Originally, selenium (Se) was viewed as a toxin, but now it is understood as an important micronutrient that supports various important cellular and organismal functions [1, 2].
Se, like other metals, is included in proteins by ionic association, and, more importantly, is covalently bonded with the 21st amino acid, selenocysteine (Sec). This amino acid is structurally similar to cysteine (Cys), the only difference is the presence of Se instead of the sulfur [3]. The first Sec found in a protein was glutathione peroxidase (GPx) 1 in 1987 [4]. Due to the discovery, cloning the proteins led to the coding codon for Secs, TGA, which also acts as a stop codon in the other genes (non-selenoprotein genes) [5]. Among the other amino acids, Sec is the only known amino acid in eukaryotes whose biosynthesis occurs on its own tRNA [2]. The tRNA for Sec (tRNASec) recognizes the codon UGA [3].
The functionally characterized Secs are located in enzyme active sites and allow them to perform catalytic redox reactions, nevertheless, Sec is implied in diverse biological roles [2]. Taking that into consideration, the physiological functions of selenoproteins strictly depend on the presence of Sec [2].
Also, studying iodothyronine deiodinases (DIO), the Sec insertion sequence, also known as SECIS, element was discovered [6]. SECIS elements are stem-loop RNA structures found in the 3’-untranslated regions (UTR) of the mRNA [2].
Previous analyses of the selenoproteome in various model organisms have revealed widely different selenoprotein sets. Aquatic organisms, like Helostoma temminkii, generally have larger selenoproteomes than terrestrial organisms. Also, it has been shown that, maybe because the conversion of Sec to Cys with a single point mutation, mammalian selenoproteomes are reducing the use of selenoproteins [7, 8].
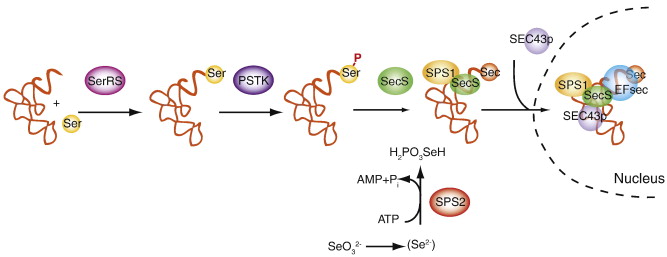
Around 53 selenoproteins and Cys-containing homologues are annotated for Danio rerio in the databases, distributed in 26 families, according to SelenoDB. In fact, Zebrafish carries in its genome the largest amount of selenoproteins with a total number of 38 known. This amount of proteins is similar in number with human selenoproteins and cysteine homologous proteins, which are also about 50 in SelenoDB, and divided 37 families. Previous literature shows that Human genome contains 25 Selenoprotein genes [8]. Nevertheless, different selenoproteins have been described between these two species. Moreover, more than 50 families of selenoproteins are known to exist along all the kingdoms [9]. Vertebrates, at least, present 45 selenoprotein subfamilies. GPx1-4, TR1, TR3, Dio1, Dio2, Dio3, SelH, SelI, SelK, SelM, SelN, SelO, SelP, MsrB1, SelS, SelT1, SelW1 and Sep15 selenoproteins appear in all vertebrates genomes [8].
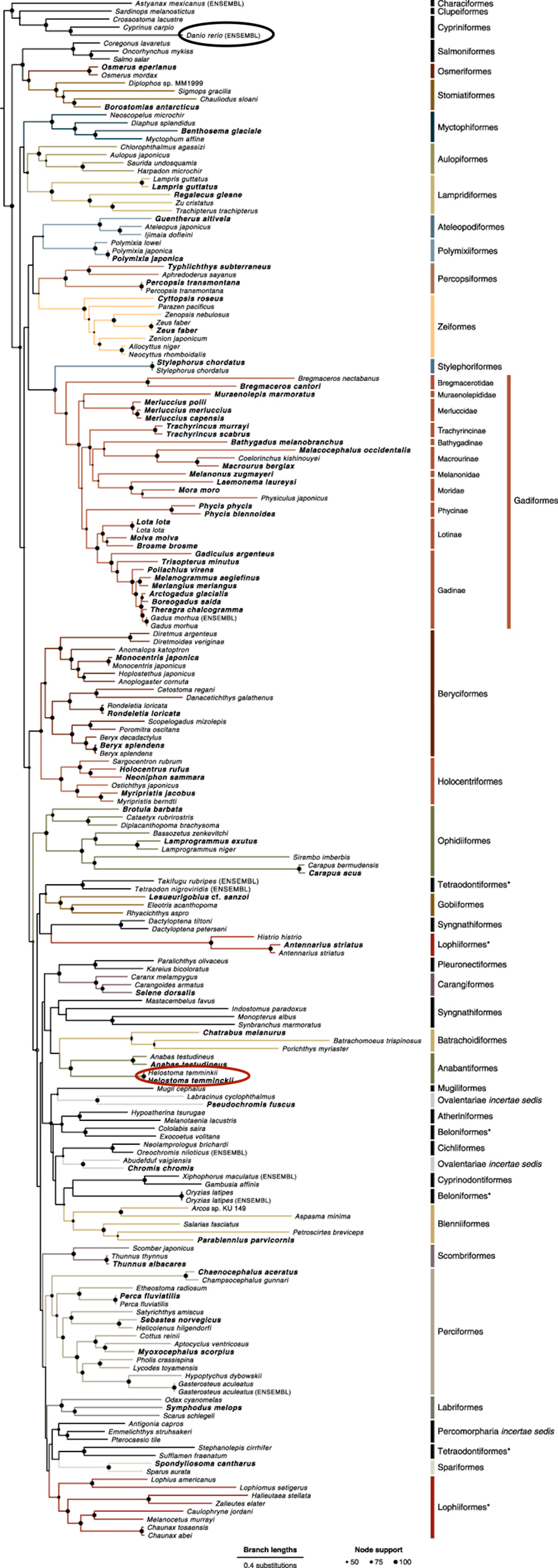
In the SelenoDB are 26 families known in Danio rerio, as previously mentioned, and three of those are the most studied. Firstly, the Glutathione Peroxidases (GPxs) family, composed of eight paralogs, five with Sec residue in the active site while the other three have the Sec replaced by Cys [2, 8]. Secondly, the thyroid Hormone Deiodinases (DIOs or DIs) family, formed by three paralogs generally, nevertheless, in Danio rerio there are four, due to a whole-genome duplication (Figure 1) [2, 8]. Thirdly, the thioredoxin Reductases (TXNRDs) family, composed of three paralogs, but in Danio rerio TXNRD1 hasn’t been found [2, 8].
As mentioned before, the main common feature that determines a selenoprotein is the Sec residues in their sequence [9]. This amino acid is located in most of the selenoproteins in the enzyme active site and it is known to perform catalytic redox reactions [9,10]. Thus, Sec is the main element in charge of the physiological functions of selenoproteins.
It is important to highlight that, a previous event of duplication that happened while bony fishes where diverging, generated duplications in all the genome of this Osteichthyes that result in DIO3b and GPx3b proteins [8].
Below, current documented selenoproteins are classified followed by an explanation of their function:
Iodothyronine deiodinases (DIO)
The thyroid hormone deiodinases or iodothyronine deiodinase consist of three paralogous proteins in mammals (DI1, DI2 and DI3) [9]. All three proteins are involved in reductive deamination which is essential for the regulation of thyroid hormone activity [9, 11]. In most vertebrates, T4 (pro-hormone) has to be metabolized in order to obtain the T3 (hormone). These proteins are widely extended, not only along eukaryotes, but also in some other kingdoms as bacteria. Danio rerio has four DIO proteins annotated: DIO1, DIO2, DIO3a and DIO3b.
DIO1, DIO2 and DIO3:
DIO1 is present in the cell membrane and is in charge of the metabolization of T4 into T3 hormone in peripheral tissues, this process can be also performed by DIO2 (localized in the endoplasmic reticulum). Thus it can bind to thyroid hormone nuclear receptors [9, 11, 12].
DIO 3 can only metabolize T4 into reverse triiodothyronine (rT3), which is biologically inactive [9, 11, 13].
Glutathione peroxidases (GPx)
Glutathione peroxidases selenoproteins are widespread in all the domains of life. In mammals 8 different paralogues are described: GPx1, GPx2, GPx3, GPx4, GPx5, GPx6, GPx7 and GPx8. GPx1, GPx2, GPx3, GPx4 and GPx6 are selenoproteins containing Sec, whereas GPx5, GPx7 and GPx8 are Cys-containing homologs [9, 14]. Danio rerio predictions include 9 GPx selenoproteins: GPx1a, GPx1b, GPx2, GPx3a, GPx3b, GPx4a, GPx4b, GPx7 and GPx8. In this species, only GPx7 and GPx8 are missing Sec in their catalytic sites.
GPx selenoproteins are involved in physiological functions and are responsible of part of hydrogen peroxide signaling, detoxification of hydroperoxides and of the maintenance of cellular redox homeostasis [9].
Specifically, GPx1 is the most abundant selenoprotein in mammals. It is in charge of the catalysis of glutathione-dependent reduction of hydrogen peroxide to water [9, 15]. As hydrogen peroxide is known to bé toxic to the cells, GPx1 is considered an important antioxidant molecule.
For the other GPx2,GPx3, GPx4, they have been shown to be enrolled in other pathways as development of cancer (GPx2), or in the regulation of tyrosine phosphatases and reduction of complex lipid hydroperoxides bound to the cell membrane (GPx4). Kidney is characterized by the expression of GPx3 [9].
Methionine sulfoxide reductases A (MsrA)
Msr A is highly conserved, and has its largest expression in kidney and liver. Its main function is to catalyze the enzymatic reduction of the amino acid methionine-S-sulfoxide by using thioredoxin [9]. It is important to understand that the MSRB family of proteins was so named because of its functional similarity to MrsA family, despite MSRB and MsrA are structurally different and they do not have any similarity in their sequences, they have complementary functions. Two MsrA Cys-containing homolog proteins have been described in Danio rerio, MsrA1 and MsrA2.
Selenoprotein R (MSRB family)
Methionine-R-sulfoxide reductase 1 (MSRB1) is a zinc-containing selenoprotein that was previously identified as Selenoprotein R (SelR) and Selenoprotein X (SelX) [16, 17]. As mentioned the relation with MsrA in the previous section, MSRB selenoproteins function is the stereospecific reduction of methionine-R-sulfoxide [9].
MSRB1 is the main MSRB protein present in mammals and is localized in the cytosol and in the nucleus. Nevertheless, two additional homologs (MSRB2 and MSRB3) show lower levels of expression in mammals but their catalytic efficiency is similar to MSRB1. They also vary in their localitzation: MSRB2 is located in mitochondrias, whereas MSRB3 is targeted to the ER [9].
MSRB1a, MSRB1b, MSRB2 and MSRB3 have been annotated in Danio rerio.
15-kDa selenoprotein (Sel15)
Sel15 is known in mammals as Sep15, which is together with Selenoprotein M (SelM) thioredoxin-like fold ER-resident protein [9]. This Sep15 is known to form a complex with an ER-resident chaperone (UGGT) which is involved in calnexin regulation cycle, which is essential for the folding process of some glycoproteins in the ER, and its expression is induced by misfolded proteins in ER [18]. This protein has been shown to be implicated in preventing some types of cancers as liver, prostate, breast and lung cancers [9].
Selenoprotein E (SELENOE)
SELENOE is a gene that is part of SEP15 family. Unlike previous proteins mentioned above, SELENOE has been detected only in fish. Probably, this gene has been arisen by gene duplication. Its location is inside the ER and its function remains unknown [8, 19].
Selenoprotein H (SELENOH)
SELENOH, known in mammals as SelH is a stress-related selenoprotein because it functions as an oxidoreductase[9, 20, 21]. This protein is widespread over different domains and encodes for a nucleolar protein from the SelWTH family. In mammals, it has been shown to protect neurons against UVB damage, promote mitochondrial biogenesis and suppress cellular senescence.
Selenoprotein I (SELENOI)
SELENOI is one of the most recent evolved selenoproteins and it can only be found in vertebrates [9]. It is a multi-pass transmembrane (transmembrane) protein that is encoded by this gene which belongs to the CDP-alcohol phosphatidyltransferase class-I family [8, 9] Its main function is to catalyse the transfer of phosphoethanolamine from CDP-ethanolamine to diacylglycerol to produce phosphatidylethanolamine. This last molecule is necessary for the formation and maintenance of vesicular membranes. It is also involved in lipid metabolism and in protein folding [8].
Selenoprotein J (SELENOJ)
SELENOJ is one of the vertebrate-specific selenoproteins [9]. Nevertheless, this SelJ is highly restricted in phylogeny, being found only in actinopterygian fishes and sea urchin, but not in other vertebrates (mammals, birds and amphibians). The potential role of of Selenoprotein J is during embryogenesis, preferentially expressed in the eye lens (it shares significant similarity with jellyfish crystallins). This role is unlike most of the selenoproteins since the majority of them are believed to have enzymatic function [8, 22, 23]. In Danio rerio two homologs have been described SELENOJ1 and SELENOJ1.2.
Selenoprotein K (SELENOK)
SELENOK is known to be related with SelS based on their topology [9]. It is a transmembrane protein located in the endoplasmic reticulum. It is involved in mechanisms in charge of endoplasmic reticulum associated degradation of proteins that are glycosylated and misfolded. It also protects cells against endoplasmic reticulum stress-induced apoptosis [8, 24].
Selenoprotein L (SELENOL)
SELENOL is encoded by a gene that belongs to a subfamily of selenoprotein L, within a superfamily of thioredoxin (Txr). It is restricted in aquatic organisms and, in vertebrates, this protein is found only in fish, but not in mammals, birds or amfibians. In SELENOL, the UxxU motif replaces the catalytic CxxC motif in thioredoxins (it possesses a thioredoxin-like fold), and that suggests a redox function for this protein family. Then it is important to notice that typically Selenoprotein L contains two Sec residues [8, 19].
Selenoprotein M (SELENOM)
SELENOM or SelM, as it has been previously mentioned, is (together with Sel15) a thioredoxin-like folds ER-resident proteins. In fact, SelM is a distant homolog of Sel15 [9] which are encoded by genes that belong to selenoprotein M/SEP15 family. It is present in vertebrates but is specially highly expressed in mammals. Its function remains unknown but it is possible that SELENOM may be enrolled in maintaining redox homeostasis. It is localized in the perinuclear region [8, 21, 25].
Selenoprotein N (SELENON)
SELENON is one of the first selenoproteins that were identified through bioinformatics [9]. This selenoprotein is a glycoprotein localized in the endoplasmic reticulum. It is enrolled in redox regulation of calcium homeostasis and in cell protection against oxidative stress[21,26]. Mutations on the orthologous gene in human are associated with early onset muscle disorders [9,27].It also have an important role in muscle organization in early development in zebrafish [28].
Selenoproteins O (SELENOO)
SELENOO was discovered more than a decade ago but no structural or biochemical characterization of this protein has been reported. SelO human homologs have been detected in a wide variety of species including different domains [9]. This protein is localized in the mitochondria, and it is the largest selenoprotein present in mammals [25, 29]. In Danio rerio two homologs have been described SELENOO1 and SELENOO2.
Selenoproteins P (SELENOP)
SELENOP or SelP is a protein that is really abundantly expressed (predominantly in liver) and it is secreted to the plasma (accounts for approximately 50% of the total Se of the plasma) [9]. This protein has recently evolved and, for that reason, SelP homologous are found predominantly in vertebrates[9]. An example of that is Danio rerio which shows two different homologs: SELENOP1 and SELENOP2. This selenoprotein is unique because it contains 17 Sec residues per polypeptide in zebrafish (and 16 in medaka). It is implicated as an extracellular antioxidant and also in the selenium transport to extra-hepatic tissues [30, 31].
Selenoprotein S (SELENOS)
As mentioned before, SELENOS (or SelS) could be assigned to a single SelK/SelS family (related by topology)[9].
This protein encodes for a transmembrane protein that is localized in the endoplasmic reticulum. As other selenoproteins, it is involved in the degradation process of misfolded proteins. It is thought that it can also have a role in inflammation control in higher eukaryotes [25].
Selenoproteins T (SELENOT)
SELENOT or SelT belongs to the Rdx family of selenoproteins together with SelW, SelH and selV [9]. This protein is found in the endoplasmic reticulum. As the name of the family already indicates, it possesses a thioredoxin-like fold and a conserved CxxU motif [32]. Some studies with mice indicate that this protein may be enrolled on protection of dopaminergic neurons against oxidative stress in Parkinson's disease and, also, in the control on glucose homeostasis in pancreatic beta-cells [33, 34]. In Danio rerio three homologs have been described: SELENOT, SELENOT1b,SELENOT2.
Selenoproteins U (SELENOU)
SELENOU or SelU proteins are a family within a peroxiredoxin-like FAM213 superfamily. SELENOU is phylogenetically restricted in vertebrates such as chicken and fish, while other vertebrate members of this family, including mammals, contain cysteine-containing homologs. SELENOU contains UxxC motif where the cysteine-containing homologs have CxxC (catalytic site) [8,26,35]. In Danio rerio three homologs have been described: SELENOU1a, SELENOU2 and SELENOU3.
Selenoproteins W (SELENOW)
SELENOW1 belongs to SelWTH family, which possesses a thioredoxin-like fold and a conserved CxxU motif, that suggests a redox function of this protein. In mammals, this protein is expressed in skeletal muscle, heart and brain, while in Zebrafish it is expressed in sensory organs. Two paralogs of this gene exist: SELENOW2, and SELENOW3 [21, 26].
Selenophosphate synthetases (SEPHS)
SEPHS is in charge of the synthesis of selenophosphate, which is the active selenium donor. Two SEPHS paralogues (SEPHS1 and SEPHS2) are common in some eukaryotes, while prokaryotes only have one form of SEPHS. In eukaryotes, only SEPHS2 shows catalytic activity during selenophosphate synthesis. Nevertheless, SEPHS1 has been shown to play an essential role in regulating cellular physiology, Prokaryotic SEPHS can contain a cysteine or selenocysteine (Sec) at the catalytic domain. Nevertheless, in eukaryotes, SEPHS1 contains other amino acids (Thr, Arg, Gly, or Leu) at the catalytic domain, and SEPHS2 contains only a Sec [36]. In Danio rerio two homologs have been annotated: SEPHS and SEPHS2.
Thioredoxin reductases (TXNRDs)
Thioredoxin reductases (TTXNRDs) are oxidoreductases that, by combining with thioredoxin (Trx), comprise the major disulfide reduction system of the cell. In mammals, there are three TR protein isoforms which contain Sec [9]. TRs are selenocysteine-containing flavoenzymes, which are enrolled in reducing thioredoxins, among other substrates, and have an important role in redox homoeostasis. Cell localization varies among the different TRs: TXNRD1 is mainly localized in the cytosol and nucleus while TXNRD2 and TXNRD3 is localized in the mitochondria. The three TRs are present in all vertebrates despite of Danio rerio and other fish species, in which TXNRD1 has not been found [8, 25, 26].
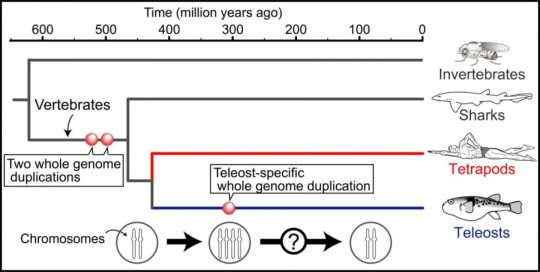
In order to incorporate selenium as a Sec into a selenoprotein it is necessary to conduct a specific mechanism to decode UGA codon (which is commonly understood as a stop codon) in mRNA. Moreover, to accomplish this purpose, it is necessary to recognize the UGA codon as a codifying codon and the loading of the Selenocysteine by a tRNA-Sec. Sec is the unique amino acid whose synthesis occurs on its own tRNA: Sec-tRNA[Ser]Sec [9, 37].
First of all, a serine is loaded on tRNA-sec by the seryl-tRNA synthetase, also known as SerRS.
Then, the PSTK, which is the enzyme O-phosphoseryl-tRNA[Ser]Sec Kinase, proceeds to phosphorylate the Ser-tRNASec [9].
The next enzyme involved in the obtention of tRNA-Sec is Sec Synthase (SecS), which can convert Ser-tRNA[Ser]Sec into Sec-tRNA[Ser]Sec. In order to accomplish this step, it has to spend ATP [9].
To incorporate the Sec from a Sec-tRNA[Ser]Sec into the protein sequence which is being codified by the ribosomes, the presence of selenocysteine insertion sequence (SECIS) is needed. This element located on 3’-UTR in eukarya and archea domains, whereas in bacteria, SECIS elements are normally localizated relatively near the UGA codon (immediately downstream). This mRNA sequence contains approximately 60 nucleotides that adopt a stem-loop secondary structure. The secondary structure acts as a cis-element that recognizes trans-acting factors and directs them to the ribosomes[9, 38].
At least, two trans-acting factors are required for efficient recording of UGA as Sec in eukaryotes:
SBP2 is stably associated with ribosomes and contain an RNA-binding domain that binds SECIS elements with high affinity and specificity. On the other hand, SBP2 not only binds SECIS elements, it also interacts with eEFSec, which is in charge of recruiting Sec-tRNA[Ser]Sec and facilitates the incorporation of this Selenocysteine in the translated peptide [9].
Finally, it is important to take into account that another protein known as SECp43 (tRNA Sec 1 associated protein 1) is known to have a role in selenoprotein synthesis through interaction with tRNA[Ser]Sec in a multiprotein complex [39]. This process can be observed in Figure 2.
None of these proteins which are involved in selenoprotein machinery contain Sec in their amino acid sequence. All of them are essential for a physiological function of the cells. Mutations on the genes that codify for these proteins lead to pathological circumstances.
Danio rerio contains in its genome 2 homologs for SBP2 and 2 homologs for SECp43.
Helostoma temminkii, commonly known as kissing gourami, is the only species of the Helostomatidae family, which is included inside the Perciformes order. This kind of fishes are highly common in Southeast Asia countries, where they are used by humans: both as food and as an aquarium fish (mainly in Japan). Even though this species is being exported for aquarium use in Japan, Europe, North America, Australia and other countries they are originated from Thailand to Indonesia [39].
| Kingdom | Animalia |
|---|---|
| Phylum | Chordata |
| Class | Actinopterygii |
| Order | Perciformes |
| Family | Helostomatidae |
| Genus | Helostoma |
ORIGIN AND NAME
H. temminkii was first described by Cuvier in 1829. Their common name, gourami, is a Javanese word which comes from Java island, Indonesia. The genus name comes from Greek: elos which means nail or claw, and stoma which means mouth [40].
DESCRIPTION
As other type of gourami, the body of H. temminkii is very compressed laterally. They have dorsal and anal fins in the end of their body, near the tail, so both fins mirror each other, their pectoral fins are rounded and large. The colour of this species may vary a little depending on the habitat of the fish but there are two main patterns: the green ones (which have dark wings and lateral stripes) and the pink and ivory ones (which have transparent fins) [41]. Kissing gourami can reach a maximum total length of 30 cm and their sexual dimorphism is non-existent, so it is nearly impossible to distinguish the sexes by their morphology.
The most distinctive feature of H. temminkii is its mouth: it is large and highly protrusible and it reminds to human lips, that is why another common name that is given to this species is the kissing fish. The specific function of these “lips” is still unknown: some hypothesis suggest that male kissing gourami use their mouth as a weapon to fight against more males to establish a hierarchy (intrasexual competition) while others suggest that this morphology facilitates H. temminkii to enhance substrate scraping when it has to search nutrients [42].
HABITAT and ECOLOGY
Kissing gourami is a fresh water fish which has a preference for slow-moving waters with thick vegetation. Because of this, it is common to find H. temminkii in temporarily flooded areas and changing water volume habitats such as lakes, backwaters and swamps (both black and clear watered) [43]. It needs a pH from 6.0 to 8.0 and a temperature from 22 to 30 ºC. All these features added to the particularities of its morphology make H. temminkii a good candidate to habit big aquariums.
Its spawning occurs under floating vegetation and it is initiated by females, this event takes place from May to October but this period varies depending on the country and the habitat. Eggs are spherical and their development is fast: they hatch after only one day, and two days later they are able to free-swim [40, 44].
NUTRITION
Helostoma temminkii is an omnivore microphagous filter-feeder whose nutrition is based in a high variety of power supplies such as little insects, alga, some kind of other species larva and other microorganisms from submerged surfaces. Its mouth, teeth and gill make this fish a well-adapted species: it can find nutrients in places where other species cannot (like algae covered surfaces, for example) [42, 45].
IMPORTANCE FOR HUMANS
Helostoma temminkii is a species which has become very commercialized in Southeast Asian countries. Large quantities of this fish are exported to Japan, Australia and North America as aquarium fish because of their harmless behaviour for humans but mainly because of their curious mouth morphology and their “kissing” behaviour [40]. In its countries of origin such as Indonesia, Thailand, Borneo, etc. they are eaten and served in many ways.
For more information visit:
The main purpose of this study was to predict all the selenoproteins of Helostoma temminkii including the selenoprotein machinery proteins, which are the ones involved in the synthesis of the first ones. For that reason, a comparison between Helostoma temminkii and Danio rerio genomes has been performed (more information in Materials and Methods). Our organism of reference contains about 38 selenoproteins described as can be seen in Figure 3. These species are part of the Osteichthyes group which, as mentioned before, experienced a duplication event. This could be noticeable in Figure 3 where bony fishes have at least 10 selenoproteins more than the rest of the vertebrates. For that reason, and assuming no other duplication event neither in Helostoma temminkii nor Danio rerio, the results we expect for our predictions should be very similar quantitatively and qualitatively to the selenoproteins present in Zebrafish.
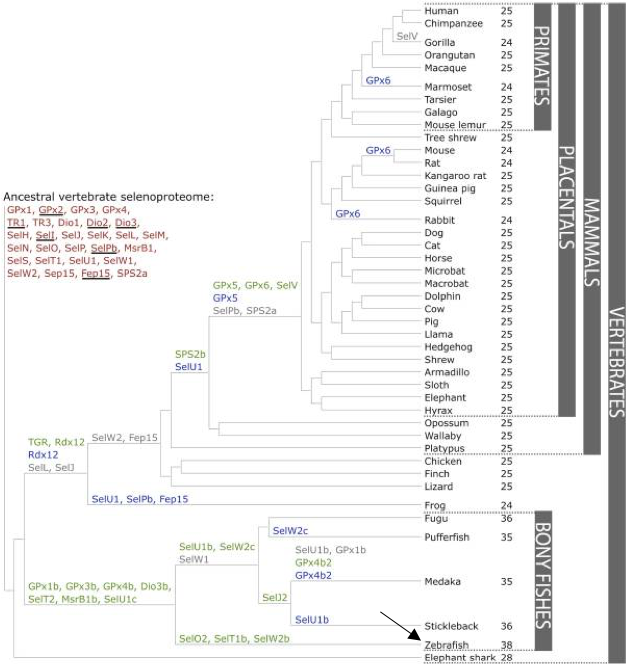
Nevertheless, while observing the comparison between teleost species in Figure 4, both species seem to be phylogenetically distant. This divergence during evolution could have led to differences between both genomes, and, therefore, between both selenoproteomes. Because of this, we cannot reject the possibility of duplication or loss of selenoproteins in Helostoma temminkii when compared to Danio rerio. According to Figure 4, Helostoma temminkii is closer related with other species as Oryzias latipes (medaka) and Gasterosteus aculeatus (stickleback) whose selenoproteome has been annotated as shown in Figure 3. Even though, the annotation of these two species may not be enough accurate, this information could be useful in doubtful results since these species should present more phylogenetic similarities with the species Helostoma temminkii.