INTRODUCTION
Selenium
Selenium (Se) is a nonmetal chemical element with symbol Se and atomic number 34. Although it is toxic in large doses, selenium is an essential micronutrient for animals [1]. Particularly, Selenium is an essential component of the unusual amino acids selenocysteine and selenomethionine.
Its main function in animals is to act as a cofactor for the reduction of antioxidant enzymes, such as glutathione peroxidases and some forms of the thioredoxine reductase [2]. It also functions as a cofactor for three of the four known thyroid hormone diodinases, which are present in the thyroid gland and in all cells that use the thyroid hormone [3].
The deficit of selenium in humans produces Keshan disease, characterised by pulmonary edema and heart problems [4]. On the other hand, accumulations of this micronutrient cause intoxication and lead to hepatotoxicity and natural killer cell impairment [5].
Selenocysteine
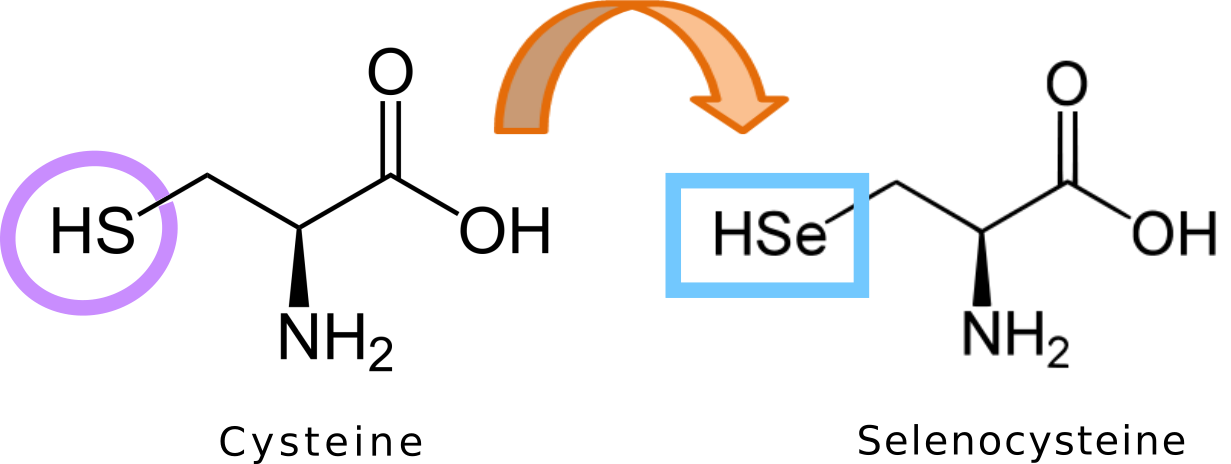
Selenocysteine (also known as Sec and U) is considered the 21st proteinogenic aminoacid of the genetic code. Sec is not found in a standard way in the genetic code, requiring specials mechanisms to be found in a protein’s structure [2,6].
Selenocysteine is an analog of cysteine with one remarkable difference: Sec presents a selenol group instead of a thiol group. Sec is encoded by the codon UGA, which normally acts as a stop codon. This is the main reason why Sec needs alternative mechanisms to be placed in proteins instead of stopping translation [7].
What are selenoproteines?
Selenoproteins conform a protein family characterized by proteins containing one or more selenocysteines [8].Selenoproteins are present in the three domains of life: eukarya, bacteria and archaea [7]. In eukarya, selenoproteins are largely present in animals, but are absent in higher plants and yeasts. In the case of bacteria and archaea, selenoproteins' presence is dependent on the lineage [9].
Selenoproteins can be broadly classified into two classes: housekeeping and stress-related. Housekeeping selenoproteins participate in functions critical to cell survival, whereas stress-related selenoproteins are not essential for survival and often show decreased expression in Se-deficient conditions [10]. A set of selenoproteins in an organism is known as the selenoproteome. The main problem surrounding this family of proteins resides in its genomic annotation: the UGA codon is normally predicted as a stop codon and not as a selenocysteine coding signal, leading to misannotations.
Known Selenoproteins
Currently there are more than 50 selenoprotein families known [7]. The majority of these proteins have been identified by computational approaches. Aquatic organisms generally have larger selenoproteomes than terrestrial organisms, and that mammalian selenoproteomes show a trend toward reduced use of selenoproteins. One process contributing to the reduction of selenoproteome is the conversion of Sec to Cys. This process is specific to selenoproteins and can be accomplished by a single point mutation can transform a Sec UGA into a Cys codon. However, it has to be noted that Sec and Cys are not functionally equivalent, and Cys conversions are not neutral, although the reasons are still unclear [10].
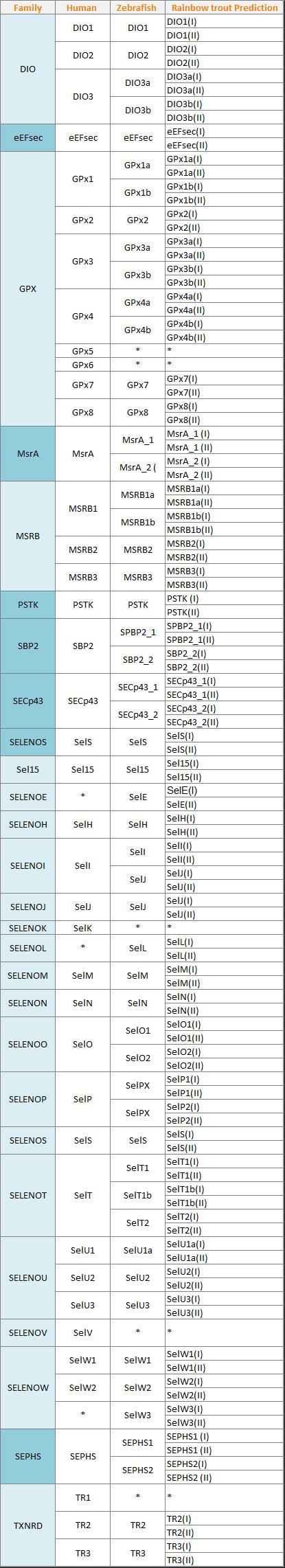
Twenty-one selenoproteins have been found in all vertebrates being: GPx1-4, TR1, TR3, Dio1, Dio2, Dio3, SelH, SelI, SelK, SelM, SelN, SelO, SelP, MsrB, SelS, SelT1, SelW1, Sep15. The selenoproteome of the rat (Rattus rattus ) and the mouse ( Mus musculus ) comprises 24 selenoproteins while the human proteome comprises 25. Particularly selenoproteins Fep15, SelI, SelJ, SelN, and SelP are only found in vertebrates while SelV is uniquely found in placental mammals [7,10].
The creation of a new selenoprotein (here always by duplication of an existing one) is indicated by its name in green.
Loss is indicated in grey. Replacement of Sec with Cys is indicated in blue.Mariotti M, Ridge PG, Zhang Y, Lobanov AV, Pringle TH, et al. (2012)
Composition and Evolution of the Vertebrate and Mammalian Selenoproteomes. PLoS ONE 7(3): e33066.
Selenoprotein families
Although some of the selenoproteins remain undocumented, current discovered selenoproteins are classified in different families, depending on their function and structure. The best characterized families of selenoproteins are: glutathione peroxidases (GPxs), iodothyronine deiodinases (DIOs) and thioredoxin reductases (TXNRDs).
Glutathione peroxidases (GPx)
GPx proteins are found in the three domains of life, being the largest family in vertebrates.The members of this family are involved in detoxification of hydroperoxides, in the maintenance of cellular redox homeostasis and in the hydrogen peroxide signaling [7]. In humans eight GPx have been identified, whereas in Danio Rerio nine have been predicted.
GPx1 is the most abundant selenoprotein in mammals. GPx1 is considererd (together with catalases and peroxiredoxins) one of the major antioxidant enzymes in cells. This cytosolic enzyme catalyzes glutathione-dependent reduction of hydrogen peroxide to water.
GPx2, which has been reported to play a role in tumour development, is found in the epithelium of the gastrointestinal tract whereas GPx3 is secreted basically from kidney, being the most abundant GPx in plasma. GPx4 is implicated in the regulation of tyrosine phosphatases and the reduction of the complex lipid hydroperoxides bound to cellular membranes [7, 11, 12].
Thioredoxin reductases (TRs)
This family participates in the maintenance of the redox state of thioredoxins proteins (Trx), which are also involved in the redox metabolism. TRs work together with Trx proteins, comprising the major disulfide reduction system of the cell.
TR1 is primarily localized in the cytosol and nucleus while TR3 is localized in the mitochondria, where it is involved in reduction of mitochondrial thioredoxin (Trx2) and glutaredoxin 2 (Grx2).
TR1 and TR3 are both present in all vertebrates with the exception of Danio rerio, in which TR1 hasn’t been found [6, 7].
Iodothyronine deiodinases (DIO)
In mammals this family is composed by three paralogous proteins: DOI1, DIO2, and DIO3. All of them are involved in deodination of the thyroid hormone which regulates its activity [3, 7]. These proteins present different subcellular locations:
- DIO1 and DIO3 are present in the cell membrane
- DIO2 is found in the endoplasmic reticulum
DIO1 and DIO2 catalyze the deiodination of T4 (thyroxine) into the active hormone T3 (triiodothyronine), whereas DIO3 converts T4 into reverse T3 (rT3) and also T3 into 3,3-diiodothyronine (T2), both being inactive forms of the thyroid hormone [10].
Homologs of these proteins have been found not only in vertebrates but also in simple eukaryotes and bacteria. In Danio rerio four have been found: DIO1, DIO2, DIO3a and DIO3b. Particularly, DIO3b is a protein found in all bony fishes as a product of their whole-genome duplication and its main function is to irreversibly inactivate the thyroid hormone.
Methionine-R-Sulfoxide Reductase (MSRB) and MsrA
The MsrB family is comprised by three different enzymes: MsrB1, MsrB2 and MsrB3. MsrB1 is a Sec-containing protein, while MsrB2 and MsrB3 are Cys-containing homologs, which maintain their catalytic efficiency.
- MsrB1 is present in the cytosol and nucleus
- MsrB2 is present in the mytochondria
- MsrB3is present in the endoplasmic reticullum
MsrB1 is a zinc-containing enzyme that was initially identified using bioinformatics tools. This protein, present in all vertebrates, functions as a Methionine-R-sulfoxide reductase, which allows the repair of oxidized methionine residues in proteins.
MsrA catalyzes the reduction of methionine residues independently if they are free or present in a protein chain. This is one of the main differences between MsrA and MsrB. As an exception, in some organisms such as bony fishes, an additional MsrB has been discovered, MsB1b which reduces free methionine-R-sulfoxide residues [7, 14].
The MsrA and MsrB proteins form the methionine sulfoxide reductase (Msr), a complex that catalyses methionine sulfoxide to methionine. This complex is involved in antioxidant defense, protein regulation and prevention of ageing-associated diseases [7].
Selenophosphate Synthetase 2
This proteins catalyzes the synthesis of the active Se donor selenophosphate, which is necessary for the selenocysteine biosynthesis. SPS2 is a Sec-containing protein in all vertebrates, whereas in low eukaryotes the Sec has been substituted by a Cys [7, 15].
Selenoprotein E (SELENOE)
Also known as Fep15. This protein is related to the other members of the selenoprotein family of 15 kDa (e.g. Sep15). It is an selenoprotein of unknown function found only in the ER of fish [16].
Selenoprotein H (SELENOH)
Selenoprotein H is localized in nucleus and has glutathione peroxidase activity. This protein is highly expressed in embryonic tissues whereas its expression is heavily reduced in adult tissues. Moreover, this protein binds to sequences containing stress and heat shock response elements [7].
Selenoprotein I (SelI)
Selenoprotein I is only found in vertebrates. It contains seven transmembrane domains and has three conserved aspartic residues within a particular motif that are required for its activity. Despite that, the physiological function of this protein remains unknown [7, 10].
Selenoprotein J (SELENOJ)
Selenoprotein J is only present in actynopterygian fishes and sea urchins, presenting some Cys homologues in cnidarians. The main feature of this protein is, in contrast with the rest of selenoproteins, to serve as a structural protein. This proteins is stated to have a role as a crystallin by comparisons with other proteins (e.g. jellyfish J1-crystallins).
In Danio rerio SELENOJ is preferentially expressed in the eye lens during early stages of development [7].
Selenoproteins K and S
Although Selenoprotein K (SelK) and Selenoprotein S (SelS) do not show any significant sequence similarity or identity, both are grouped in the same family because of their similar topology. Both are located in the ER and are implicated in ER-associated degradation (ERAD) of misfolded proteins.
The SelK/SelS-like protein family is the most widespread eukaryotic selenoprotein family, whose members are present in nearly all known Se-utilizing organisms ranging from humans to unicellular eukaryotics [7].
Selenoprotein L (SELENOL)
Particularly, Selenoprotein L contains two Sec residues and is present only among aquatic eukaryotes such as fish, invertebrates and marine bacteria [17].
15-kDa selenoprotein (Sep15) and selenoprotein M (SelM)
The 15-kDa selenoprotein (Sep15) and the selenoprotein M (SelM) are thioredoxin-like proteins found in the ER. Sep15 regulates redox homeostasis in the ER and mediates the cancer preventive effect of dietary Selenium. SelM and Sep15 share 31% identity, demonstrating their distant homology and similar distribution [7, 18].
Selenoprotein N (SELENON)
Selenoprotein N is an ER-resident transmembrane glycoprotein which is highly expressed in the embryonic development, being necessary for muscle development, differentiation and maintenance of satellite muscle cells. It is also expressed in a variety of adult tissues with an unclear function, probably related to regeneration after stress or injury [7].
Selenoprotein O
Selenoprotein O contains a Sec residue located in C-terminal end. Homologs of human Selenoprotein O have been detected in a wide variety of species, even though, the majority of homologs contain a Cys residue in place of a Sec. The function of selenoprotein O and of its homologues is yet unknown [7, 10].
Selenoproteins W, T, H, and V
Selenoprotein W (SelW), T (SelT), H (SelH), and V (SelV) belong to the Rdx family of selenoproteins and are characterized by the presence of a conserved Cys-x-x-Sec motif. All of them are considered thiol-based oxireductases, even though their exact function remains unclear [7, 19].
Selenoprotein machinery
The UGA codon requires specific mechanisms to induce the loading of a Sec residue into the protein sequence instead of stopping the polypeptide translation. For this purpose, it is necessary the recognition of the UGA codon as a codificating codon and then, the loading of Sec by a tRNA carrying Sec (tRNA-Sec).
tRNA-Sec synthesis in eukarya
Firstly, the tRNA-Sec is loaded with a serine (Ser) by a seryl-tRNA synthetase (SerRS), followed by its phosphorylation by the enzyme O-phosphoseryl-tRNA-Sec kinase (PSTK), providing the phosphorylated intermediate PSer-tRNA-Sec [7, 20].
Then, the conversion of Ser-tRNASec into Sec-tRNA-Sec is catalyzed by the SecS synthase (SecS), consuming selenophosphate and ATP [15].
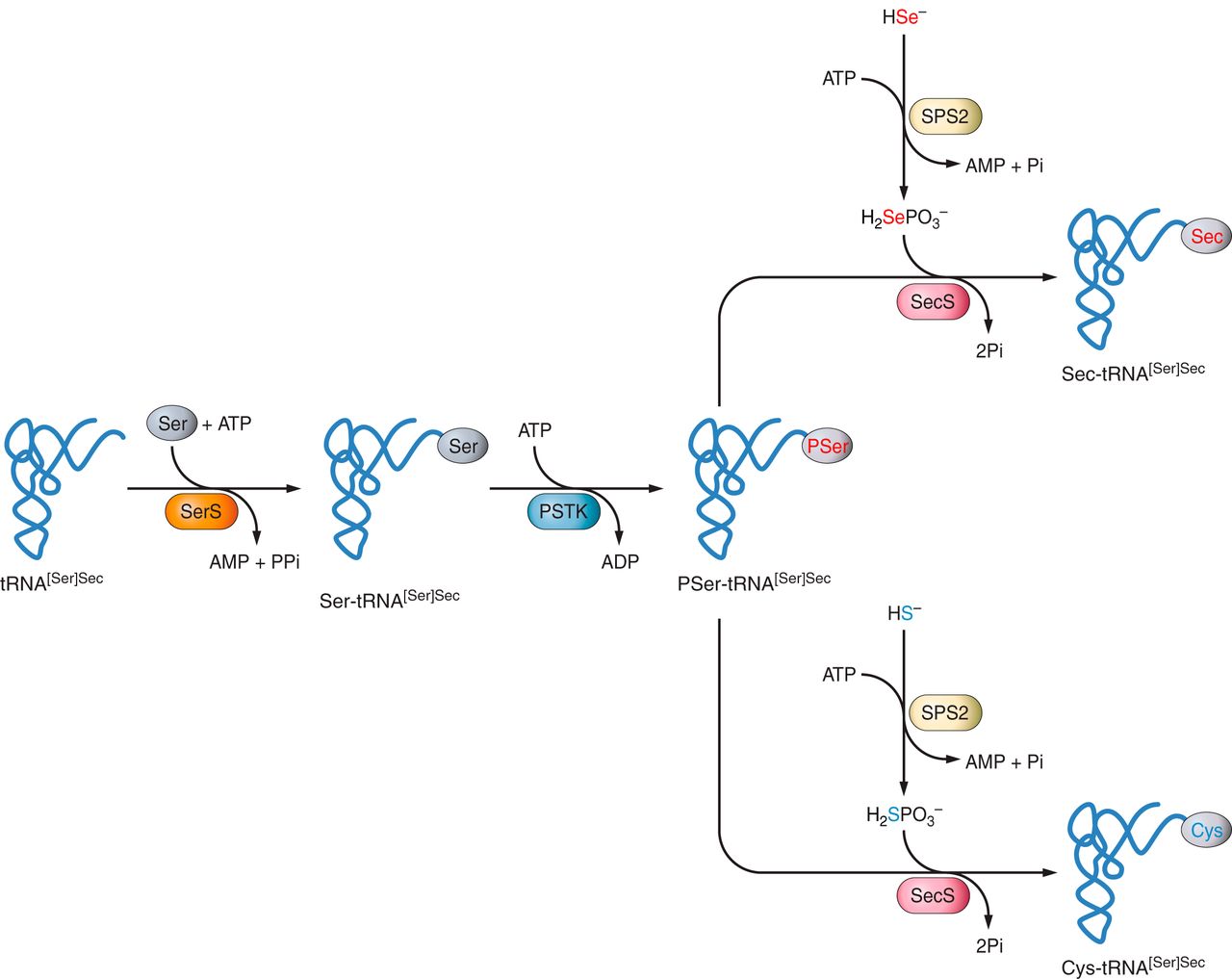
Loading of Sec by tRNA-Sec
The incorporation of Sec from a tRNA-Sec into the protein sequence requires a SECIS (SElenoCysteine Insertion Sequence). This element consist in a 60-nucleotide sequence found in the protein mRNA that adopts a stem-loop secondary structure, acting as a cis-element that recognize trans-acting factors, directing them to the ribosomes [6, 21]. SECIS forms complexes with two different elements: the Sec specific elongation factor (eEF-Sec) and the SECIS binding protein 2 (SBP2).
The protein SBP2 is constitutively associated with ribosomes and binds SECIS elements through a RNA-binding domain. Additionally, SBP2 interacts with eEFSec, which recruits the Sec-tRNASec and induces the incorporation of Sec into the new nascent protein [22].
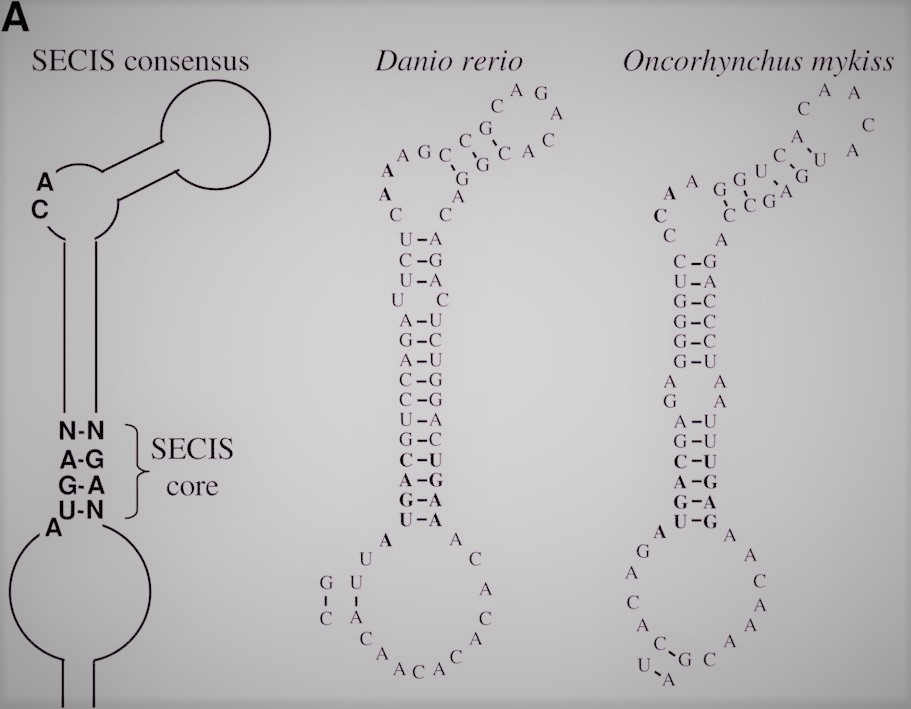
Therefore, the prediction of SECIS elements becomes an important tool in the annotation of selenoproteomes. In bioinformatics, different algorithms has been described in order to predict these structures within a genome. However, it is worth pointing out that SECIS location depends on the domain of life we refer to. In case of eukarya and archaea it is located in 3’ UTR. In bacteria, SECIS normally occur relatively near the UGA codon, immediately downstream [6].
The importance of this specific machinery can be observed in specific human diseases. One example is the mutation of SBP2, which is characterized by the abnormal thyroid hormone metabolism.
Oncorhynchus mykiss
Oncorhynchus mykiss, commonly known as rainbow trout is a salmonid native to the cold-water affluents of the Pacific Ocean in Asia and North America. There are two forms: freshwater resident and anadromous. The commonly called Steelheadis the anadromous form of the coastal rainbow trout (O. Irideus) or the red trout (O. M. Gairdneri) that returns to freshwater after living two or three years in the ocean [23, 24].
| Kingdom: | Animalia |
| Phylum: | Chordata |
| Class: | Actinopterygii |
| Order: | Salmoniformes |
| Family: | Salmonidae |
| Genus: | Oncorhynchus |
| Specie: | Oncorhyncus mykiss |
Origin and name
The species was originally named by German naturalist Johann Julius Walbaum in 1792 based on specimens found in the Kamchatka Peninsula of Siberia.
The species name, mykiss, was derived from the local Kamchatkan name used for the fish, mykizha. The name of the genus is from the Greek onkos ("hook") and rynchos("nose"), in reference to the hooked jaws of males in the mating season [23].
Description
Adult freshwater stream rainbow trouts weight between 0.5 and 2.3 kg, while lake-living and anadromous forms may reach up to 9 kg. Adult fish are distinguished by a red stripe along the lateral line, which is most vivid in males.The maximum recorded length is 120 cm, although the normal length is 60 cm.
They are torpedo-shaped and generally blue-green or yellow-green in color with a pink streak along their sides, white underbelly, and small black spots on their back and fins.
The anadromous forms usually live for about 11 years, while the lake-dwelling and freshwater forms usually live a maximum of 6 years [23,24,25].
Habitat
Adults inhabit in cold headwaters, estuaries, small to large rivers, and lakes. Anadromous forms live in coastal streams in depths from 0 to 200 meters.
The native habitat of Oncorhynchus mykiss is in the coastal waters and tributary streams of the Pacific basin. The range of coastal rainbow trout (O. m. irideus) extends north from the Pacific basin into tributaries of the Bering Sea, while forms of the redband trout (O. m. gairdneri) extend east into the upper Mackenzie River and Peace River which eventually drain into the Arctic Ocean.
Nowadays, Oncorhynchus mykiss has been introduced for food or sport in every continent except Antarctica. Particularly, it was introduced in the Catalan Countries at the beginning of the 20th century [26].
Alimentation

The rainbow trout is a predator with a varied diet. They feed on aquatic insects, fish eggs and adult forms of terrestrial insects (crickets, ants, grasshoppers and beetles) that fall into the water. Other prey include: small fish, squid, shrimp, and other crustaceans. They can even eat decomposing flesh of other fish. Some lake-dwelling forms may eat plankton.
Importance for humans
Oncorhynchus mykiss is a specie with large commercial importance and highly appreciated in gastronomy. It is also widely used in sport fishing, because of that, the species has been introduced in many water courses for this purpose. In fact, it is considered an invasive species that is creating an ecological problem due to its introduction. Furthermore, it is included in the list of the top 100 globally invasive species. Furthermore, in 2003 it was included in the Spanish Catalog of Invasive Alien Species, being prohibited its introduction into the spanish natural environment both in the national territory and in the marine jurisdictional zones [27, 28].
Phylogeny
The ancestral genome of all teleost fish underwent a whole-genome duplication, termed Ts3R and dated 225 to 333 million years ago. This duplication was preceded by two older whole-genome duplications common to all bony vertebrates. Commonly after this kind of event, the resulting genomes eventually retain only a small proportion of the duplicated genes by a process termed gene fractionation.
Salmonids such as Oncorhynchus mykiss are of particular interest to study these events because they underwent an additional and recent whole-genome duplication termed Ss4R and dated 25 to 100 million years ago.
As reported by Berthelot C et al. (2014), even after 100 million years of evolution the two ancestral genomes of Oncorhynchus mykiss have retained half of the protein-coding genes as duplicated copies. Genes have been lost mostly by pseudogenization. Surprisingly, almost all miRNA been retained as duplicated copies. Interestingly, those genes retained as duplicated copies after the successive whole-genome duplication that occurred during vertebrate evolution were also more likely to be retained as duplicates following the Ss4R [29].
The red stars show the position of the teleost-specific (Ts3R) and the salmonid-specific (Ss4R) whole-genome duplications.
Groups of species in which a genome sequence is available are shown in red.
Berthelot C, Brunet F, Chalopin D, Juanchich A, Bernard M, Noël B et al.
The rainbow trout genome provides novel insights into evolution after whole-genome duplication in vertebrates.
Nat Commun 2014;5:3657.
For more information visit the wikipedia page in Catalan or in English.
Purpose and expected results
The purpose of this study is to predict the entire selenoproteome of Oncorhynchus mykiss, including those proteins related to it synthesis and incorporation of selenocystein. In order to achieve this, we have compared Oncorhynchus mykiss's genome with Danio rerio (see Methodology).
As said before, marine organisms normally present larger selenoproteomes than terrestrial ones. As commented through the introduction, bonny fishes present duplicated segments as a result of a whole-genome duplication event termed Ts3R. This fact can be easily observed comparing the 24 selenoporteins of Homo sapiens against the 38 present in Danio reiro.
Moreover, it is worth pointing out that salmonids such as Oncorhynchus mykiss underwent a recent whole-genome duplication event (termed Ssr4). Normally after this kind of events a big part of the duplicated genes disappear because of a process called gene fractionation, but, particularly, Oncorhynchus mykiss has been reported to retain half of the protein-coding genes as duplicated copies.
For this reason, we expect to find some duplicated selenoproteins in our results. We expect that the final number of selenoproteins predicted will be proximal to the double of the number present in Danio rerio. In this case, as we selected 53 different proteins form Danio rerio we expect to obtain approximately 106 different predictions in Oncorhynchus mykiss.
In the table below (Figure 8) we present a summary of our expected results.