
These selenoproteins activate or inactivate thyroid hormones through a reductive deodination. Iodothyronine deiodinases are expressed in all tissues. Their main function is to control activation and inactivation of thyroid hormones. Tetraiodothyronin (T4) is activated by removing a iodine to convert it to T3. However, other reactions lead to reverse T3 and to T2, which are inactive forms. It is highly expressed in the thyroid, and may contribute significantly to the relative increase in thyroidal T3 production in patients with Graves disease and thyroid adenomas.
The protein encoded by this gene is a thiol-requiring propylthiouracil-sensitive oxidoreductase. It activates thyroid hormone by converting the prohormone thyroxine (T4) by outer ring deiodination (ORD) to bioactive 3,3',5-triiodothyronine (T3). It also degrades both hormones by inner ring deiodination (IRD). Alternative splicing results in multiple transcript variants encoding different isoforms. All three DI isoforms contain at least one selenocyesteine. In the case of DI2 contains three Sec in humans.
DI1
The DI1 protein of Anolis carolinensis has 114 aminoacids in its sequence. We predicted 3 genes of this protein in Vipera berus genome. In the t-coffee output between Anolis carolinensis protein and our 3 predictions, selenocystein was aligned in all of them. The first predicted gene was found in scaffold KN631148.1 between positions 10201-10545, with only 1 exon of 344 nucleotides, which codes for 114 aminoacids. The second prediction was in scaffold KN631311.1 between positions 23511-24524, with 2 exons that in total have 340 nucleotides, which in total code for 113 aminoacids. The last predicted gene was found in scaffold KN637869.1 between positions 26908-27264, with one exon of 356 nucleotides, coding for 119 aminoacids. As our results suggest, Vipera berus genome contains 3 paralogous copies of DI1 gene.
For our first gene, we obtained 4 hits in the Secis prediction, three of which were in the correct strand and in the 3' region of the gene. Of these, only one was at a reliable distance from the gene, so it was our choice (location: 11118-11196). For the second prediction, we had 5 hits. 2 of them were on the correct frame and downstream the gene, so we chose the one with the most feasible distance (location: 22198-22266). For the third prediction, out of four hits we obtained two that were in the at 3' and in the correct frame so, again, we chose the closest one to our gene (location: 28700-28779).
.png)
.png)
.png)
DI2
The DI2 protein of Anolis carolinensis has 266 aminoacids in its sequence. We predicted 2 genes of this protein in Vipera berus genome. When running a t-coffee between Anolis carolinensis protein and our 2 predictions, selenocystein was aligned in all of them. The first predicted gene was found in scaffold KN637869.1 between positions 10735-27339, with only 2 exons that in total have 793 nucleotides, which code for 264 aminoacids. The second prediction was in scaffold KN631148.1 between positions 10177-10602, with 1 exon with 425 nucleotides, which code for 142 aminoacids. In fact, this last prediction corresponds and overlaps to our first predicted DI1 gene, so we discarded it. In consequence, we conclude that Vipera berus genome contains 1 DI2 gene, which is our first prediction.
We obtained 4 hits in the Secis prediction, two of which were in the correct strand and in the 3' region of the gene. Of these two, only one was at a reliable distance from the gene, so it was our choice (location: 28700-28779).

Glutathione peroxidase (GPx) are the largest and best-known selenoprotein family. GPx is the general name of an enzyme family with peroxidase activity whose main biological role is to protect the organism from oxidative damage. The biochemical function of glutathione peroxidase is to reduce lipid hydroperoxides to their corresponding alcohols and to reduce free hydrogen peroxide to water.This family is formed by 8 GPx homologs (GPx1-GPx8). GPx1-4 and GPx6 are selenoproteins in humans and in macaque, whereas GPx5, GPx7 and GPx8 are cysteine-containing homologues.
Gpx
The GPx protein of Anolis carolinensis has 193 aminoacids in its sequence. We predicted 3 genes of this protein in Vipera berus genome. When running a t-coffee between Anolis carolinensis protein and our 3 predictions, selenocysteine was aligned only in the first one, although in the other two it was aligned against a cysteine. The first predicted gene was found in scaffold KN618059.1 between positions 62217-64867, with only 2 exons that in total have 577 nucleotides, which code for 192 aminoacids. The second prediction was in scaffold KN638969.1 between positions 43692-43964, with 1 exon with 272 nucleotides, which code for 91 aminoacids. The last predicted gene was found in scaffold KN632563.1 between positions 92280-101503, with 2 exons of 214 nucleotides, coding for 71 aminoacids. In consequence we discard the last two predictions as their lengths are too short and conclude that Vipera berus genome contains 1 GPx gene.
Anolis carolinensis also presents a cysteine containing GPx homologous with 92 aminoacids and our prediction in Vipera berus genome, locates the gene in scaffold KN632563.1 between positions 92175-92441, with 1 exon with 266 nucleotides, coding for 89 aminoacids and the cysteines aligned. Moreover, it coincides with the first exon of the third GPx prediction described previously.
For the selenoprotein GPx gene prediction, we obtained 10 hits in the Secis prediction, three of which were in the correct strand and in the 3' region of the gene. Of these, only one was at a reliable distance from the gene, so it was our choice (location:61677-61745). For the cysteine-homologous GPx gene prediction, we predicted 12 SECIs elements, but only one accomplished our conditions (location: 90371-90444).


Gpx2
This gene is a member of the glutathione peroxidase family and encodes a selenium-dependent glutathione peroxidase that is one of two isoenzymes responsible for the majority of the glutathione-dependent hydrogen peroxide-reducing activity in the epithelium of the gastrointestinal tract. The protein encoded by this locus contains a selenocysteine (Sec) residue encoded by the UGA codon, which normally signals translation termination.
The GPx2 protein of Anolis carolinensis has 189 aminoacids in its sequence. We predicted 1 gene of this protein in Vipera berus genome. Moreover, when running a t-coffee between Anolis carolinensis protein and our prediction, selenocystein was aligned in it. The predicted gene was found in scaffold KN633694.1 between positions 46754-52287, with only 2 exons that in total have 565 nucleotides, which code for 188 aminoacids. In consequence, we suggest that Vipera berus genome contains 1 GPx2 gene.
We obtained 6 hits in the Secis prediction, three of which were in the correct strand and in the 3' region of the gene. Of these three, only one was at a reliable distance from the gene, so it was our choice (location: 46492-46556)

GPx3
This gene product belongs to the glutathione peroxidase family, which functions in the detoxification of hydrogen peroxide. It contains a selenocysteine (Sec) residue at its active site. The selenocysteine is encoded by the UGA codon, which normally signals translation termination. The 3' UTR of Sec-containing genes have a common stem-loop structure, the sec insertion sequence (SECIS), which is necessary for the recognition of UGA as a Sec codon rather than as a stop signal.
The GPx3 protein of Anolis carolinensis has 191 aminoacids in its sequence. We predicted 1 gene of this protein in Vipera berus genome. When running a t-coffee between Anolis carolinensis protein and our prediction, selenocysteine was aligned in it. The predicted gene was found in scaffold KN638526.1 between positions 16865-20932, with 4 exons that in total have 563 nucleotides, which code for 188 aminoacids. As our result shows, Vipera berus genome contains 1 GPx3 gene.
We obtained 2 hits in the Secis prediction, but none of them accomplished our requirements, because one was in the other frame and the other was not on the 3' region of the gene.

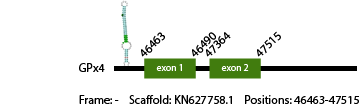
GPx4
This gene encodes a member of the glutathione peroxidase protein family. Glutathione peroxidase catalyzes the reduction of hydrogen peroxide, organic hydroperoxide, and lipid peroxides by reduced glutathione and functions in the protection of cells against oxidative damage. Human plasma glutathione peroxidase has been shown to be a selenium-containing enzyme and the UGA codon is translated into a selenocysteine.The encoded protein has been identified as a moonlighting protein based on its ability to serve dual functions as a peroxidase as well as a structural protein in mature spermatozoa. Through alternative splicing and transcription initiation, rat produces proteins that localize to the nucleus, mitochondrion, and cytoplasm. In humans, alternative transcription initiation and the cleavage sites of the mitochondrial and nuclear transit peptides need to be experimentally verified. Alternative splicing results in multiple transcript variants.
The GPx4 protein of Anolis carolinensis has 64 aminoacids in its sequence and it is a cysteine containing homologous. We predicted 1 gene of this protein in Vipera berus genome. The cysteine was not aligned. The predicted gene was found in scaffold KN627758.1 between positions 46463-47515, with 2 exons that in total have 178 nucleotides, which code for 59 aminoacids. As our result shows, Vipera berus genome contains 1 GPx4 gene.
We obtained 3 hits in the Secis prediction, but only one was in the correct strand and in the 3' region of the gene. We consider it as a feasible hit, although is a bit too far from our gene (location: 42858-42932).
GPx8
It is a surface protein.
The GPx8 protein of Anolis carolinensis has 132 aminoacids in its sequence and it is a cysteine containing homologous. We predicted 2 genes of this protein in Vipera berus genome. When running a t-coffee between Anolis carolinensis protein and our 2 predictions, only in the first one, the cysteine was aligned with the selenocysteine of the GPx8 selenoprotein. The first predicted gene was found in scaffold KN638969.1 between positions 43704-43967, with 1 exon that in total have 263 nucleotides, which code for 88 aminoacids. The second prediction was in scaffold KN635910.1 between positions 65237-65368, with 1 exon with 131 nucleotides, which code for 44 aminoacids. We discard the last prediction and conclude that Vipera berus genome contains 1 GPx8 gene.
No Secis elements were found in this gene.

This selenoprotein is an extracellular glycoprotein, and is unusual in that it contains 10 Sec residues per polypeptide. It is a heparin-binding protein that appears to be associated with endothelial cells, and has been implicated to function as an antioxidant in the extracellular space. Several transcript variants, encoding either the same or different isoform, have been found for this gene.
SelP1
The SelP1 selenoprotein of Anolis carolinensis has 169 aminoacids in its sequence. We predicted 1 gene of this protein in Vipera berus genome. When running a t-coffee between Anolis carolinensis protein and our prediction, the selenocysteine was aligned. The predicted gene was found in scaffold KN620947.1 between positions 816892-818519, with 3 exon that in total have 498 nucleotides, which code for 166 aminoacids. As our results show, Vipera berus genome contains 1 SelP1 gene.
We obtained 4 hits in the Secis prediction, two of which were in the correct strand and in the 3' region of the gene. Of these, only one was at a reliable distance from the gene, so it was our choice (location: 812208-812290).

SelP2
The SelP2 selenoprotein of Anolis carolinensis has 177 aminoacids in its sequence. We predicted 1 gene of this protein in Vipera berus genome. When running a t-coffee between Anolis carolinensis protein and our prediction, the selenocysteine was aligned. The predicted gene was found in scaffold KN623297.1 between positions 96916-99220, with 3 exon that in total have 519 nucleotides, which code for 173 aminoacids. As our results show, Vipera berus genome contains 1 SelP2 gene.
We obtained 5 hits in the Secis prediction, two of which were in the correct strand and in the 3' region of the gene. Of these, only one was at a reliable distance from the gene, so it was our choice (location: 101813-101880).

The thioredoxin reductase are involved in a sequence of reactions which ultimately leads to the formation of reduced disulfide bonds. TRs reduce thioredoxin, taking its electrons in order to give them to a protein and transform it from the S2 (oxidized) form to the SH2 (reduced) form. This process is an antioxidant mechanism present in all cells. TR1 is cytosolic, TR2 mitochondrial and TR3 is testes specific.
The TR selenoprotein of Anolis carolinensis has 559 aminoacids in its sequence. We predicted 1 gene of this protein in Vipera berus genome and the selenocysteine was not aligned. The predicted gene was found in scaffold KN632363.1 between positions 26882-41340, with 12 exons that in total have 1461 nucleotides, which code for 487 aminoacids. However, we discard this prediction as it overlaps the TR3 one, which contains the selenocysteine aligned and more exons were predicted, so we suggest that Vipera berus does not present a TR gene.
TR3
This gene encodes a member of the family of pyridine nucleotide oxidoreductases. This protein catalyzes the reduction of thioredoxin, and is implicated in the defense against oxidative stress. It contains a selenocysteine (Sec) residue (which is essential for catalytic activity), encoded by a UGA codon, at the penultimate C-terminal position. The 3' UTR of Sec-containing genes have a common stem-loop structure, the sec insertion sequence (SECIS), which is necessary for the recognition of UGA as a Sec codon rather than as a stop signal. Alternatively spliced transcript variants encoding different isoforms have been found for this gene.
The TR3 selenoprotein of Anolis carolinensis has 609 aminoacids in its sequence. We predicted 1 gene of this protein in Vipera berus genome and the selenocysteine was aligned. The predicted gene was found in scaffold KN632363.1 between positions 9900-42843 , with 16 exons that in total have 1808 nucleotides, which code for 608 aminoacids. As our result shows, Vipera berus presents a TR3 gene in its genome.
We obtained 4 hits in the Secis prediction, but none of them matched our requirements.

Studies in mouse suggest that this selenoprotein may have redox function and may be involved in the quality control of protein folding. This gene is localized on chromosome 1p31, a genetic locus commonly mutated or deleted in human cancers. Two alternatively spliced transcript variants encoding distinct isoforms have been found for this gene.
The Sel15 selenoprotein of Anolis carolinensis has 137 aminoacids in its sequence. We predicted 1 gene of this protein in Vipera berus genome and the selenocysteine was aligned. The predicted gene was found in scaffold KN622219.1 between positions 113416-132255, with 4 exons that in total have 407 nucleotides, which code for 136 aminoacids. As our result shows, Vipera berus presents a Sel15 gene in its genome.
We obtained 2 hits in the Secis prediction, but none of them suited our requirements.

The function is not known, however, is thought to be responsible for most biomedical effects of dietary selenium. Overexpression in neurons is protective again UV effects. Several papers report that its overexpression (in neurons and other cells) promotes formation of mitochondria.
The SelH selenoprotein of Anolis carolinensis has 79 aminoacids in its sequence. We predicted 1 gene of this protein in Vipera berus genome and the selenocysteine was aligned. The predicted gene was found in scaffold KN632251.1 between positions 178900-182678, with 2 exons that in total have 232 nucleotides, which code for 77 aminoacids. As our result shows, Vipera berus presents a SelH gene in its genome.
We obtained 10 hits in the Secis prediction, two of which were in the correct strand and in the 3' region of the gene. Of these, only one was at a reliable distance from the gene, so it was our choice (location: 177376-177451).

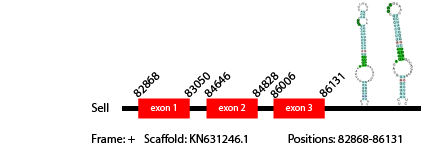
Its function is not known.
The SelI selenoprotein of Anolis carolinensis has 167 aminoacids in its sequence. We predicted 1 gene of this protein in Vipera berus genome and the selenocysteine was aligned. The predicted gene was found in scaffold KN631246.1 between positions 82868-86131, with 3 exons that in total have 489 nucleotides, which code for 163 aminoacids. As our result shows, Vipera berus presents a SelI gene in its genome.
We obtained 5 hits in the Secis prediction, two of which were in the correct strand and in the 3' region of the gene. They both were at a reliable distance from the gene, so we consider them as equally good predictions (locations: 87910-87977, 86174-86255).
This selenoprotein is localized to the endoplasmic reticulum and is highly expressed in the heart, where it may function as an antioxidant.
The SelK selenoprotein of Anolis carolinensis has 93 aminoacids in its sequence. We did not predict any gene for SelK in Vipera berus genome.
It is expressed in several tissues and its location is perinuclear.
The SelM selenoprotein of Anolis carolinensis has 94 aminoacids in its sequence. We did not predict any gene for SelK in Vipera berus genome.
Is expressed in the membrane of the endoplasmic reticulum of several tissues. Mutations in this gene cause the classical phenotype of multiminicore disease and congenital muscular dystrophy with spinal rigidity and restrictive respiratory syndrome.
The SelN selenoprotein of Anolis carolinensis has 492 aminoacids in its sequence. We predicted 1 gene of this protein in Vipera berus genome and the selenocystein was aligned. The predicted gene was found in scaffold KN614981.1 between positions 26068-33905, with 7 exons that in total have 878 nucleotides, which code for 293 aminoacids. As our result shows, Vipera berus presents a SelN gene in its genome, although our prediction is too short.
We obtained 5 hits in the Secis prediction, but none of them suited our expectations.

SelR1
This protein belongs to the methionine sulfoxide reductase (Msr) protein family which includes repair enzymes that reduce oxidized methionine residues in proteins. The protein encoded by this gene is expressed in a variety of adult and fetal tissues and localizes to the cell nucleus and cytosol.
The SelR1 selenoprotein of Anolis carolinensis has 94 aminoacids in its sequence. We predicted 1 gene of this protein in Vipera berus genome and the selenocystein was aligned. The predicted gene was found in scaffold KN635457.1 between positions 48530-49734, with 2 exons that in total have 262 nucleotides, which code for 87 aminoacids. As our result shows, Vipera berus presents a SelR1 gene in its genome.
We obtained 4 hits in the Secis prediction, but only one was in the correct frame, was in the 3' region and was at a suitable distance from the gene (location: 50989-51066)

SelR2
SelR2 function is unknown. Overexpression of mitochondrial methionine sulfoxide reductase B2 protects leukemia cells from oxidative stress-induced cell death and protein damage.
The SelR2 protein of Anolis carolinensis has 140 aminoacids in its sequence and it is a cysteine containing homologous. We predicted 1 gene of this protein in Vipera berus genome. When running a t-coffee between Anolis carolinensis protein and our prediction, the cystein was aligned with the selenocystein of the SelR2 selenoprotein. The predicted gene was found in scaffold KN633105.1 between positions 32289-32474, with 1 exon that in total has 185 nucleotides, which code for 62 aminoacids. Our result suggests thatVipera berus genome contains 1 SelR2 gene, although our hit is a bit too short.
We obtained 6 hits in the Secis prediction, but none of them suited our requirements so we discarded them.

SelR3
The protein encoded by this gene catalyzes the reduction of methionine sulfoxide to methionine. This enzyme acts as a monomer and requires zinc as a cofactor. Several transcript variants encoding two different isoforms have been found for this gene. One of the isoforms localizes to mitochondria while the other localizes to endoplasmic reticula.
The SelR3 protein of Anolis carolinensis has 184 aminoacids in its sequence and it is a cysteine containing homologous. We predicted 1 gene of this protein in Vipera berus genome. When running a t-coffee between Anolis carolinensis protein and our prediction, the cystein was aligned with the selenocystein of the SelR2 selenoprotein. The predicted gene was found in scaffold KN628141.1 between positions 8521-67112, with 6 exons that in total have 546 nucleotides, which code for 182 aminoacids. Our result suggests that Vipera berus genome contains 1 SelR3 gene.
We obtained 2 hits in the Secis prediction, but none of them suited our requirements, so we discarded them.

Play a key role in the control of the inflammatory response and it has been associated with cardiovascular disease in diabetes[16].
The SelS selenoprotein of Anolis carolinensis has 164 aminoacids in its sequence. We did not predict any gene for SelS in Vipera berus genome.
Along with SelT, SelW, SelH and Rdx12; this selenoprotein has been proposed as a member of a novel thioredoxin-like family[17].
The SelT selenoprotein of Anolis carolinensis has 153 aminoacids in its sequence. We predicted 1 gene of this protein in Vipera berus genome and the selenocystein was aligned. The predicted gene was found in scaffold KN635297.1 between positions 59408-61861, with 4 exons that in total have 455 nucleotides, which code for 152 aminoacids. As our result shows, Vipera berus presents a SelT gene in its genome.
We obtained 5 hits in the Secis prediction, 3 of which were in the correct strand and in the 3' region of the gene. Of these, only one was at a reliable distance from the gene, so it was our choice (location: 57993-58067).

SelU1
Although SelU1 is expressed in bone, brain, liver and kidney, its function is unknown.
The SelU1 selenoprotein of Anolis carolinensis has 233 aminoacids in its sequence. We predicted 1 gene of this protein in Vipera berus genome and the selenocystein was aligned. The predicted gene was found in scaffold KN635350.1 between positions 170571-180081, with 4 exons that in total have 552 nucleotides, which code for 184 aminoacids. As our result shows, Vipera berus presents a SelU1 gene in its genome.
We obtained 5 hits in the Secis prediction, but none of them was in the correct frame, in the 3' region and at a suitable distance from the gene at the same time.

SelU2
SelU2 function is unknown.
The SelU2 protein of Anolis carolinensis has 225 aminoacids in its sequence and it is a cysteine containing homologous. We predicted 1 gene of this protein in Vipera berus genome and it does not have the cysteine aligned. The predicted gene was found in scaffold KN632497.1 between positions 8609-10556, with 2 exons that in total have 256 nucleotides, which code for 85 aminoacids. Therefore, our result suggests that Vipera berus genome does not contains 1 SelU2 gene.
This protein encodes an enzyme that synthesizes selenophosphate from selenide and ATP.
The SPS selenoprotein of Anolis carolinensis has 432 aminoacids in its sequence. We predicted 1 gene of this protein in Vipera berus genome and the selenocystein was aligned. The predicted gene was found in scaffold KN626389.1 between positions 80733-90051, with 7 exons that in total have 1169 nucleotides, which code for 390 aminoacids. As our result shows, Vipera berus presents a SPS gene in its genome.

The SPS1 is a machinery protein related to the synthesis of selenoproteins. In Anolis carolinensis genome, it has 392 aminoacids. We predicted 2 genes of this protein in Vipera berus genome. The first prediction was in scaffold KN636817.1 between positions 39126-51377, with 7 exons that in total have 977 nucleotides, which in total code for 326 aminoacids. The second predicted gene was found in scaffold KN626389.1 between positions 80772-88750 and, as we can see, it corresponds to the SPS selenoprotein prediction. As our results suggest, Vipera berus genome contains 1 copy of SPS1 gene.
We only found one Secis element, and it matched our requirements (location: 79442-79516). For the machinery no Secis elements were predicted because it is not a selenoprotein.

Sel0(1)
Its function is unknown.
The Sel0(1) protein of Anolis carolinensis has 262 aminoacids in its sequence and it is a cysteine containing homologous. We predicted 1 gene of this protein in Vipera berus genome. The predicted gene was found in scaffold KN616814.1 between positions 126855-127061, with 1 exon that in total have 206 nucleotides, which code for 69 aminoacids. Our result suggests that Vipera berus genome does not contain Sel0(1) gene.
Sel0(2)
Its function is unknown.
The Sel0(2) protein of Anolis carolinensis has 66 aminoacids in its sequence and it is a cysteine containing homologous. We predicted 1 gene of this protein in Vipera berus genome. The predicted gene was found in scaffold KN624046.1 between positions 121870-139588, with 9 exons that in total have 256 nucleotides, which code for 486 aminoacids. Our result suggests that Vipera berus genome does not contain 1 Sel0(2) gene.
.png)
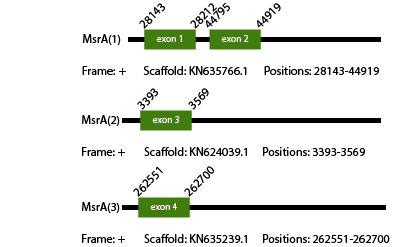
The Methionine Sulfide Reductase A is an enzyme that carries out the enzymatic reduction of methionine sulfoxide to methionine. Human and animal studies have shown the highest levels of expression in kidney and nervous tissue. Its proposed function is the repair of oxidative damage to proteins to restore biological activity. Three transcript variants encoding different isoforms have been found for this gene. It does not contain a Sec.
The MsrA protein of Anolis carolinensis has 209 aminoacids in its sequence and it is a cysteine containing homologous We had 5 predictions of the MrsA gene that, on their own, were too small to be a complete gene. But when looking at each of the t-coffee's output, it was clear that the first three hits were part of the same protein, because in the alignment where one finished the other started, completing this way the whole protein. Therefore, our gene is split between 3 scaffolds, in the following order: KN635766.1 (positions: 28143-44919), KN624039.1 (3393-3569) and KN635239.1 (positions: 262551-262700). The final gene has 4 exons, 518 nucleotides and 173 aminoacids, so we can conclude that Vipera berus contains one MsrA gene in its genome.
It is highly expressed in skeletal and cardiac muscle. It may be involved in anti oxidation processes, as most selenoproteins. Its specific function is still unknown.
The SelW selenoprotein of Anolis carolinensis has 48 aminoacids in its sequence. We did not predict any gene for SelW in Vipera berus genome.
Eukaryotic elongation factor has a glutathione peroxidase functions in the detoxification of hydrogen peroxide and it is one of the most important antioxidant enzymes in humans.
The eEFsec is a machinery protein related to the synthesis of selenoproteins. Anolis carolinensis genome contains 3 copies of it. The first one, eEFsec(1), has 215 aminoacids. We predicted 1 gene of this protein in Vipera berus genome. The prediction was in scaffold KN617171.1 between positions 36097-36741, with 1 exons that in total have 644 nucleotides, which in total code for 215 aminoacids. The second one, eEFsec(2), has 79 aminoacids. We didn’t predict any genes of this protein in Vipera berus genome. The last one, eEFsec(3) has 168 aminoacids. We predicted 1 gene of this protein in Vipera berus genome. The prediction was in scaffold KN617171.1 between positions 2901-10232, with 3 exons that in total have 492 nucleotides, which in total code for 164 aminoacids. As our results suggest, Vipera berus genome contains 2 copies of eFsec gene.
.png)
.png)
SBP2 encodes a nuclear protein that functions as a SECIS binding protein. Mutations in this gene have been associated with a reduction in activity of a specific thyroxine deiodinase, a selenocysteine-containing enzyme, and abnormal thyroid hormone metabolism.
Anolis carolinensis has two paralogous copies of the SBP2 gene. The first one, SBP2(1), has 1055 aminoacids. We predicted 1 gene of this protein in Vipera berus genome. The prediction was in scaffold KN618877.1 between positions 581837-599039, with 15 exons that in total have 2997 nucleotides, which in total code for 999 aminoacids. The second one, SBP2(2), has 854 aminoacids. We predicted 1 gene of this protein in Vipera berus genome. The prediction was in scaffold KN632477.1 between positions 82268-108573, with 14 exons that in total have 2170 nucleotides, which in total code for 723 aminoacids. Therefore, we predicted both copies of the SBP2 gene. No Secis elements were predicted, as it is not a selenoprotein or a homologous.
.png)
.png)
The amino acid selenocysteine is the only amino acid that does not have its own tRNA synthetase. Instead, this amino acid is synthesized on its cognate tRNA in a three step process. The protein encoded by this gene catalyzes the third step in the process, the conversion of O-phosphoseryl-tRNA(Sec) to selenocysteinyl-tRNA(Sec).
The SecS is a machinery protein related to the synthesis of selenoproteins. In Anolis carolinensis genome, it has 466 aminoacids. We predicted 1 gene of this protein in Vipera berus genome. The prediction was in scaffold KN615039.1 between positions 4537-76572, with 11 exons that in total have 1333 nucleotides, which in total code for 444 aminoacids. As our results suggest, Vipera berus genome contain 1 copy of SecS gene.

Phosphoseryl-tRNA kinase is found to be highly conserved in evolution, hence suggesting that it plays an important role in selenoprotein biosynthesis and/or regulation.
The PSTK is a machinery protein related to the synthesis of selenoproteins. In Anolis carolinensis genome, it has 352 aminoacids. . We predicted 1 gene of this protein in Vipera berus genome. The prediction was in scaffold KN633361.1 between positions 47138-54632, with 6 exons that in total have 1047 nucleotides, which in total code for 349 aminoacids. As our results suggest, Vipera berus genome contains 1 copy of PSTK gene. As it is a machinery protein related to the synthesis of selenoproteins, we did not predict any Secis.

This protein regulates selenoprotein expression.
The SECp43 is a machinery protein related to the synthesis of selenoproteins. In Anolis carolinensis genome, it has 268 aminoacids. . We predicted 1 gene of this protein in Vipera berus genome. The prediction was in scaffold KN636726.1 between positions 201915-204816, with 6 exons that in total have 621 nucleotides, which in total code for 207 aminoacids. As our results suggest, Vipera berus genome contains 1 copy of SECp43 gene. As it is a machinery protein related to the synthesis os selenoproteins, we did not predict any Secis.
