|
INTRODUCTION
What are selenoproteins?
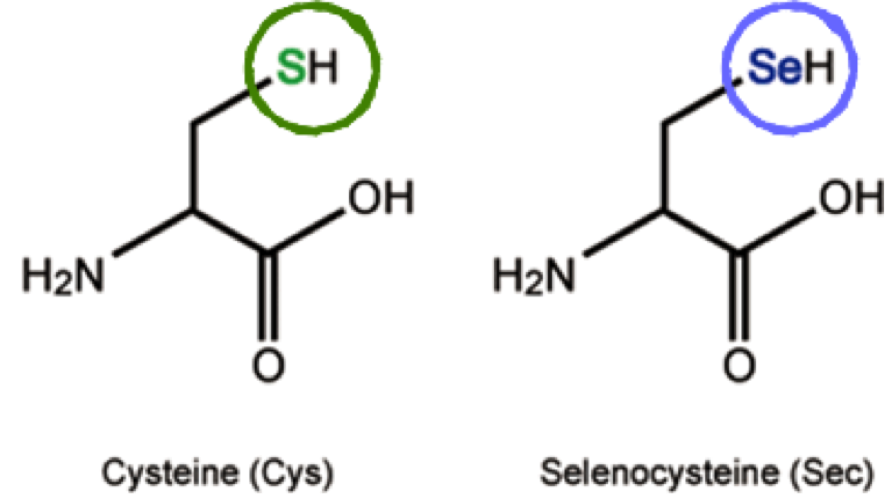
Selenium (Se) is an essential micronutrient which plays its main biological function by forming selenoproteins [1]. Se is present in selenocysteine (Sec; U), known as the 21st amino acid (aa), which selenoproteins incorporate in their amino acidic sequence. It is very similar to cysteine (Cys), but it has a Se atom instead of the sulfur (S) atom present in Cys [2]. Differently from other aa, Sec is coded by UGA codon, which typically acts as a stop codon.
Selenoprotein synthesis
» Enzymes involved in selenoprotein synthesis
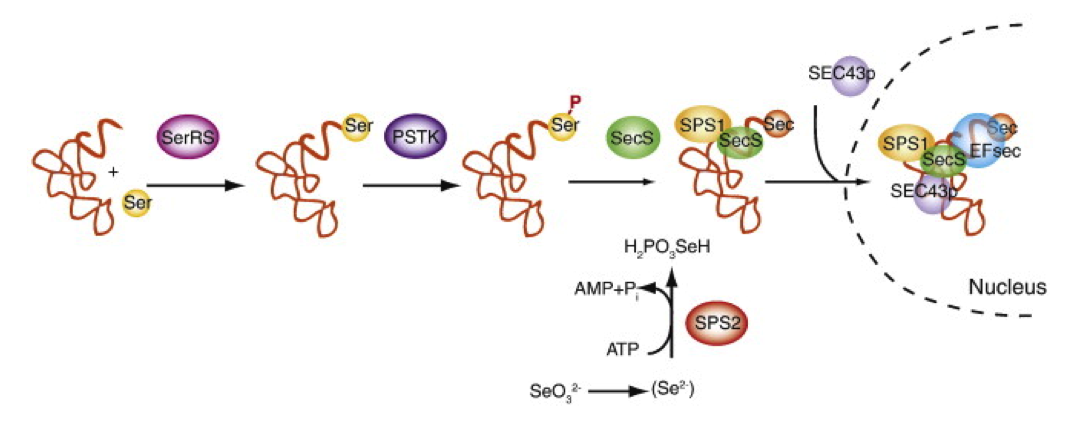
Sec has to be loaded into tRNASec in order to be incorporated into proteins. In mammals, this process starts with a serine (Ser) being charged to a tRNASec by seryl-tRNA synthetase, followed by Ser phosphorylation by O-phosphoseryl-tRNASec kinase (PSTK). Then, SecS (Sec synthase) uses monoselenophosphate in order to convert Ser-tRNASec into Sec-tRNASec. Once Sec-tRNASec is formed, it recognises the UGA codon and Sec is incorporated in that position, creating a selenoprotein.
SPS1 and SPS2 (selenophosphate synthetases) are two other proteins known to catalyse the formation of monoselenophosphate from selenide, but only SPS2 is essential for selenoprotein synthesis. Moreover, in some organisms such as humans or mice, SPS2 is a selenoprotein by itself [3].
There are also other factors involved in selenoprotein biosynthesis, such as SECp43, which interacts with different proteins involved in this pathway and helps redistributing SecS/Sec-tRNASec complexes to the nucleus [4].
 Allmang et al. (2009)
» SECIS elements
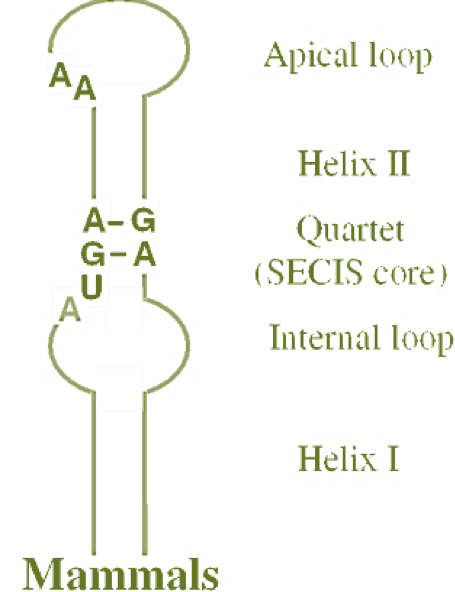 The incorporation of Sec also requires a specific element, the SECIS (SElenoCysteine Insertion Sequence) element, in order to recode the UGA codon [5]. This element has to be located in the 3'UTR of the mRNA and has a helix-loop structure with a binding site for SBP2 (SECIS-binding protein 2). Selenoprotein biosynthesis requires the binding of SBP2 to the SECIS element and the recruitment of the specific Sec-tRNASec by the elongation factor eEFsec (eukaryotic Sec tRNA-specific elongation factor) [6][7] The incorporation of Sec also requires a specific element, the SECIS (SElenoCysteine Insertion Sequence) element, in order to recode the UGA codon [5]. This element has to be located in the 3'UTR of the mRNA and has a helix-loop structure with a binding site for SBP2 (SECIS-binding protein 2). Selenoprotein biosynthesis requires the binding of SBP2 to the SECIS element and the recruitment of the specific Sec-tRNASec by the elongation factor eEFsec (eukaryotic Sec tRNA-specific elongation factor) [6][7]
Therefore, in order to correctly describe the selenoproteome of Mus spretus, we need to identify not only its selenoproteins but also those proteins needed for their synthesis, including the ones involved in Sec-tRNASec formation and the ones involved in SECIS recognition.
Evolution of selenoproteins
Selenoproteins have a scattered distribution but are present in the three domains of life: bacteria, archaea and eukaryotes. However, in each lineage, they perform different functions. In bacteria, selenoproteins carry out redox homeostasis and compound detoxification, among others. In contrast, archaeal selenoproteins are involved in hydrogenotrophic methanogenesis, whereas eukaryotic selenoproteins are involved in redox regulation and energy metabolism [8][9].
Selenoproteins can be absent in a particular taxonomic group but, at the same time, present in a sister lineage. Some organisms such as Droshophila willistoni, plants or fungi [10] have lost their capacity to synthesize selenoproteins. However, vertebrates seem to have a strong purifying selection across Sec sites. Over the past years, several analyses have been performed to provide a better insight in selenoproteome description. It has been shown that selenoprotein distribution has a wide variation among groups. In mammals, the selenoproteome is not as large as the one in other species, such as bony fishes. This difference may be due to the change mammals experienced to a terrestrial environment during evolution, when they could have reduced Sec function. A total of 21 selenoproteins have been found in all vertebrates: GPx1-4, TR1, TR3, DIO1, DIO2, DIO3, SelH, SelI, SelK, SelM, SelN, SelO, SelP, MsrB1 (methionine-R-sulfoxide reductase 1), SelS, SelT1, SelW1 and Sep15 [8][11]. Apart from this set, other selenoproteins have been found in some lineages, suggesting that duplication events may have occurred. Particularly in mammals, 25 selenoproteins are known to exist, but the quantity varies among species.
Because the similarity between Cys and Sec, it is common to find selenoprotein orthologous genes with a Cys instead of a Sec [11].

Selenoprotein families
Selenoproteins with similar functions can be grouped together into different families. The most important and best characterized families of selenoproteins are glutathione peroxidases (GPxs), iodothyronine deiodinases (DIOs) and thioredoxin reductases (TXNRDs).
» Glutathione Peroxidases (GPxs)
Glutathione peroxidases are involved in antioxidant processes. Its main role is to protect cells from oxidative damage through elimination of hydrogen peroxide (H2O2), which reacts with glutathione to produce glutathione disulfide and water. [12]. In humans, 8 GPx have been identified, whereas in Mus musculus only 6 GPxs isoforms have been predicted.
» Iodothyronine Deiodinases (DIOs)
This family is involved in the activation and inactivation of thyroid hormones. In mammals, 3 DIOs have been described: DIO1, DIO2 and DIO3, all of them being selenoproteins. DIO1 and DIO2 catalyze the deiodination of T4 (thyroxine) into the active hormone T3 (triiodothyronine), whereas DIO3 converts T4 into reverse T3 (rT3), which is its inactive form, and also T3 into 3,3-diiodothyronine (T2), which is inert. DIO1 and DIO2 can also convert rT3 into T2 [13].
DIO1 is found in the liver, kidney and thyroid, while DIO2 is located in the brain, pituitary, thyroid, skeletal muscle and brown adipose tissue. Regarding DIO3 location, it is found in the cerebral cortex and skin and is expressed at a very high level in the placenta and the uterus of pregnant women. Looking into the subcellular level, DIO1 and DIO3 are found at the plasma membrane, whereas DIO2 is localized in the ER membrane [14][15].
» Thioredoxin Reductases (TXNRDs)
These selenoproteins are members of the pyridine nucleotide-disulfide oxidoreductase family. In mammals, 3 TXNRDs have been identified: TXNRD1, TXNRD2 and TXNRD3, also known as thioredoxin-glutathione reductase. These are located in cytosol or nucleus, mitochondria and testis, respectively [16]. TXNRD1 and TXNRD2 are essential during development. TXNRD1 is the most abundant selenoprotein in mouse macrophages and it maintains the redox tone in immune cells, therefore it is particularly important [17].
This family is able to reduce oxidized thioredoxin which, in turn, provides electrons to ribonucleotide reductase, which is essential to transform ribonucleotide to deoxyribonucleotide in order to synthesize DNA. These proteins are also involved in other signalling pathways regulating the activity of transcription factors [14].
» Other selenoprotein families
Apart from the ones described above, other selenoprotein families have been identified. For more information about the role of theses proteins, see Discussion):
- Methionine sulfoxide reductases (Mrs)
- 15 kDa selenoprotein (Sel15)
- Selenoprotein H (SELENOH)
- Selenoprotein I (SELENOI)
- Selenoprotein K (SELENOK)
- Selenoprotein M (SELENOM)
- Selenoprotein N (SELENON)
- Selenoprotein O (SELENOO)
- Selenoprotein P (SELENOP)
- Selenoprotein S (SELENOS)
- Selenoprotein T (SELENOT)
- Selenoprotein U (SELENOU)
- Selenoprotein V (SELENOV)
- Selenoprotein W (SELENOW)
» Proteins involved in selenoprotein biosynthesis
As explained before, there are also some proteins from the selenoprotein synthesis machinery that we need to include in order to correctly describe the selenoproteome. Depending on their function, these can be separated into 2 categories:
- Synthesis of Sec (on tRNASec):
- Selenophosphate synthetase (SPS1 or SPS2): only SPS2, which is also a selenoprotein, is essential
- Sec synthase (SecS)
- O-phosphoseryl-tRNASec kinase (PSTK)
- Incorporation of Sec into selenoproteins:
- SECIS binding protein 2 (SBP2)
- Eukaryotic Sec-specific elongation factor (eEFsec)
There are also other factors, such as SECp43, which are important in this process.
» Other classifications
Another way of classifying selenoproteins is according to the location of Sec in their sequence [14]. Following this, we can group the different families named above in 2 categories:
- Sec located closely to the C-terminus end: TXNRDs, SelS, SelR, SelO, SelI and SelK.
- Sec at the N-terminus end: GPxs, DIOs, SelH, SelM, SelN, SelT, SelV, SelW, SPS2 and Sep15.
Mus spretus
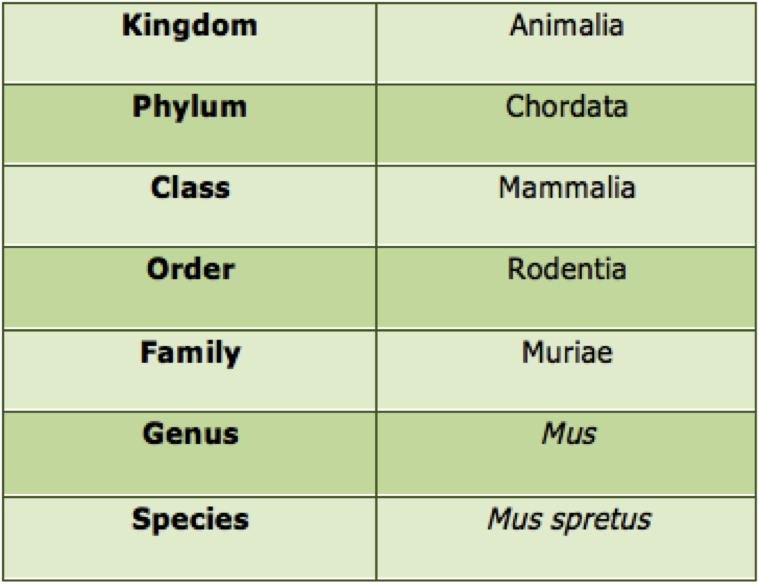
» Taxonomy
» Characteristics
 Mus spretus is a wild specie of mouse that inhabits the western Mediterranean. It is also known as the Algerian mouse or the western Mediterranean mouse.
Mus spretus is a wild specie of mouse that inhabits the western Mediterranean. It is also known as the Algerian mouse or the western Mediterranean mouse.
The main characteristics are a short tail and a brown fur that covers most part of its body except from some white underparts.
Its predators are owls, mammalian carnivores and snakes.
» Habitat
Mus spretus is found in southern France, practically entire Iberian Peninsula including the whole continental Portugal. In Spain its distribution correlates with the Mediterranean climate. Also it is found in the Balearic Islands of Mallorca, Menorca and Ibiza but it is absent in the Canary Islands.
Moreover, it is distributed in North Africa from Morocco to Algeria and Tunisia.
[18].
» Alimentation
It is a nocturnal and opportunistic omnivore animal, feeding himself with seeds, fruits or insects, mainly in the form of larvae.
» Phylogeny
Here, we can see Mus spretus phylogeny [19]
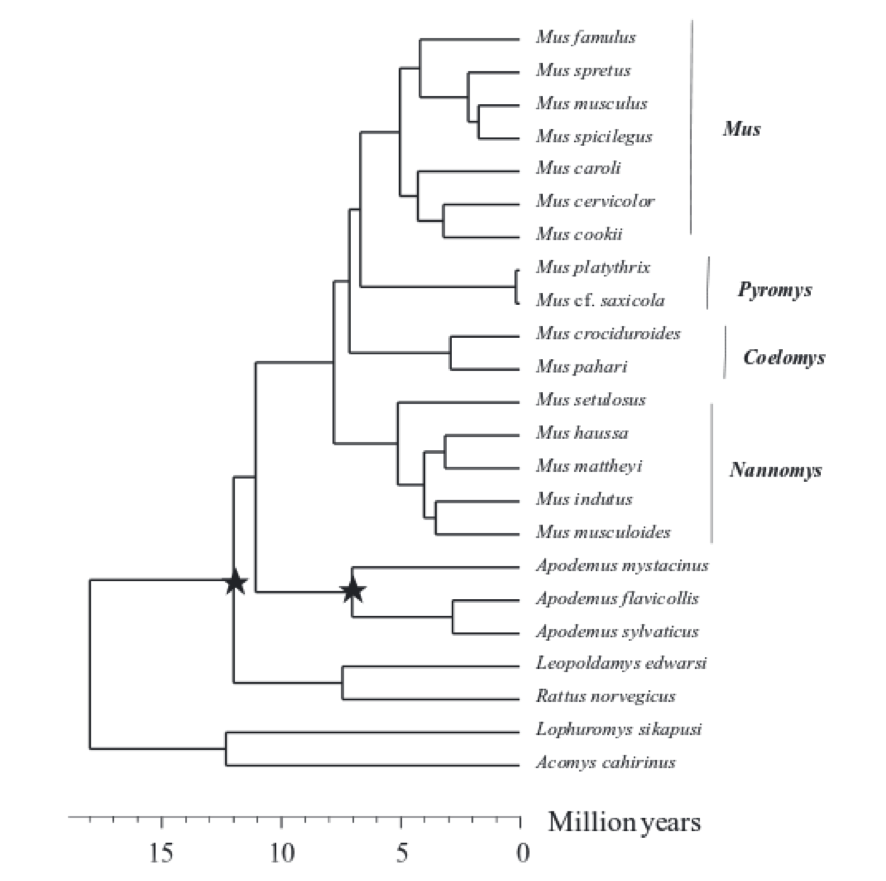 Chevret P, et al. (2005)
More information can be found at:


| |
