|
CONCLUSIONS
Selenium is an important micronutrient which is incorporated into proteins through Sec, the 21st amino acid. Proteins with a Sec residue in its structure are called selenoproteins and are known to have important biological functions, most of them involving oxidative damage protection. However, the exact function of many selenoproteins is not known.
Selenoproteomes are not well described among the different species. Sec is coded by UGA, which is classically recognised as a stop codon. For this reason, many bioinformatic programs fail in recognising selenoproteins.
The aim of this project was to identify selenoproteins in Mus spretus genome. To develop this task we made a comparison with two model organisms: Mus musculus and Homo sapiens, since in close species these proteins are habitually well-conserved.
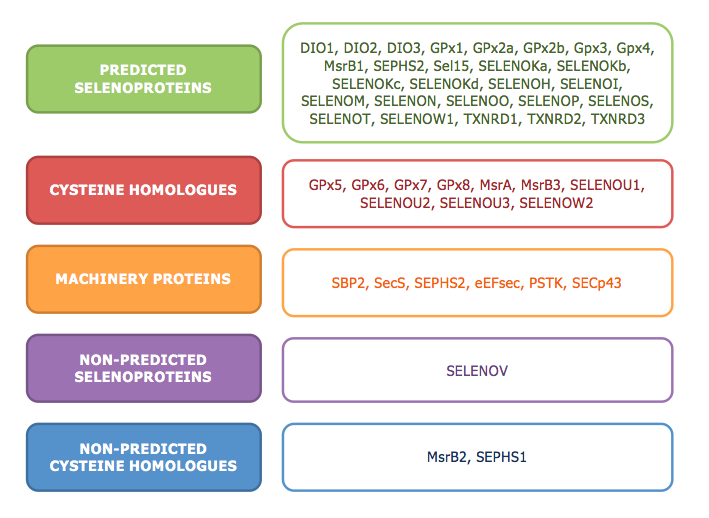
In this study, we could predict a total of 26 selenoproteins, 10 cysteine homologues of existing selenoproteins and 6 machinery proteins involved in the biosynthesis of selenoproteins. However, there are few proteins that we were not able to predict, which are mostly Cys homologues, and another which is a selenoprotein.
The scheme below pretends to illustrate in a visual manner the results obtained:
The main limitation we faced during this study was the poor annotation of Mus musculus selenoproteins in SelenoDB 2.0, as in some cases Met was not the first amino acid of the protein. For this reason we had to compare the genome of Mus spretus with the human genome, better annotated but phylogenetically further distant. Then we found a different protein than the mouse-predicted.
Other limitations were related to the possibility of sequencing problems in Mus spretus genome, since in some cases there were multiple N instead of the corresponding nucleotide (A, C, T or G), which did not allow us to properly predict some of the proteins. Furthermore, in some cases we found insertions/deletions in the nucleotide sequence that caused frameshifts in the predictions when compared to the model organisms annotated selenoproteins. These might also be sequencing problems. After finding these errors, we also thought that Mus musculus genome might not be perfectly accurate.

| |
