
Kingdom: Animalia
Phylum: Chordata
Class: Sauropsida
Order: Squamata
Family: Viperidae
Genus: Vipera
Species: V. berus
Vipera berus, the common European adder[1] or common European viper[2] is a venomous snake that can be found throughout most of Western Europe and as far as East Asia[3]. The Vipera berus is found in different terrains, habitat complexity being essential for different aspects of its behavior. It feeds on small mammals, birds, lizards, and amphibians, and in some cases on spiders, worms, and insects. Like most other vipers, is ovoviviparous and females breed once every two or three years, with litters usually being born in late summer to early autumn in the Northern Hemisphere. Adults grow to a total length (including tail) of 60 to 90 cm and a mass of 50 to 180 grow.
Description
The Vipera berus is easily identified by the dark zigzag line passing along the back bordered by rows of spots, by the dark mark which takes the form of an 'X' 'V' or 'H' located on the rear of the angular head and the vertical pupils[4].


Males are greyish, whitish, pale yellow or cream in colour with very dark contrasting markings, whereas females are typically a brownish or reddish colour with brown markings[5]. Females also tend to be longer and wider than males, and have shorter tails. Male and female juveniles are reddish in colour. In all cases, the belly is grey, greyish-brown or bluish, and the throat is dirty yellow or white[4].
Size

Average adult length: up to 65 cm[5]
Maximum adult length: 90 cm[5]
Biology and Behaviour
The common European adder usually hibernates in September or in October[4] and usually uses the same hibernation site for life[9].The adder emerges from hibernation in March, with males emerging before females[4] and for the first few weeks after emergence still quite inactive and spends much of its time basking [6]. After the male has shed its skin in April it becomes more active and will begin to search for potential mates by following scent trails. The female sheds its skin a month later than the male, and both sexes shed again later on in summer. The adder does not feed until after it has mated, and so during the time before mating, both the male and female adder live off fat reserves that are built up during the previous year[4].
The Vipera berus is typically active during the day, when it hunts mainly for small mammals, including voles, shrews and mice. Grasshoppers and locusts, lizards, young birds and frogs may also be taken[6]. In warm conditions, the adder will actively hunt its prey, but it often uses a sit and wait technique. The adder strikes prey animals with its fangs to inject venom, then releases the prey and follows the scent trail it leaves behind. Upon finding the dying or already dead animal, the adder begins to swallow it head first[4].
Upon discovering a receptive female, the male begins a courtship display in which the tongue is flicked over the female’s body. The male and female may vibrate their tails briefly and bouts of body quivering may ensue. If the courtship is a success, copulation takes place, after which the pair may remain together for two hours or so. If another male approaches the pair at any point, the first male will defend the female aggressively, and a fight may result[4]. These fights are known as the dance of the adders as the males partly raise their bodies off the ground and may become entwined, often repeatedly falling to the ground and rising up again. More than two males may be involved in such a contest[7].
The female adder usually reproduces once every two years, returning to the site of hibernation towards the end of August or early September to give birth. The adder is viviparous, giving birth to between 3 and 18 live young which are initially encased in a membrane[8]. After giving birth the female must feed intensively in order to build up sufficient reserves for hibernation[4]. The young adders do not feed until the following year, but live off the yolk sac and fat reserves that they are born with. The adder reaches sexual maturity at three to four years of age[4].
Although adders are poisonous, they are not aggressive and rarely bite humans or domestic animals, preferring to retreat into thick vegetation instead. Most adder bite incidents result when they are picked up or trodden upon, and in most cases they are not serious. The elderly, the very young or people in ill health are at most risk[4].
Geographic Range
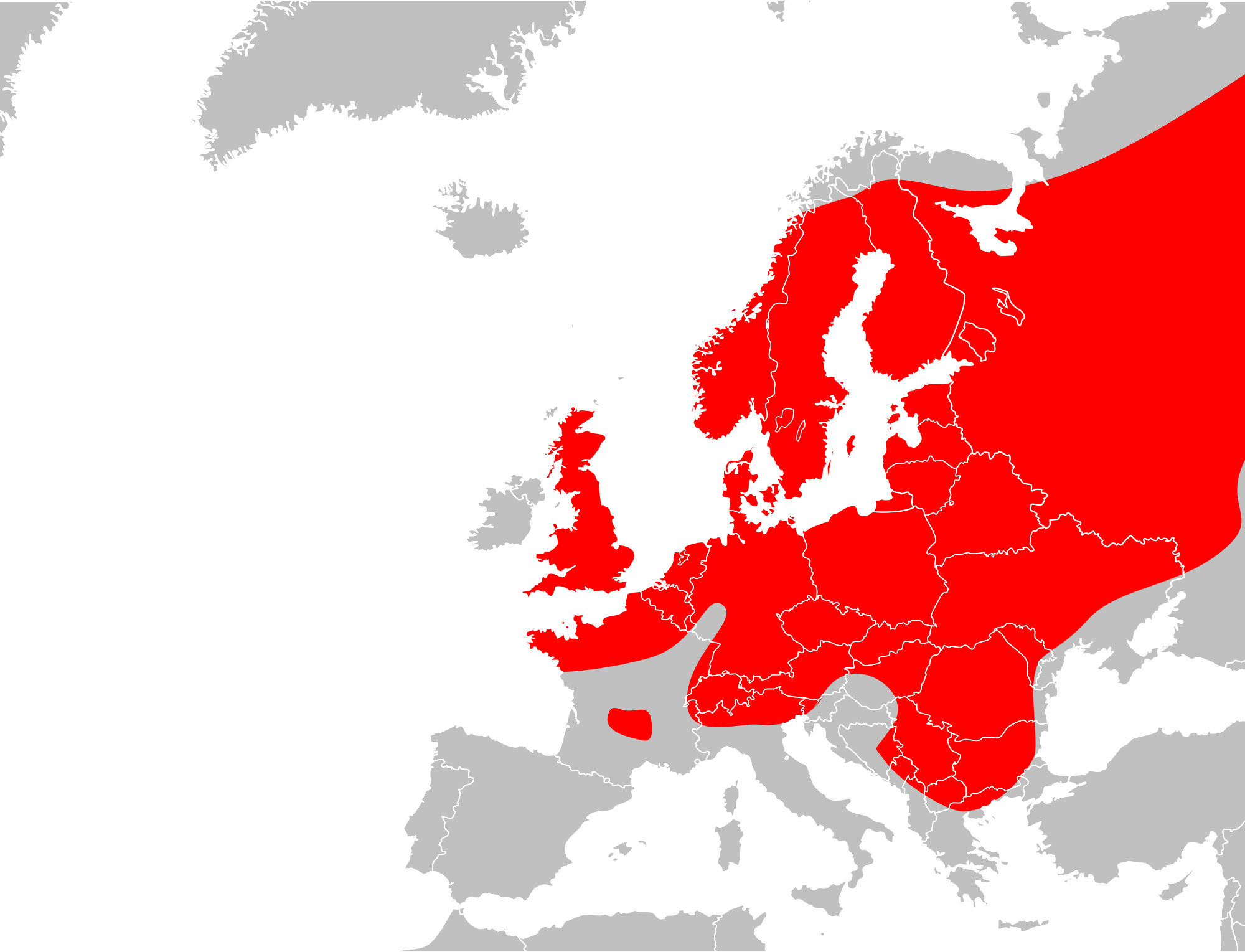
Vipera berus has a wide range. It can be found across the Eurasian land-mass; from northwestern Europe (Great Britain, Scandinavia, Germany, France) across southern Europe (Italy, Serbia, Albania, Croatia, Montenegro, Bosnia and Herzegovina, Republic of Macedonia, Bulgaria, and northern Greece) and eastern Europe to north of the Arctic Circle, and Russia to the Pacific Ocean, Sakhalin Island, North Korea, northern Mongolia and northern China. It is found further north than any other snake species.
Status
The adder is classified as Least Concern (LC) on the IUCN Red List[10].
The adder is threatened by habitat loss in a number of areas throughout its European range. The open habitats it needs, particularly heathland, has been locally lost as a result of scrub encroachment, development, agriculture and afforestation[11].
A further threat to the adder is the persecution of this species by humans, and potentially collection for the pet trade. In Romania, the adder is threatened by illegal collection for its venom[10]. However, the exceptionally wide distribution of this species, combined with its tolerance to a very large variety of habitats, mean that this species is not currently threatened on a global scale.
In molecular biology, a selenoprotein is any protein that includes a selenocysteine (Sec, U, Se-Cys) amino acid residue. This is an strange aminoacid known as the 21st aminoacid.
This rare aminoacid consists in a cysteine which have an atom of selenium instead of sulfur and it is codified by an UGA codon, which is normally a stop codon.
Selenium
Selenium is a scarce element that constitutes an essential nutrient in the diet of animals, and some eukaryotic microorganisms.
UGA Codon
UGA codon is the codon which defines the stop codon and the selenocysteine codon, so we are in a unique case. The UGA codon is the first that has two different meanings within the same specie. For this reason, selenoproteins are poorly annotated, since most programs assume that UGA is a stop codon and, therefore, are not recognized selenoproteins.
Biosynthesis of Selenoproteins
The synthesis of selenoproteins is an evolutionarily conserved process where the selenocysteine tRNA is synthesized from the serine tRNA, thanks to the intervention of the following factors:
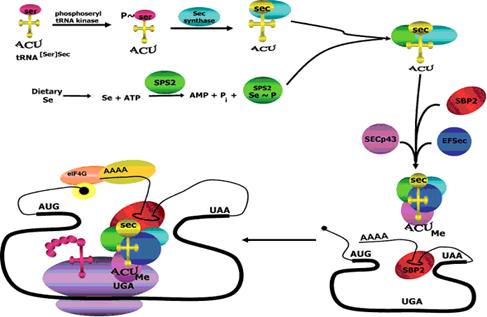
- SPS1 and SPS2 (Selenofosfat Syntase 1 and 2).
- eEFSec (Selenocysteine specific elongation factor).
- SBP2 (SECIS Binding Protein 2) is responsible SECIS joining element and the ribosome.
- tRNASec: tRNA specific selenocysteine.
- Ribosomal protein L30: ribosomal component is also involved in the binding element SECIS.
- Sec43p: part of the complex formed by SCP2 / tRNAsec / eEFSec, but their exact function is unknown.
- Dried (SLA / LP) (Eukaryotic Selenocysteine Syntase): tRNASec to-be converts Sec-tRNASec.
- PSTK (Phosphoseryl tRNA kinase): phosphorylates Ser-tRNASec.
The first step in this byosynthesis is the charge of serine on the specific tRNAsec, which will be phosphorylated by PSTK in order to allow the next reaction, meanwhile selenophosphate synthetase 2 will be in charge of preparing the dietary selenium to be incorporated. Eventually, selenocysteine synthase (SecS) will add the phosphorylated selenium to phosphoserine to produce Sec.
Once the sec-tRNAsec is formated, the protein SBP2 will bind to the SECIS element and at the same time will bin eEFsec. This elongation factor will eventually bind the sectRNAsec, thus allowing the whole complex to get closer to the specific region of the UGA codon. All together sets the stage for decoding UGA codon as Sec. Additional cofactors may contribute to this synthesis, such as SECp43, which has been shown to bind tRNAsec, although its function has not been elucidated yet.
Species Distribution
Selenoproteins exist in all major forms of life, eukaryotes, bacteria and archaea. Among eukaryotes, selenoproteins appear to be common in animals, but rare or absent in other phyla (one has been identified in the green alga Chlamydomonas, but none in other plants or in fungi). Among bacteria and archaea, selenoproteins are only present in some lineages, while they are completely absent in many other phylogenetic groups. These observations have recently been confirmed by whole genome analysis, which shows the presence or absence of selenoprotein genes and accessory genes for the synthesis of selenoproteins in the respective organism.
SECIs Elements
There is a mechanism to skip the stop codon and amino acid selenocysteine incorporated in the polypeptide chain that is synthesizing the element SECIS (SElenoCysteine Insertion Sequence). SECIS element is a three-dimensional structure in a loop and formed by 60 nucleotides. Archea and eukaryotes, the SECIS are located in the extreme 3 'UTR of mRNA coding for selenoproteins, while bacteria is placed downstream after the UGA codon, also in third position.