 was found in one contig. JAEUCK010005122.1 is located in positions 16663-16842(+). e value 1.13e-23. One gene comprising five exons was found in positions 10946-19980 (+). 0 insertions, 0 deletions. Proteins were not predicted per this gene, but SECIS elements were.
SelU2 was found in one contig. JAEUCK010010378.1 is located in positions 385443-385844(+). e value 9.09e-21. One gene comprising six exons was found in positions 48425-57216 (+). 0 insertions, 2 deletions. Proteins were not predicted per this gene, but SECIS elements were.
SelU3 was found in one contig.JAEUCK010004025.1 is located in positions 211916-212122(+). e value 1.62e-30. One gene comprising six exons was found in positions 49754-53387 (+). 0 insertions, 0 deletions. Neither SECIS elements nor proteins were predicted per this gene.
SelW was found in two contigs. The first one JAEUCK010010366.1 is located in positions 142755-142829(+). e value 6.45e-09. One gene comprising fifteen exons was found in positions 50166-48009 (+). 0 insertions, 10 deletions and 4 framshifts. Neither SECIS elements nor proteins were predicted per this gene.
The second contig JAEUCK010003396.1was found between positions 988242-988475(+) e value 2.40e-14 One gene comprising one exon was found between positions 47997-48050 (+). 0 deletion and 3 insertions. SECIS elements and proteins were predicted per this gene.
TR1 was found in four different contigs. The first one JAEUCK010009245.1 is located in positions 65546 - 65815 (+). e value 3.37e-36. One gene comprising two exons was found in positions 45017-45319 (+). o insertions. Neither SECIS elements nor proteins were predicted per this gene.
The second contig JAEUCK010006363.1 was found between positions 939956 - 940201 (+) e value 1.50e-34. One gene comprising two exons was found between positions 47997-48050 (+). 0 deletions. SECIS elements and proteins were predicted per this gene.
The third contig JAEUCK010004863.1 was located in positions 101722-102489 (-). e value 1.05e-39. One gene comprising one exon was found between positions 49997 - 50161 (+). Neither SECIS elements nor proteins were predicted per this gene.
The fourth contig JAEUCK010001568.1 was located between positions 328306-328464(-) e value 9.47e-15. One gene comprising one exon was found between positions 106867-106936(+). 3 insertions. Neither SECIS elements nor proteins were predicted per this gene.
TR2 was found in four different contigs. The first one JAEUCK010009245.1 is located in positions 65549-65752 (+). e value 2.18e-14. One gene comprising two exons was found in positions 45017-45319 (+). o insertions. Neither SECIS elements nor proteins were predicted per this gene.
The second contig JAEUCK010006363.1 was found between positions 939947-940189(+) e value 4.99e-17. One gene comprising two exons was found between positions 58031-58273 (+). 9 deletions. SECIS elements and proteins were predicted per this gene.
The third contig JAEUCK010004863.1 was located in positions 101725-102492(-). e value 1.67e-42 One gene comprising one exon was found between positions 49997 - 50161 (+). Neither SECIS elements nor proteins were predicted per this gene.
The fourth contig JAEUCK010001568.1 was located between positions 328306 - 328464(-) e value 7.87e-20. One gene comprising one exon was found between positions 38719-87853(+). 0 insertions. SECIS elements and proteins were predicted per this gene.
TR3 was found in four different contigs. The first one JAEUCK010009245.1 is located in positions 65546- 65788(+). e value 3.32e-26. One gene comprising twelve exons was found in positions 46536-76715 (+). o insertions. Neither SECIS elements nor proteins were predicted per this gene.
The second contig JAEUCK010006363.1 was found between positions 939956-940201(+) e value 4.65e-34.6. One gene comprising sexteen exons was found between positions 49949-101937 (+). 9 deletions. SECIS elements and proteins were predicted per this gene.
The third contig JAEUCK010004863.1 was located in positions 49949-101937(+). e value 2.18e-14One gene comprising two exons was found between positions 50133 - 49764 (+). Neither SECIS elements nor proteins were predicted per this gene.
The fourth contig JAEUCK010001568.1 was located between positions 328306- 328464(-) e value 2.38e-14. One gene comprising one exon was found between positions 80423-6749(-). 0 insertions. SECIS elements and proteins were predicted per this gene.
No data regarding selenoprotein SECp43 could be found in Otocyon megalotis genome when compared to Mus musculus’ genome using the homology-based methods mentioned.
The aim of this study was to characterize the selenoproteome and the machinery in charge of synthesizing them in the animal species Otocyon megalotis. The selenoproteome consists of all the selenoproteins present in a genome. Selenoproteins contain at least one selenocysteine (Sec). Sec residue is codified by the UGA codon, which normally acts as a stopping translation signal, and thus requires a specific machinery to be produced. To predict the Otocyon megalotis selenoproteome and the proteins related to its synthesis, we have studied the homology between the already described selenoproteins of Mus musculus and the proteins encoded by the genes of the fox. To do this prediction, we designed and executed an automatic program that gave us the files that allowed us to analyse through Seblastian the selenoproteome. SECISearch3 was used to predict if the sequence produced by our gene could contain any SECIS.
The 15-kDa selenoprotein 15 is a thioredoxin-like protein located in the endoplasmic reticulum (ER). It is involved in the quality control of glycoprotein folding through its interaction with the UDP-glucose glycoprotein-glucosyltransferase[19]. Sel15 was found in one contig, JAEUCK010002617.1. The predicted gene was located between positions 42041-84773 in the + strand, containing five exons. In the T-Coffee output, a Sec residue was aligned with Sec from the mouse query. This protein was also found in another contig, JAEUCK010006653.1. The predicted gene was located between positions 38160-38426 in the + strand, containing five exons. In the T-Coffee output, a Sec residue was aligned with Sec from the mouse query. In conclusion, Sel15 was found in Otocyon megalotis' genome as a selenoprotein.
Eukaryotic elongation factor is a machinery protein related to the synthesis of selenoproteins. It is essential in the elongation process, during the translation process. Specifically, it guides the Sec-tRNA to the UGA codon and allows its translation into a Sec[19]. eEFSec was found in one contig, JAEUCK010003379.1. The predicted gene was located between positions 7411-101126 in the negative strand, containing four exons. In the T-Coffee output, one SECIS element was found but no protein was found. We also found this protein in the contig JAEUCK010005760.1 and JAEUCK010010429.1, in the positions 70682-70936 and 5664-51385, respectively. Considering all the information above, it was concluded that eEFSec was not found in Otocyon megalotis' genome.
Glutathione peroxidases are the largest selenoprotein family in vertebrates. widespread in all three domains of life [18] GPxs play a wide range of physiological functions in organisms and are involved in hydrogen peroxide (H2O2) signaling, detoxification of hydroperoxides and maintaining cellular redox homeostasis. Selenoproteins of the glutathione peroxidase (GPx) family are widespread in all three domains of life. In mammals, there are eight GPx paralogs, from which five (GPx1, GPx2, GPx3, GPx4, and GPx6) contain a Sec residue in their active site. In the other three GPx homologs (GPx5, GPx7, and GPx8), the active-site Sec is replaced by Cys. Moreover, GPx6 homologs in some mammals are not selenoproteins and have a Cys in the active site[18] GPx are highly conserved, and this is even more evident in Sec-containing GPx (in the case of mammalian GPx, they share approximately 80% of identity) [4]
It is the most abundant selenoprotein in mammals. It was the first selenoprotein discovered in the class Mammalia. Its expression is abundant in the liver and the kidney. It is located in the cytosol and catalyses GSH-dependent reduction of hydrogen peroxide to water. For this reason it has a protective role under oxidative stress and signalling cascades. However, in some situations, it can also act as a pro-oxidative agent. [4] However, Gpx1 couldn’t be found in Otocyon megalotis' genome when compared to Mus musculus’ genome using the homology-based methods mentioned.
It is only found in vertebrates. GPx2 is primarily found in the epithelium of the gastrointestinal tract It is also implicated in the GSH-dependent detoxification of hydrogen peroxide. It is thought to have a role in the development of cancer. [4] According to our results the GPx2 protein is not part of the selenoproteome of the Otocyon megalotis. .
GPx3 is secreted primarily from the kidney and is the major GPx form in plasma in mammals. It is also implicated in the GSH-dependent detoxification of hydrogen peroxide. [4] GPx3 was not found in Otocyon megalotis’ genome by using a homology-based approach.
It was created by GPx3 duplication in placental organisms. It immediately suffered a replacement of Sec with Cys. [18,22] GPx5 was found in three contigs. In JAEUCK010000467.1 the predicted gene was located between 50111 and 50266 positions in the forward strand, comprising one exons. In the T-Coffee output, Cys located in Mus musculus’ genome was aligned with Cys from the Otocyon sequence. Despite this fact, neither SECIS elements nor selenoprotein were predicted by Seblastian. The protein was found in other two contigs, JAEUCK010002264.1, and JAEUCK010003412.1. The selenoprotein prediction offered by the Seblastian for the first contig was the glutathione peroxidase 6 precursor (GPx6). In the case of the second contig, it was related with glutathione peroxidase 1 (GPx1). Therefore, it is concluded that GPx5 doesn’t form part of the Otocyons megalotis’ selenoproteome.
GPx6 is only found in olfactory epithelium and during embryonic development. It was created by duplication in placental. It has independly suffered replacement of Sec with Cys in many species, such as the marmoset, the rat, the mouse and the rabbit. [18]. It is also implicated in the GSH-dependent detoxification of hydrogen peroxide. [4] The Tblastn prediction only showed four different contigs. For two of them neither SECIS elements nor proteins were predicted. A third one didn’t have related selenoprotein prediction results and the fourth one had relation with the glutathione peroxidase 2 (GPx2) as the Seblastian results showed. Thus, we can conclude that the GPx6 is not present as a selenoprotein in the Otocyon megalotis genome.
GPx7 evolved from a GPx4 ancestor, however it did lost the selenocysteine residue.[4] We decided to study four Tblastn predictions for GPx7 gene. However only the contig JAEUCK010003412.1 contained one gene that allowed us to predict one SECIS element that was related with the glutathione peroxidase 1 (GPx1). All the three other contigs weren’t related with neither SECIS elements nor selenoproteins. With this information, we can conclude that GPx7 isn’t a selenoprotein of the Otocyon megalotis’ genome.
GPx was found in two different contigs. The fist one JAEUCK010007861.1 was related with one gene comprising one exon between positions 14351 - 14085 (-) The second also related with one gene that comprised one exon at positions 50000 - 50265 (+). However, neither of the two contigs were related with selenoproteins nor SECIS elements. We can conclude that the protein GPxd isn’t an Otocyon megalotis’ selenoprotein.
Iodothyronine deiodinases (DIs) are implicated in the regulation of the activation and inactivation of thyroid hormone. There are three DIs with different functions. D2 is directed almost exclusively to the efficient conversion of T4 to T3 by 5′-deiodination (an activating reaction)[21]. D3 inactivates T4 and T3 by converting them to rT3 and T2 [21]. In contrast, D1 is able to catalyze both 5- and 5′-deiodination and can act on a variety of different iodothyronines[21]. All three proteins were found in the vertebral ancestor and in mammals they all have Sec[22]. The deiodinases possess a thioredoxin-fold and show significant intra-family homology[22].
DI1 was found in three contigs, JAEUCK010006202.1. The first predicted gene was located between positions 657946 - 658506 in the positive strand, containing one exon. In the T-Coffee output, neither SECIS element nor protein was found. We also found this protein in the contig JAEUCK010003376.1 and JAEUCK010003714.1, in the positions 1066794 - 1053594 and 10358 - 10864, respectively, with several exons each one.Considering all the information above, it was concluded that DI1 was not found in Otocyon megalotis' genome.
DI2 is found in Otocyon megalotis’ genome; its gene has three contigs. The first one, JAEUCK010003714.1 is located between positions 2114 - 10879 in the positive strand. The second contig, JAEUCK010006202.1, is located between positionsns 657769 - 658506 in the positive strands. And, the third one, JAEUCK010003376.1 is found between positions 1058928 - 1058764. All of them contain several exons. The SECI analysis revealed that there are neither SECI elements nor proteins.
When studying DI3, we found a similar situation to DI2. We discovered three different contigs are found in Otocyon megalotis’ genome. The first one, JAEUCK010006202.1 is located betwen positions 657769 - 658506. The second one, JAEUCK010003714.1, is located between positions 10304 - 10864. And the last one, JAEUCK010003376.1, is located between positions 1053806 - 1058782. After analysing them individually, none of them showed any SECI elements nor protein, thus they are not present in Otocyon megalotis’ genome.
Methionine sulfoxide reductases (Msrs) are thiol-dependent enzymes which catalyze conversion of methionine sulfoxide to methionine[21,25]. MsrA was found in one contig JAEUCK010000992.1 located between the positions 607153 - 986481 in the positive strand. Two genes were found, the first one was located between positions 175845 and 429328 and the other one was located between positions 50000 and 50149. Two SECIS elements were predicted for this gene, but according to our results the MsRA protein is not part of the selenoproteome of the Otocyon megalotis.
PSTK is machinery protein related to the synthesis of selenoproteins. It is a kinase that phosphorylates the seryl-tRNA[Ser]Sec complex, so that it can serve as a substrate for the SecS[7].
PSTK was found in two contigs. The first one, JAEUCK010007423.1, is located between the positions 58323 - 69062, in the positive strand, while the second one (JAEUCK010008664.) is located in positions 204306 - 203686, in the negative strand. The first one contains six exones, whereas the second one only contains one. After analysing them, no SECI elements were found, nor proteins.
SECIS binding protein 2 is a machinery protein related to the synthesis of selenoproteins. It binds to the SECIS element, preventing the reading of the UGA codon as a stop codon.
SBP2 was found in two different contigs. The first one JAEUCK010005358 was located between positions 249015 and 297610 in the negative strand and the second oneis located between the positions 98116 - 49521 in the negative strand too, The first contig contains sexteen exons while the second contig contains one exone. The SECI analysis revealed that there are neither SECI elements nor proteins, thus they are not present in Otocyon megalotis’ genome.
SecS only showed one contig, JAEUCK010009065.1 located between the positions 204629- 172385, in the negative strand. It contained eleven exons. A SECI element was found in the position 48935 - 49003. However, no protein was found. Thus, we can conclude that this protein is not found in Otocyon megalotis' genome.
SPS2 is a paralog of SPS1, is a selenoprotein related to the synthesis of selenoproteins. [28] According to our results the GPx2 protein is not part of the selenoproteome of the Otocyon megalotis.
Selenophosphate syntetase is a paralog that has an unknown rol in the synthesis of selenoproteins. [26] SPS was not found in Otocyon megalotis’ genome by using a homology-based approach.
SelH only showed one contig that contained eleven exons: JAEUCK010009065.1. This contig is found in the positions: 240560 - 241123, concretely in the positive strand. After examining its SECI elements, three candidates were found and so did an protein. Thus, SelH is present in Otocyon megalotis’ genome.
SelI is a selenoprotein found only in vertebrates. Even though it is a recently evolved selenoprotein and most of its functions are still unknown, we know that it is a transmembrane protein that contains a highly conserved CDP-alcohol phosphatidyltransferase domain[4, 24]. SelI was found in three different contigs: JAEUCK010009754.1, JAEUCK010009040.1 and JAEUCK010009754.1. Once they had been analysed, none of them showed a protein, however, JAEUCK010009754.1, contained a SECI element.
Selk is a selenoprotein transmembrane located in the ER and the plasma membranes. It contains a single transmembrane domain, and is implicated in ER-associated degradation of misfolded proteins. We discovered three different contigs are found in Otocyon megalotis’ genome. The first contig JAEUCK010006495.1 was located between positions 13322 and 13420 in the positive strand and has a gene with three exones. The second contig JAEUCK010008780.1 was located between positions 7002 and 7142 also in the positive strand, in this contig comprising two exons was founded. The third contig JAEUCK010010833.1 was located between positions 1241368 and 1241466 in the positive strand and it contained four exons. After examining its SECI elements, three candidates were found and so did an protein. Thus, SelK is present in Otocyon megalotis’ genome.
SelM is a distant homolog of Sel15, a thioredoxin-like protein located in the ER. It is highly expressed in the brain, where it is may play a neuroprotective role[23].SelM was found in one contig JAEUCK010003309.1 located between positions 43608 - 43925 (+). One gene comprising five exons was found between positions 41530 - 43924 (+). Two SECIS elements were predicted for this gene at positions 43966 - 44039 (+) and 91231 - 91301(+). The selenoprotein predicted matched the protein studies: selenoprotein M precursor. So we can conclude that SelM is present in the Otocyon megalotis’ genome as a selenoprotein.
SelN is a transmembrane glycoprotein located in the ER. It is highly expressed during embryonic development, but its function remains unknown. In adult skeletal muscle it is essential for muscle regeneration and satellite cell maintenance. No data regarding selenoprotein SelU3 could be found in Otocyon megalotis' genome when compared to Mus musculus genome using the homology-based methods mentioned.
Selenoprotein O (SelO) is one of the least characterized human selenoproteins and is part of the ancestral vertebrate selenoproteome. The majority of SelO homologs contains a Cys residue in place of Sec (Sec-containing SelO sequences are present only in vertebrates). Analysis of vertebrate SelO protein sequences revealed the presence of a mitochondrial targeting peptide and a putative protein kinase but its function remains unknown [24]. SelO was found in two contigs JAEUCK010006337.1 and JAEUCK010004982.1, both with two genes comprising multiple exons. However, neither of these genes were related with any selenoproteins prediction. We can conclude that SelO isn’t present on the Otocyon megalotis’ genome.
SelP is an abundant glycoprotein that has multiple Sec residues and SECIS elements, which can vary between species. It is responsible for the transport and delivery of selenium to peripheral tissues and also, it is linked to an antioxidant function, this is the reason why it is very abundant in the plasma[29, 30].
SelP was found in one contig JAEUCK010004288.1, located between positions 132498 - 139507 and, as expected, it showed SECI elements and protein, this, we can confirm that this protein is found in Otocyon megalotis’ genome.
SelR1 is a zinc-containing Methionine sulfoxide reductase located in the cell nucleus and in the cytosol. It has a key role as an antioxidant enzyme with Sec that participates in oxidized protein repair[4, 30]. This protein was found in two different contigs: JAEUCK010007266.1 and JAEUCK010002698.1, with three exons and two, respectively. After the Seblastian analysis, the first contig showed a SECI element and a protein, whereas, the second contig showed a SECI element, but not a protein.
SelR2 is a cys-containing homolog in human and lizard species[32]. SelR2 was found on three different contigs JAEUCK010009932.1, JAEUCK010002140.1 and JAEUCK010001505.1. each of them comprehends one gene with one exon. The Seblastian results didn’t show any relation with SECIS or selenoproteins for none of the three genes which suggests that SelR2 does not form part of the Otocyon megalotis’ selena proteome.
SelR3 is a Cys-containing homologue with two isoforms located both in the mitochondria and ER[31]. SelR3 was found in two different contigs. The first one JAEUCK010002140.1 was located between positions 82362 and 206095 in the positive strand, The second one JAEUCK010009932.1 was located between positions 2372716 and 2372811 in the positive strand too, The first contig contains five exons while the second contig contains two exons. However, neither of the two contigs were related with selenoproteins nor SECIS elements. We can conclude that the protein SelR3 is not an Otocyon megalotis’ selenoprotein.
SelS is as a binding partner of p97 and Derlin-1; together with SelS form a retrotranslocation channel. In higher eukaryotes, two additional Derlin-1-related proteins are encoded in the genome (Derlin-2 and Derlin-3). More recently, SelS was found to also bind human Derlin-2 and a long form of Derlin-3. SelS differs from SelK by the presence of an additional coiled-coil domain in the cytosolic portion of the protein, which has been proposed to mediate the interaction with other proteins or oligomerization of SelS.[4,30] SelS was found in two different contigs, JAEUCK010008033.1 and JAEUCK010000639.1. The first contig was related with one gene that comprises one exon, the second one was related with another gene with five exons. The first gene was related with one SECIS candidate but the gene of the second contig didn’t. Seblastian results showed that only the second contig had a selenoprotein prediction and it did match the protein studied. We can conclude that SelS is a selenoprotein in the Otocyon megalotis’ genome.
SelT belongs to the Rdx family of selenoproteins[34]. The members of this protein family are characterized by the presence of a conserved Cysx-x-Sec motif. It was proposed that the Rdx family proteins are thiol-based oxidoreductases, but the exact function of any of these proteins remains unknown. [25] A gene in the contig JAEUCK010000881.1 with five exons was found between positions 49952-70035 (+). However, this gene wasn’t related with any selenoprotein predictions. The selenpoproteome of the Otocyon megalotis doesn't have the SelT protein.
Selenoprotein U (SelU) was firstly found in fish and also reported in birds and unicellular eukaryotes, such as Chlamydomonas reinhardtii. In high mammalian species, such as humans and mice, all SelU proteins exist in Cys form, due to the Sec to Cys event that occurred in the early period of mammalian history for the SelU lineage. Three subfamilies of SelU family, SelU1, 2 and 3 are found in humans. The Prx-like2 structure domain presented in these proteins implies that they belong to the thioredoxin-like superfamily [33]
SelU1 was found in one contig. JAEUCK010005122.1 is located in positions 16663-16842(+). One gene comprising five exons was found in positions 10946-19980 (+). Proteins were not predicted per this gene, but SECIS elements were. We can conclude that SelU1 isn’t a selenoprotein in the Otocyon megalotis’ genome.
SelU2 was found in one contig. JAEUCK010010378.1. One gene comprising six exons was found in positions 48425-57216 (+). No selenoproteins were predicted for this gene, but SECIS elements were. We can conclude that SelU2 isn’t a selenoprotein in the Otocyon megalotis’ genome.
SelU3 was found in one contig.JAEUCK010004025.1 is located in positions 211916-212122(+). One gene comprising six exons was found in positions 49754-53387 (+). Neither SECIS elements nor proteins were predicted per this gene. Thus, we can conclude that SelU3 isn’t a selenoprotein in the Otocyon megalotis’ genome.
SelW was found in two contigs. The first one JAEUCK010010366.1. Neither SECIS elements nor proteins were predicted per the gene contained in the contig. The second contig JAEUCK010003396.1 it was also not related with any selenoprotein predictions. Thus, we can conclude that SelW isn’t a selenoprotein i the Otocyon megalotis’ genome.
Thioredoxin reductase 1 is primarily localized in the cytosol and nucleus. TR1 reduces a variety of low-molecular-weight compounds, mainly cytosolic thioredoxin (Trx1). Among other physiological roles, TR1 has been implicated in DNA repair, maintaining redox homeostasis and regulation of cell signaling, as well as activating the p53 tumor suppressor. [20] TR1 was found in four different contigs. each one contained one gene with one or multiple exons
TR1 was found in four different contigs JAEUCK010009245.1 , JAEUCK010006363.1, JAEUCK010004863.1 and JAEUCK010001568.1. Although all contigs did contained candidate genes, none of them were related with possible selenoproteins. With this information we can conclude that TR1 isn't part of the proteome of the Otocyon megalotis’.
TR2 (also known as TGR) differs from TR1 and TR3 in that TGR contains an additional glutaredoxin (Grx) domain, which suggests that this protein is involved in both Trx and GSH systems. However, its physiological function is still unknown. [19] TR2 was found in four different contigs. Only the gene at JAEUCK010001568.1 was located between positions 328306 - 328464 (-) was related with SECIS elements and selenoproteins. The selenoprotein predicted for this gene was AF166126_1 selenoprotein Zf1, it didn’t match the protein studied.
TR3 is localized in the mitochondria, where it is involved in reduction of mitochondrial thioredoxin (Trx2) and glutaredoxin 2 (Grx2). [19] TR3 was found in four different contigs. Only two of them, JAEUCK010006363.1 and JAEUCK010001568.1 had genes that were related with possible selenoproteins. In the case of JAEUCK010006363.1 the predicticon matched the protein studied: thioredoxin reductase 3. In the case of JAEUCK010001568.1 the protein predicted was the same as the one related with TR2: the AF166126_1 selenoprotein Zf1. We can conclude that TR3 is a selenoprotein located in the genome of the Otocyon megalotis.
No data regarding selenoprotein SECp43 could be found in Otocyon megalotis genome when compared to Mus musculus’ genome using the homology-based methods mentioned.
After collecting all the information obtained from the multiple computer analysis and subsequent data processing, we can affirm that of the 35 coding sequences for selenoproteins obtained from the reference organism Mus musculus, several have been found in the genome of our organism, Otocyon megalotis .
Specifically, we have been able to identify that the Otocyon megalotis selenoproteome is made up of a total of 12 selenoproteins: Gpx5, Gpx6, Gpx7, SelH, SelK, SelP, SelR1, SelS, SelW, TR1, TR2 and TR3.
Regarding the proteins involved in the machinery that allow the formation of selenoproteins, neither PSTK, SBP2, eEFsec, nor SecS have been conserved in this species.
On the one hand, regarding the pros of this project, we must highlight that it has been really useful to have databases such as SelenoDB 2.0 to obtain the different querys needed for the homology-based approach. Another advantage that has made a big difference, has been being able to count on Mus musculus as a reference organism, since this has greatly facilitated the analysis due to the fact that it is a phylogenetically close to Otocyon megalotis and, in addition, its selenoproteome is very well annotated and characterized. The automation of bioinformatics commands has also been a key advantage for the development of this analysis, since it has allowed us to obtain results more quickly and efficiently.
On the other hand, regarding the cons, due to the short time to perform this project, this study only took into account Mus musculus’ genome as referential, which is why some selenoproteins of Otocyon megalotis can not have been predicted if they were not in the reference genome. In that way, the results obtained would be much closer to the complete selenoproteome of Otocyon megalotis if more reference organisms had been considered. What is more, despite having an automation to obtain the results, our knowledge in the area of bioinformatics is limited, so a manual review has been necessary for a more accurate results. In the same way, there have been parts of the methodology, such as the search for the SECIS elements or the use of the Seblastian Server, that we have not been able to carry out in an automated way, thus favouring the possibility of human factor errors.
Finally, despite the fact that this project has catalogued an exhaustive analysis of Otocyon megalotis ’ selenoproteome, we can conclude that the categorization of selenoproteins in a wide variety of living beings continues to be a topic that, even today, remains quite unknown and novel. It is for all these reasons that we believe that more research in this area is necessary to obtain more reliable and extensive results.
We would like to thank the Bioinformatics team for providing us with the mounted directory containing the genome of Otocyon megalotis that made this project also. We would also like to thank the Bioinformatics teachers for the guidance and help.
We would also like to express our gratitude to Vasilis Ntasis, for being our mentor during this project. Specially, we are very grateful for the patience that has shown during the development of this project, as well as his collaboration in the development of the script that has been a key step for the final result of this project.
1. Clark, Howard O. (2005). Otocyon megalotis. Mammalian Species, 766(), 1–5. doi:10.1644/1545-1410(2005)766[0001:OM]2.0.CO;2.
2. Hoffmann, M. (2014). Otocyon megalotis. The IUCN Red List of Threatened Species 2014: e. T15642A46123809.
3. Kasaikina M, Hatfield D, Gladyshev V. Understanding selenoprotein function and regulation through the use of rodent models. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research. 2012;1823(9):1633-1642.
4. Labunskyy V, Hatfield D et al. Selenoproteins: Molecular Pathways and Physiological Roles. Physiological Reviews. 2014;94(3):739-777.
5. Lobanov A, Fomenko D, Zhang Y, Sengupta A, Hatfield D, Gladyshev V. Evolutionary dynamics of eukaryotic selenoproteomes: large selenoproteomes may associate with aquatic life and small with terrestrial life. Genome Biol. 2007;8(9):R198.
6. Lu J, Holmgren A. Selenoproteins. J Biol Chem. Jan 2009; 284 (2): 723-7.
7. Mariotti M, Guigo R et al. Composition and Evolution of the Vertebrate and Mammalian Selenoproteomes. PLoS ONE. 2012;7(3):e33066.
8. Mariotti M, Guigó R, et al. Lokiarchaeota Marks the Transition between the Archaeal and Eukaryotic Selenocysteine Encoding Systems. Mol Biol Evol. 2016;33(9):2441–53.
9. Schoch CL, et al. NCBI Taxonomy: a comprehensive update on curation, resources and tools.
10. Squires J, Berry M. Eukaryotic selenoprotein synthesis: Mechanistic insight incorporating new factors and new functions for old factors. IUBMB Life. 2008;60(4):232-235.
11. Thomson, P. 2002. "Otocyon megalotis" (Online), Animal Diversity Web. Accessed December 02, 2021 at https://animaldiversity.org/accounts/Otocyon_megalotis/.
12. Trofast J. Berzelius’ Discovery of Selenium. Chem Inter. Oct 2011; 33(5).
13. Turanov AA, Xu XM et al. Biosynthesis of selenocysteine, the 21st amino acid in the genetic code, and a novel pathway for cysteine biosynthesis. Adv Nutr. 2011; 2(2): 122-128.
14. Wikipedia. Cysteine structure. https://es.wikipedia.org/wiki/Ciste%C3%ADna.
15. Wikipedia. Selenocysteine structure. https://es.wikipedia.org/wiki/Selenociste%C3%ADna.
16. Wikipedia. Geographic distribution. https://es.wikipedia.org/wiki/Otocyon_megalotis.
17. Wikipedia. Otocyon megalotis. https://es.wikipedia.org/wiki/Otocyon_megalotis.
18. Bhabak KP, Mugesh G. Functional mimics of glutathione peroxidase: Bioinspired synthetic antioxidants. Acc Chem Res. 2010;43(11):1408-19.
19. Labunskyy, V., Hatfield, D. and Gladyshev, V. (2014). Selenoproteins: Molecular Pathways and Physiological Roles. Physiological Reviews, 94(3), pp.739-777.
20. St Germain DL, Galton VA, Hernandez A. Minireview: Defining the roles of the iodothyronine deiodinases: current concepts and challenges. Endocrinology. 2009;150(3):1097-107.
21. Mariotti M, Lobanov A V, Guigo R, Gladyshev VN. SECISearch3 and Seblastian: new tools for prediction of SECIS elements and selenoproteins. Nucleic Acids Res. 2013;41(15):e149.
22. Labunskyy V, Hatfield D, Gladyshev V. The Sel15 protein family: Roles in disulfide bond formation and quality control in the endoplasmic reticulum. IUBMB Life. 2007;59(1):1-5.
23. Hatfield DL, Schweizer U, Tsuji PA, Gladyshev VN. Selenium: Its Molecular Biology and Role in Human Health. Switzerland; Springer:2012
24. Reeves M, Bellinger F, Berry M. The Neuroprotective Functions of Selenoprotein M and its Role in Cytosolic Calcium Regulation. Antioxid Redox Signal. 2010;12(7):809-818.
25. Reeves M, Bellinger F, Berry M. The Neuroprotective Functions of Selenoprotein M and its Role in Cytosolic Calcium Regulation. Antioxid Redox Signal. 2010;12(7):809-818.
26. Han S, Lee B, Yim S, Gladyshev V, Lee S. Characterization of Mammalian Selenoprotein O: A Redox-Active Mitochondrial Protein. PLoS ONE. 2014;9(4):e95518.
27. Burk R, Hill K. SELENOPROTEIN P: An Extracellular Protein with Unique Physical Characteristics and a Role in Selenium Homeostasis. Annu Rev Nutr. 2005;25(1):215-235.
28. Wrobel J, Power R, Toborek M. Biological activity of selenium: Revisited. IUBMB Life. 2015;68(2):97-105.
29. Lee BC, Dikiy A, Kim HY, Gladyshev VN. Functions and Evolution of Selenoprotein Methionine sulfoxide reductase s. Biochim Biophys Acta. 2009;1790(11):1471-1477.
30. Kim H, Gladyshev V. Methionine Sulfoxide Reduction in Mammals: Characterization of Methionine-R-Sulfoxide Reductases. Mol Biol Cell. 2004;15(3):1055-1064.
31. Jiang Y, Huang J, Lin G, Guo H, Ren F, Zhang H. Characterization and Expression of Chicken Selenoprotein U. Biol Trace Elem Res. 2015;166(2):216-224.
32. Whanger P. Selenoprotein expression and function—Selenoprotein W. Biochim Biophys Acta. 2009;1790(11):1448-1452.
33. Gromer S, Johansson L, Bauer H, Arscott D, Rauch S, David P, et al. Active sites of thioredoxin reductases: Why selenoproteins?. PNAS. 2003;100(22):12618-12623.
34. Low S. SECIS-SBP2 interactions dictate selenocysteine incorporation efficiency and selenoprotein hierarchy. EMBO J. 2000;19(24):6882-6890.
35. Lobanov A, Hatfield D, Gladyshev V. Selenoproteinless animals: Selenophosphate synthetase SPS1 functions in a pathway unrelated to selenocysteine biosynthesis. Protein Sci. 2007;17(1):176-182.
We are a group of four students of the Human Biology degree at Universitat Pompeu Fabra, in Barcelona. This project was developed during the subject of Bioinformatics.
If you have any doubt, don't hesitate to ask us!

elaia.bercianos01@estudiant.upf.edu

alba.carus01@estudiant.upf.edu

j.pinyolsans@gmail.com

julia.terzulli01@estudiant.upf.edu