
Selenoproteins of Mus spretus |
DISCUSSION
This section contains the reasoning by which we predicted the Mus spretus selenoproteome and associated machinery. Note that most proteins were predicted with an homology-based approach using the mouse (Mus musculus) selenoproteins described in SelenoDB2.0 and hereafter referred with their ID in the database (see Methods). However, any protein not starting with methionine (Met), finishing without a stop codon or having any other particular feature that would make us be concerned about the mouse SelenoDB 2.O annotation was analyzed in parallel using the human homologs as query proteins. It also includes a brief introduction about the role of each of these proteins, grouped by protein families. The color of the protein title indicates if it is a selenoprotein (green), a cysteine homologue of existing selenoproteins (red) or a protein involved in the biosynthesis of selenoproteins (orange). Here are the links to the identified proteins: GPX, DIO, TXNRD, Msr, Sel15, SelenoH, SelenoK, SelenoI, SelenoM, SelenoN, SelenoO, SelenoP, SelenoS, SelenoT, SelenoU, SelenoV, SelenoW, SECp43, SEPHS, SEPHS2, SecS, PSTK, SBP2, eEFsec. GPxGlutathione peroxidases catalyse the reduction of H2O2using monomeric glutathione as a cofactor [12]. Since H2O2 causes protein oxidation and is toxic to cells, GPxs are important in dealing with oxidative damage.According to SelenoDB 2.0, the mouse genome has 6 selenoproteins and 4 cysteine homologues in this family. GPx1, GPx2 are GPx3 selenoproteins, GPx5, GPx6, GPx7 are homologues, and there are also 4 unclassified GPx proteins (3 selenoproteins and 1 Cys homologue). Due to the high homology between them, almost all of them present predictions against each other, which results in a complex analysis. We will discuss their Mus spretus relatives individually, making the relevant associations. On the other hand, there are 8 subfamilies of human GPx proteins annotated (GPx1-GPx4, selenoproteins, and GPx5-GPx8, Cys homologues), so this difference with the mouse annotation raised our concern and made us run the analysis for all human proteins to make the relevant associations. First, we present a discussion about mouse proteins that could not be found in Mus spretus: The mouse protein SPP00001552_2.0 does not start with Met, so we compared it to different human GPx proteins' predictions. We found that GPx4 has nucleotide homology to the mouse genomic region in which this protein is contained. However, the predicted Mus spretus protein does not start with Met and lacks an N-terminal part relative to the human protein. We cannot be sure if this protein is part of the Mus spretus protein, so we discarded it from our prediction. Below, we show the proteins predicted in the Mus spretus genome: GPx1 In this case, we got a good prediction when the mouse query was used, and the human prediction was similar but with less homology, so we chose the mouse one. Exonerate, Seblastian and Genewise found the same gene and a SECIS element was present in the 3'UTR. We conclude that this protein is found in the Mus spretus genome in contig CM004102.1 between 109207309-109208129 in the forward strand. This is the detailed exon prediction: GPx2a We found one Mus spretus homolog with the human and mouse search (completely overlapping), which we defined as GPx2A. Exonerate and Seblastian found the same protein and a SECIS could be identified in the 3'UTR. We conclude that it can be found in the Mus spretus genome in contig CM004106.1 between 70218431-70221164 in the reverse strand. This is the detailed gene map: GPx2b In addition, another sequence came out with high homology from both queries (human and mouse), exactly in the same position, which suggests a duplication. However, this protein has an insertion/deletion (in/del) mutation with respect to the human and mouse protein, something that raises three hypothesis about this prediction:
GPx3 All human GPx3 isoforms and the mouse protein give rise to gene predictions in a region of the Mus spretus contig CM004105.1. The mouse protein does not start with Met; however, a Seblastian run with the genomic region of Mus spretus results in a protein starting with Met, very different from the mouse annotated protein. Then, due to these differences between predictions, we decided to run the prediction based on human isoforms. We found that the human isoform SPP00000021_2.0 gives a selenoprotein prediction, with both Exonerate and Genewise, that is equivalent to the one found by Seblastian. Therefore, we took the Seblastian prediction as definite. In addition, a SECIS element was found in the 3'UTR. We conclude that this protein can be found in the Mus spretus genome in contig CM004105.1 between positions 53888501-53895197 in the forward strand. Below, there is the detailed gene map: GPx4 The mouse SPP00001553_2.0 and SPP00001555_2.0 unclassified GPx proteins have high homology to the same region of contig CM004103.1 in Mus spretus (in a way that the second is included in the first). However, none of these mouse queries start with Met, so we decided to use the human GPx4 to make our prediction. The two human isoforms of GPx4 point to the same region and both start with Met. We next ran Seblastian in this region and found a protein that has a SECIS element in the 3'UTR and is very similar to the human GPx4. We therefore conclude that GPx4 can be found in Mus spretus in contig CM004103.1 between 78895105-78898745 in the forward strand. This is the detailed gene map: GPx5 In this case, we got a good prediction from the mouse query only with Exonerate, and the human prediction was similar but with a lower homology, so we chose the mouse one. We conclude that this protein is found in the Mus spretus genome in contig CM004107.1 between 17270357-17275565 in the reverse strand. This is the detailed exon prediction: GPx6 In this case, we obtained a good prediction using the mouse query with both Exonerate and Genewise softwares. Using the human GPx6 protein we obtained a similar prediction but with a lower degree of homology. However, human GPX6 is a selenoprotein, whereas the mouse predicted protein is a Cys homolog, so we chose the mouse one. We conclude that it is a Mus spretus Cys homolog which can be found in contig CM004107.1 between 17297628-17304819 in the forward strand. This is the detailed exon prediction: GPx7 The mouse GPx7 predicted protein from SelenoDB 2.0 does not start with Met, whereas the human does. This raised our concern about the mouse annotation. We found that both human and mouse predictions pointed to the same genomic region of Mus spretus, but the human prediction resulted in a protein that starts with Met. In the human analysis, Exonerate and Genewise predicted the same gene. We conclude that the Mus spretus genome contains GPx7 in contig CM004097.1 between positions 105404803-105410611 in the reverse strand. This is the detailed gene map: GPx8 The human GPx8 isoforms and the unclassified mouse protein SPP00001554_2.0 gave rise to important gene predictions in a region of the Mus spretus contig CM004107.1. The mouse protein does not start with Met and it is quite different from the human one, so we decided to take the human protein as query, as the human selenoproteome is the best annotated one. We chose the human SPP00000042_2.0 isoform because the protein predicted was in the same region than the one obtained when comparing against mouse. Regarding gene prediction, both Genewise and Exonerate using the human query found the same gene. We conclude that we can find it in the Mus spretus genome in contig CM004107.1 between position 110154109-110157254 in the reverse strand. This is the detailed gene prediction: DIOIodothyronine deionidases (DIO) are a subfamily of deionidase enzymes involved in regulating the activity of thyroid hormones. They can both activate or inactivate thyroid hormones by deionidation of the outer or inner ring, respectively. There are 3 known DIO proteins described in mammals: DIO1, DIO2 and DIO3 [11].DIO 1 Regarding the gene prediction in Mus spretus, Exonerate, Genewise and Seblastian predicted the same protein, and a SECIS element can be found in the 3'UTR. Therefore, we conclude that this protein can be found in the contig CM004097.1 between positions 104286841-104301651 in the reverse strand. This is the detailed exonic prediction: DIO 2 Regarding the gene prediction in Mus spretus, Exonerate and Genewise predicted the same protein, and a SECIS element can be found in the 3'UTR. However, Seblastian was not able to predict any protein in this case. Despite the failure of Seblastian predicting a selenoprotein, we consider that Exonerate and Genewise predictions are accurate, so we conclude that this protein can be found in the contig CM004106.1 between positions 84555859-84564721 in the reverse strand. Here, we present the detailed exonic prediction: DIO 3 Regarding the gene prediction in Mus spretus, Exonerate found an identical gene to the mouse query. However, Seblastian predicts a protein that has 26 more amino acids in the N-terminus (N-term) end and 3 more amino acids in the C-terminus end (C-term). We looked for the mouse transcript in SelenoDB 2.0 and found that the annotated mouse DIO3 does not have a stop codon where it is annotated. Moreover, the 3 additional amino acids in C-term predicted by Seblastian correspond to the first 9 3'UTR mouse bases annotated in SelenoDB 2.0. We therefore conclude that this selenoprotein is incorrectly annotated in mouse SelenoDB 2.0 and that these 9 bp correspond to a C-terminal amino acid fragment that Exonerate failed to predict. A SECIS element could be found in the 3'UTR. All in all, this selenoprotein can be found in Mus spretus genome in the contig CM004106.1 between positions 105018284-105019117 in the forward strand. Below is the detailed exonic prediction: TXNRDsThioredoxin reductase (TXNRD) proteins are a well-characterised family of selenoproteins that produce reduced thioredoxin (TXN), which also reduces other oxidized proteins. This system is one of the most important mechanisms involved in the regulation of cellular redox balance. There are three proteins in this family described in mammals: TXNRD1, TXNRD2 and TXNRD3, also called thioredoxin-glutathione reductase [16].Some of the predicted Mus spretus proteins do not start with Met, so we decided to run the human analysis. TXNRD1 Both human and mouse analysis point to the same Mus spretus genomic region, but the homology is much higher for the mouse prediction, so we chose this one. Exonerate and Genewise performed the same prediction, and a SECIS element could be found in the 3'UTR. We conclude that TXNRD1 can be found in the Mus spretus genome in contig CM004103.1 between positions 81689812-81714116 in the forward strand. This is the detailed gene prediction: TXNRD2 The mouse annotated protein does not start with Met, whereas the human isoforms do. Both of them have predicted a homolog in the same Mus spretus genomic region, but we chose the human as it is a better query for further analysis. Exonerate performed a proper prediction from the human query, and a SECIS element could be found in the 3'UTR. Moreover, the predicted protein also started with Met. We conclude that TXNRD2 can be found in the Mus spretus genome in contig CM004110.1 between positions 15356078-15410943 in the forward strand. This is the detailed gene prediction: TXNRD3 The mouse protein starts with a Pro, and so does the corresponding Mus spretus prediction. On the other hand, all three human isoforms do not start with Met (the starting amino acid is Gly or Val). We ran all these proteins (human and mouse) against the Mus spretus genome and found a region in contig CM004099.1 with many gene predictions. A Seblastian run on this region found a Met-starting selenoprotein with a SECIS in the 3'UTR, which we took as correct. We conclude that TXNRD3 can be found in the Mus spretus genome in contig CM004099.1 between positions 88803548-88833559 in the forward strand. This is the detailed gene prediction: MsrThe methionine sulfoxide reductase (Msr) system comprises MsrA and MsrB proteins and catalyses conversion of methionine sulfoxide to methionine and is involved in antioxidant defense, protein regulation and prevention of ageing-associated diseases [20]. MsrA is involved in the reduction of methionine-S-sulfoxide residues in proteins, in addition to also reducing free methionine-S-sulfoxide. MsrB is responsible for the reduction of protein-based methionine-R-sulfoxide and also, but with a lower efficiency, of free methionine-R-sulfoxide. To counteract ROS damage, organisms developed some defense mechanisms, including antioxidant enzymes such as Msrs family [21].MsrA Regarding the gene prediction in Mus spretus, Exonerate and Genewise predicted the same protein. Thus, we conclude that this protein can be found in the contig CM004108.1 between positions 54488572-54815934 in the reverse strand. This is the detailed exonic prediction: MsrB We can find 3 proteins in this family in both mouse and human. However, MsrB2 (Cys homologue) presented some particularities in Mus spretus that we discuss below. The mouse MsrB2 annotated protein does not start with Met, whereas the human does. We ran the analysis with the mouse and the human queries and found, in the same location, a Mus spretus protein that has an in/del mutation (similar to the GPx2B protein). We came up with three hypothesis to explain this:
Regarding Mus spretus prediction, we could only find the following proteins: MsrB1 Regarding gene prediction, Exonerate, Genewise and Seblastian performed a correct and matching prediction, and a SECIS element could be found in the 3'UTR. We conclude that it can be found in the Mus spretus in contig CM004111.1 between positions 21502796-21509320 in the forward strain. This is the detailed gene map: MsrB3 The mouse MsrB3 does not start with Met, whereas the four human isoforms do. We ran the analysis from both organisms and found a candidate region in Mus spretus for containing this gene. The mouse query and two of the human isoforms (SPP00000092_2.0 and SPP00000094_2.0) predicted a Ser-starting protein in Mus spretus, and two other human queries (SPP00000093_2.0 and SPP00000095_2.0) predicted a Met-starting gene. We chose the latter two human proteins as valid queries, with which Genewise predicted the same Mus spretus protein. We conclude that MsrB3 can be found in contig CM004103.1 between positions 121537637-121634122 in the reverse strand. This is the detailed gene map: Sel15Sel15 (15 kDa selenoprotein) function is related with glycoprotein folding quality control [22].Mice have one Sel15 selenoprotein, but we found 2 homolog selenoproteins in Mus spretus (hereafter called as Sel15A and Sel15B). Genewise and Exonerate softwares predicted slightly different N-term sequences for both of them, but Sec was correctly aligned and the similarity to the query was also very high in both of them. We could also find SECIS elements in both 3'UTRs. However, Seblastian was not able to predict any selenoprotein. Analyzing the Mus spretus genomic sequence, we conclude that one Sel15B had sequencing errors in the 3' end of the ORF and start of the 3'UTR (repetition of undefined nucleotides: N), which compromised the exact prediction of the ORF end. Thus, we were only able to predict one Sel15 homolog (corresponding to Sel15A). This is the predicted Mus spretus protein: Sel15 Both Exonerate and Genewise predictions of Sel15 were very good, but slightly different, since we observed that Exonerate included one more final exon quite distant from the rest of them. However, the SECIS element found in the region was located before this last exon so we decided not to include it in our prediction and therefore used the Genewise prediction. We conclude that Sel15 can be found in contig CM004096.1 between positions 144217792-144243497 in the forward stand, with the following exonic structure: SELENOHSELENOH, or SelH, is a nucleolar oxidoreductase involved in the regulation of redox homeostasis. Through genome maintenance and redox regulation, it has been shown to inhibit apoptotic pathways, suppress cellular senescence and promote mitochondrial biogenesis [23]. Moreover, it is important in organogenesis and tumorigenesis [24].Regarding the gene prediction in Mus spretus, Exonerate and Genewise predicted the same protein, and a SECIS element can be found in the 3'UTR. However, Seblastian was not able to predict any protein in this case. Despite this fact, we conclude that this protein can be found in the contig CM004095.1 between positions 84546376-84546943 in the reverse strand. Here, we present the detailed exonic prediction: SELENOISELENOI, also known as SepI or SelI, is a membrane protein with seven transmembrane domains which has not been studied in depth. Selenoprotein I helps in the formation and stabilization of protein complexes needed for protein trafficking [25][11].Regarding the gene prediction in Mus spretus, Exonerate, Genewise and Seblastian predicted the same protein, and a SECIS element can be found in the 3'UTR. Thus, we conclude that this protein can be found in the contig CM004098.1 between positions 27268100-27306511 in the forward strand. This is the detailed exonic prediction: SELENOKSeleno-K is located in the endoplasmatic reticulum (ER) and highly expressed in the heart, where it might have an antioxidant function. It is also suggested that it is involved in degradation mechanisms of glycosylated misfolded proteins [26].The human genome has 1 SELENOK protein, but there are 12 proteins of this family in mouse according to SelenoDB 2.0. Because of this, we put special concern in the predictions obtained from mouse and we also used the human SelK as a query in order to make proper predictions. Below, there are some comments about mouse-predicted proteins that we have not found in Mus spretus: Two of the mouse-predicted proteins (SPP00001568_2.0 and SPP00001573_2.0) cannot be found in the Mus spretus genome (we only got gene predictions with ~50 % homology), so we discarded them from the final prediction. SPP00001565_2.0, SPP00001574_2.0 and SPP00001575_2.0 mouse proteins (homologous to the human SELENOK, thus being putative duplications in mouse) were found in the Mus spretus genome when analyzing both human and mouse alignments. However, the Mus spretus predicted proteins have an in/del that compromises our prediction and generates a frameshift that results in a very different protein. We have three hypothesis about this results:
SPP00001566_2.0 of mouse does not start with a Met and does not end with a stop codon. It also lacks a Sec. Thus, we conclude that the annotation of this protein is incorrect and it does not exist in the Mus spretus genome. The SPP00001564_2.0 and SPP00001571_2.0 mouse proteins are within the SPP00001569_2.0 (exact homolog of the human protein), so we discarded them from our prediction and conclude that they are incorrectly annotated in mouse. On the other hand, we were able to identify 4 SELENO-K proteins in Mus spretus: SELENOKa This is the actual homolog of human SELENOK and corresponds to the protein SPP00001569_2.0 of mouse . Both mouse and human homology-based approaches predicted the same protein in terms of Genewise, Exonerate and Seblastian softwares. We can also find a SECIS element in the 3'UTR. We conclude that it can be found in the Mus spretus contig CM004108.1 between positions 22613900-22618809 in the forward strain. This is the detailed gene map predicted: SELENOKb This protein is homologous to the mouse SPP00001567_2.0 protein. Genewise and Exonerate predicted the same gene from mouse prediction and a SECIS could be found in the 3'UTR. This protein starts with a leucine (Leu) amino acid (UUG codon) in mouse and Mus spretus and is very similar to the human SELENOK but with a gap in the middle of the Mus spretus sequence. Leu has been described as an alternative initiation codon for CUG and UUG codons [27], so we conclude that this is a duplication of the human SELENOK. It can be found in Mus spretus in contig CM004097.1 between positions 132604011-132604226 in the reverse strand. This is the detailed gene map: SELENOKc This protein is homologous to the mouse SPP00001570_2.0 protein. Genewise and Exonerate predicted the same gene with the mouse prediction and a SECIS could be found in the 3'UTR. This is very similar to the human SELENOK but it has a gap in the middle of the Mus spretus sequence, as it happened in SELENOKb. Therefore, we conclude that it is a duplication of the human SELENOK. It can be found in Mus spretus in contig CM004095.1 between positions 169487792- 169487989 in the forward strand. This is the detailed gene map: SELENOKd Homolog of the mouse SPP00001572_2.0, this protein constitutes another duplication relative to the human protein. It can also be found in Mus spretus with a high homology, although no SECIS could be found. This SECIS lack could be due to the fact that the contig is very short (13677 bp). SeciSearch3 might have failed to predict these elements, as the distance between the stop codon and the end of the contig (corresponding to the 3'UTR) is of only 9521bp, and the SECIS element could be in a further location. We conclude that it is in contig LVXV01025633.1_8 between positions 9521-9799 in the reverse strand. This is the detailed gene map: SELENOMSELENOM or SelM is an ER-retained protein that plays a role in the protection from oxidative stress-induced apoptosis. Selenoprotein M is highly expressed in the brain, where it acts as a neuroprotective agent. Some studies suggested that SelM also promotes intracellular calcium homeostasis [28].Regarding the gene prediction in Mus spretus, Exonerate, Genewise and Seblastian predicted the same protein, and a SECIS element can be found in the 3'UTR. Thus, we conclude that this protein can be found in the contig CM004105.1 between positions 417415-419622 in the forward strand. This is the detailed exonic prediction: SELENONThis protein is expressed during mouse embryogenesis and it is restricted to specific areas, including muscle precursors and maturating myocytes. However, it has been found that SELENON deficiency does not alter somitogenesis in embryos, suggesting that this selenoprotein is dispensable for these processes in mouse. Moreover, several functions of SELENON implicated during maturation of cells and organs have been hypothesized [29].Both mouse and human annotated SELENON proteins predict an homologous protein in the same genomic region of Mus spretus. However, the mouse and human proteins are different, and the mouse protein does not start with Met. We took the human protein for further analysis, as the human selenoproteome is better annotated. Exonerate and Genewise predicted the same gene and a SECIS could be found in the 3'UTR. In this case Seblastian predicted a shorter protein starting with Arg, so that we discarded its prediction. We conclude that it can be found in the Mus spretus genome in contig CM004097.1 between positions 131159360-131172024 in the reverse strand. This is the detailed gene map: SELENOOAlthough its function has been unknown since recently, SELENOO has now been identified as a redox-active mitochondrial selenoprotein. It is therefore localized in mitochondria, and expressed along mouse tissues [30].Regarding the gene prediction in Mus spretus, Exonerate, Genewise and Seblastian predicted the same protein, and a SECIS element can be found in the 3'UTR. Thus, we conclude that this protein can be found in the contig CM004109.1 between positions 89147087- 89157897 in the forward strand. This is the detailed exonic prediction: SELENOPSelenoprotein P (SelP) is an extracellular glycoprotein that contains a 40-50% of the total Se in plasma, suggesting that it might be in charge of Se transport. It is the preferred source of Se for embryonic neurons. In addition, it is also thought to be involved in protection against oxidative injuries [31][32].This protein contains 10 Sec in both humans and mouse, and we can also find this in Mus spretus. Exonerate and Seblastian softwares gave a good prediction and a SECIS element was found in the 3'UTR. We conclude that this protein can be found in CM004109.1 between positions 192628-197787 in the forward strand. This is the detailed gene map: SELENOSSELENOS is localized in the ER membrane and was identified as a component of mammalian ER-associated protein degradation (ERAD) machinery. This pathway is involved in the transport of misfolded proteins from the ER to the cytosol, leading to a degradation of these proteins by the ubiquitin-proteasome system [33].Regarding the gene prediction in Mus spretus, Exonerate, Genewise and Seblastian predicted the same protein, and a SECIS element can be found in the 3'UTR. Thus, we conclude that this protein can be found in the contig CM004100.1 between positions 53733901- 53743134 in the forward strand. This is the detailed exonic prediction: 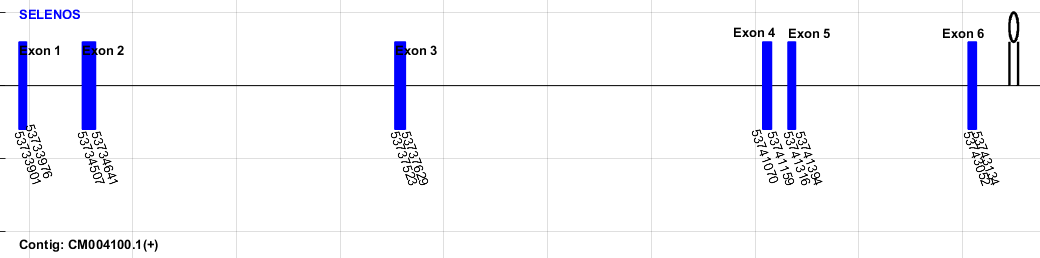 SELENOTThis selenoprotein is found in the cytosol, ER and Golgi. Although its function is not well characterised, its deficiency alters cell adhesion, enhances the expression of several oxidoreductase genes and decreases the expression of genes involved in cell structure organisation [34].Regarding the gene prediction in Mus spretus, Exonerate predicted the same protein and a SECIS element can be found in the 3'UTR. However, the mouse protein does not start with Met but with Leu, so we decided to run prediction with the human protein, which starts with Met, and we found a prediction with high homology. Thus, we conclude that this protein can be found in the contig CM004096.1 between positions 56410343-56427006 in the forward strand. This is the detailed exonic prediction: SELENOUThe function of selenoprotein U (SelU) is unknown, but it may regulate certain biological processes through its redox function [35]. In mammals, no SelIU containing Sec residues has been found, but homologues with Cys instead of Sec have been identified[36].We can find 3 SELENOU proteins in the mouse genome. The human genome also has them, but with several isoforms of each of them described. SELENOU1 Regarding the gene prediction in Mus spretus, Exonerate and Genewise predicted the same protein. Thus, we conclude that this protein can be found in the contig CM004108.1 between positions 34057328-34066959 in the reverse strand. This is the detailed exonic prediction: SELENOU2 This protein starts with Ala in mouse and in Mus spretus (according to SelenoDB 2.0 and our prediction, respectively) and the human protein has a poorly annotated Met-starting isoform and a well-annotated Gly-starting isoform. We used the human Met-starting isoform to make our predictions as it might be the most reliable and well-annotated one. Regarding gene prediction, Exonerate and Genewise predicted the same gene. Thus, we conclude that this protein can be found in Mus spretus in contig CM004107.1 between 61511789-61530095 in the reverse strand. Below, the detailed exonic prediction can be found: SELENOU3 Gene predictions from Exonerate and Genewise match completely in the Mus spretus genome. We can find this protein in contig CM004097.1 between positions 151529697-151532124 in the reverse strand. Below, there is the detailed gene map: SELENOVSelenoprotein V is a globular protein expressed in the testicles with its maximum level of mRNA expression during puberty and a gradual decrease of expression in adult mice. Although its function remains unclear, it has been shown that SELENOV possesses glutathione peroxidase and thioredoxin reductase activities [37][38] This protein can be found in humans and it is not annotated in the mouse SelenoDB 2.0. However, we found a mouse described SELENOV from NCBI, suggesting a poor SelenoDB 2.0 annotation. We ran the analysis for both human and mouse queries, and found the same predicted Mus spretus protein. However, Genewise and Exonerate failed to predict a Sec-containing protein, although the sequence homology was very high. In addition, Seblastian failed to predict any selenoprotein in this regio, so that we were not able to confirm the presence of SELENOV in Mus spretus. SELENOWSelenoprotein W works as a glutathione-dependent antioxidant and is highly expressed in proliferating myoblasts. It plays an important role in the normal function of the heart. Moreover, by protecting myoblasts from oxidative stress, it is suggested to be involved in muscle growth and differentiation [39].Regarding gene prediction, there are five unclassified proteins annotated in SelenoDB 2.0 for mouse and 2 annotated proteins in humans: SELENOW1, which is a selenoprotein, and SELENOW2, which is a Cys homologue. This raised again our concern about the mouse annotation and we made the analysis with the human proteins as well. Two of the mouse annotated proteins (SPP00001590_2.0 and SPP00001594_2.0) are very short and gave rise to thousands of BLAST hits in our analysis without any potential candidate genes in Mus spretus. None of them has a similar sequence to the human proteins and they do not start with Met. We discarded them from the prediction because it might exist an annotation error in mouse. On the other hand, SPP00001592_2.0 is a predicted mouse selenoprotein, but it is different from any human SELENOW protein. In addition, we were not able to find homologous selenoproteins in the Mus spretus genome. Thus, we could not predict this protein in the Mus spretus genome. In addition, we found that this protein has some homology (homologous C terminal part) to the human SELENOV (which has not an annotated mouse homolog). These findings further confirm that this protein is not a SELENOW protein in Mus spretus. Finally, SPP00001593_2.0 is a short mouse annotated peptide that has no Sec, does not start with Met and has no sequence homology to any of the human SELENOW proteins. We conclude that it results from a poor mouse annotation, so that we discarded it from our Mus spretus analysis. All in all, these are the predicted SELENOW proteins found in Mus spretus: SELENOW1 We ran the analysis using all three human SELENOW1 isoforms and found exactly the same genomic region in Mus spretus. In addition, the analysis with the mouse protein SPP00001591_2.0 (very similar to the human SELENOW1) pointed to the same region. The Mus spretus human-predicted homolog lacks the Sec and N-terminal part with both Exonerate and Genewise, and the mouse-predicted homolog (with Exonerate) has no Sec. In addition, the mouse Exonerate prediction includes a strange 5' splice site (CA instead of GT) that generates this aberrant Sec-lacking part. All this indicates that none of them is the Mus spretus SELENOW1 protein. We then ran Seblastian on this Mus spretus genomic region and found a selenoprotein that, although having a different N-terminal domain, includes an exon that contains the mouse Sec-containing amino acid and nucleotidic sequence (according to the SPP00001591_2.0 annotated transcript sequence). Furthermore, when we substituted this region by the mouse Exonerate Sec-lacking region, we found that the exonic assembly results in a correct 5' splice site. We finally decided to build the resulting Mus spretus SELENOW1 from the Exonerate mouse prediction combined with the Seblastian part commented above. This results in a selenoprotein that is highly homologous to the mouse and human one and with a SECIS element in the 3'UTR. We conclude that this protein can be found in the Mus spretus genome in contig CM004100.1 between positions 8372322-8374762 in the reverse strand. This is the detailed gene prediction: SELENOW2 When we ran the prediction for all SELENOW2 isoforms we found a genomic region in Mus spretus that gave a protein with high-homology. However, none of the mouse annotated pointed to this region, suggesting an incomplete mouse annotation. Genewise and Exonerate found the same gene in Mus spretus almost identical to the human isoform SPP00000127_2.0. We conclude that this protein can be found in the Mus spretus genome in contig CM004105.1 between positions 99815374-99816255 in the reverse strand. This is the detailed gene map: SECp43The tRNA Sec 1 associated protein 1 (SECp43) is an essential factor for selenoprotein biosynthesis. It interacts with several factors involved in this pathway, helps redistributing SecS/Sec-tRNASec complexes to the nucleus and promotes interaction between eEFsec and SBP2. SECp43 may also assist in the decoding of multiple UGA-Sec codons in SelP and might also prevent the degradation of selenoprotein mRNAs by the nonsense-mediated-decay pathway [4]. The mouse annotated protein does not start with Met, so that we ran the analysis with the human query (which starts with Met). Both human and mouse analysis found a gene in the same Mus spretus genomic region (with Exonerate), but we chose the human homolog because it had a better annotation. We conclude that SECp43 can be found in the Mus spretus genome in contig CM004097.1 between 128762881- 128781272 in the reverse strand. This is the detailed gene location:SEPHSThere are 2 selenophosphate synthetases, SEPHS1 and SEPHS2. Se needs to be as monoselenophosphate in order to be incorporated to tRNASec. SEPHSs are the enzymes in charge of catalysing the formation of this monoselenophosphate [3]. Despite two SEPHSs are identified in mammals, only SEPHS2 is known to be essential for selenoprotein biosynthesis. Moreover, SEPHS2 can have a Sec in its amino acidic sequence [40].According to SelenoDB 2.0,the human SEPHS1 has 4 isoforms, but some of them are poorly annotated (the promoter is annotated in the 3' end of the coding region in one of them). On the other hand, the mouse SEPHS1 is poorly annotated (lots of # and % symbols in the amino acid sequence) and our homology analysis failed to predict any gene in Mus spretus. Considering that it is not a selenoprotein and that it is not indispensable for their synthesis, we decided not to include it in our prediction. SEPHS2Regarding the gene prediction in Mus spretus, Exonerate and Seblastian predicted the same protein, and a SECIS element can be found in the 3'UTR. Thus, we conclude that this protein can be found in the contig CM004100.1 between positions 117216271-117217623 in the reverse strand. This is the detailed exonic prediction:SecSSec synthase (SecS) uses monoselenophosphate to convert Ser-tRNASec into Sec-tRNASec to allow Sec to be incorporated into proteins [41].Regarding the gene prediction in Mus spretus, Exonerate and Genewise predicted the same protein. Thus, we conclude that this protein can be found in the contig CM004098.1 between positions 50454061-50480812 in the reverse strand. This is the detailed exonic prediction: PSTKIn selenoprotein biosynthesis, the Ser in Ser-tRNASec has to be phosphorylated in order to be recognised by SecS. This phosphorylation is mediated by a kinase known as O-phosphoseryl-tRNASec kinase (PSTK) [42].Regarding gene prediction, the mouse protein does starts with Lys, so that we also ran the human analysis (as the human PSTK starts with Met). Both analysis (from human and mouse) predict a gene in the same Mus spretus genomic region, and the human-predicted gene starts with Met, so we chose it as the Mus spretus PSTK. Both Genewise and Exonerate made the same prediction. We conclude that PSTK can be found in the Mus spretus genome in contig CM004100.1 between 121562581-121571150 in the forward strand. This is the detailed gene prediction: SBP2SECIS binding protein 2 (SBP2) is important in the process of selenoprotein biosynthesis since it binds to the SECIS stem-loop and recruits eEFsec/Sec-tRNASec complex. Thus, it is necessary in order to incorporate Sec into proteins. It also interacts with the 60S ribosomal subunit [43].There are 3 predicted SBP2 proteins in mouse and 3 annotated isoforms in humans. Both of the human isoforms correspond to just one SBP2 protein in mouse, which in turn is the only one that we found in Mus spretus. The other 2 mouse SPB2 proteins correspond respectively to a peptide which contains K and E repeats, so it might be inaccurately annotated, and a protein that starts with a Lys and is very different from any human proteins. We conclude that there is only one SBP2 protein in Mus spretus, predicted correctly by Exonerate, that can be found in contig CM004107.1 between positions 48925499-48962157 in the forward strand. The exonic prediction can be seen below: eEFsecThe eukaryotic Sec-specific elongation factor (eEFsec) is an essential translation factor needed to incorporate Sec into proteins, since it recruits Sec-tRNASec. eEFsec specifically binds Sec-tRNASec and delivers it to the ribosome, therefore the presence of this protein is mandatory for selenoprotein biosynthesis [44].Regarding gene prediction, the problem with mouse was that the annotated protein does not start with Met, so we ran the human analysis. Both human isoforms start with Met and point to the same genomic region of Mus spretus. One of them predicts, just via Genewise, the same Mus spretus C-terminal part as the mouse analysis, so we chose it for the gene prediction in Mus spretus. We conclude that it can be found in the Mus spretus genome in contig CM004099.1 between positions 87343010-87537215 in the reverse strand. This is the detailed gene map: |