The main aim of this research project was to identify all the selenoproteins and selenoprotein machinery genes from Haplochromis burtoni. Danio rerio’s (most recently common ancestor known as Zebrafish) was used as a reference species in order to find these proteins based on homology. We chose the genome of Danio rerio because it is the most well characterized and studied fish genome, and its selenoproteins are thoroughly annotated. Nevertheless, Homo sapiens proteins were also used when Danio rerio proteins were not properly annotated, due to the fact that they did not begin with methionine.
For all the identified proteins in Haplochromis burtoni, the analysis and discussion of the results are performed individually. Predicted SECIS, obtained using SEBLASTIAN or SECISearc3, are also annotated. We must note that all protein predictions have been analysed by looking at the beginning of each sequence (whether it contained a methionine amino acid) and the end (if there was a stop codon). Results are shown in a table below.
In these analyses we will compare the results obtained with Danio rerio, to study cases of orthologous genes, paralogs, conservation or loss of selenocysteine and duplications.
As a starting point, we would like to highlight the large amount of proteins we have obtained, around 100. This is due to the fact that an ancestor of the teleost fish, the group in which our species is found, suffered a whole genome duplication event. These duplicated genes have different fates. The most likely outcome is the non-functionalization of a duplicated gene, due to the lack of selective restriction to preserve both. The mechanisms acting on the preservation of duplicates are sub-functionalization (partitioning of ancestral gene functions into duplicates), neo-functionalization (assigning a novel function to one of the duplicates) and dosage selection (preserving genes to maintain dosage balance between interconnected components).
We will now start with the individual analysis of the different gene families.
This is a subfamily of deiodinase enzymes that have an important role in the activation and deactivation of thyroid hormones. Iodothyronine Deiodinases Family is a paralogous gene family consisting of D1, D2 and D3 genes.
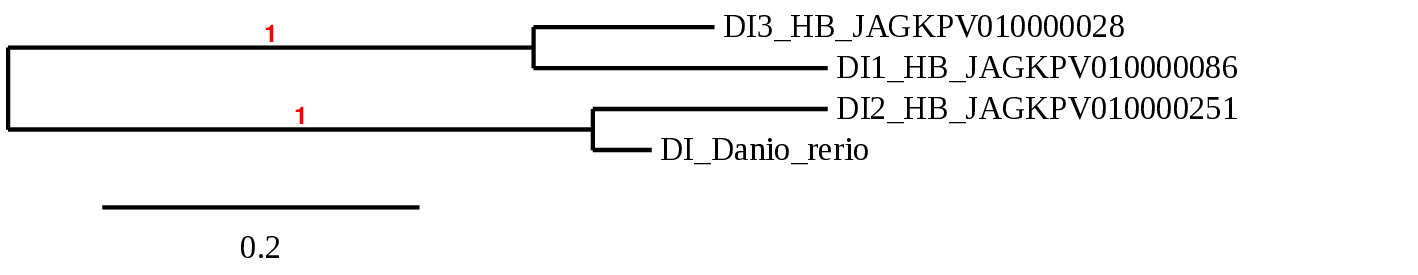
For each of the queries, we have found the same 4 scaffolds repeated within each alignment, and with similar genomic coordinates in each. After phylogenetic tree analysis (shown below) we determined that one of these 4 predicted proteins was actually a duplication of one of the other three.
The fact of all 3 query proteins belonging to the same family explains that the remaining three scaffolds are conserved and shown after aligning each separate query: they are actually paralogous genes, and as such, they are very similar, so for each one we predict the same scaffolds. Interestingly, we have seen a scaffold whose alignment is much higher, with lower e-value and higher t-coffee score values.
Nevertheless, as in Dario rerio we only observed two copies of DI, it may also be that they have not developed this family of paralogs, or that it hasn’t been preserved.
DI1
DI2 DI3
We observed that all scaffolds preserve the selenocysteine amino acid, which is also found in the different proteins of the query species (human and zebrafish). Therefore, we can conclude that the role of the enzyme is relevant, and the conservation of selenocysteine is a key factor because it is maintained throughout evolution.
As we can see in this phylogenetic tree, the DI of Danio rerio could be orthologous to the one we have classified in our species as DI2, and from here it has been duplicated twice in our species, while in Danio rerio we only observed one duplication.
For D1 we obtain the scaffold selected for the alignment features: JAGKPV010000086.1, between positions 678892 and 779506 on the positive strand with a single exon. We also found a SECIS sequence. It conserves selenocysteine.
While for D2 we obtain JAGKPV010000251.1, between 735977 and 841340 on the negative strand with four exons and for which we predict a SECIS sequence. It retains selenocysteine.
Finally, the best alignment for D3 is that in the JAGKPV010000028.1 scaffold, between positions 970733-1071461 on the positive strand with 3 exons. We have also obtained a SECIS sequence. Conserved selenocysteine.
GPx selenoproteins have a role in physiological processes such as hydrogen peroxide signaling, hydroperoxide detoxification, and the maintenance of cellular redox equilibrium. The Glutathione Peroxidases family contains multiple paralogs, some of which have a selenocysteine residue and others have lost it and now they are cysteine homologues.
When aligning each protein of the GPx family with the genome of c, we found multiple scaffolds, this is because glutathione peroxidases are very similar to each other, so the reference protein can align with other genomic regions encoding different GPx of the same family. In order to specifically identify each one, an analysis of the alignments between the reference and predicted proteins has been carried out. In addition, we observed many duplications of the different genes of this family.
We have observed that Danio rerio does not present GPx1, GPx5 and GPx6 so we used the human queries to know if it is due to a zebrafish-specific loss or common ancestor (between zebrafish and our species) loss. Once the analysis is done, we can conclude that these proteins have been lost specifically in Danio rerio because we observe that there are good alignments of the scaffolds in the Haplochromis burtoni. When aligning them with other queries of the family, the alignment of higher quality in the case of GPx5 and GPx6 came from the human query, and not from the GPx3 zebrafish orthologue, so we can deduce that our species has kept this protein. In addition, GPx3 may have been lost, as its alignment is identical to that of GPx5 and GPx6.
However, when looking at literature, we saw that it has been previously described that GPx5 and GPx6 are mammalian-specific duplications of GPx3, and as such should not be present in our fish. As such, this could be consistent with our analysis, as the alignment for these 3 genes in our species is practically the same.
We have observed that Danio rerio does not present GPx1, GPx5 and GPx6 so we used the human queries to know if it is due to a zebrafish-specific loss or common ancestor (between zebrafish and our species) loss. Once the analysis is done, we can conclude that these proteins have been lost specifically in Danio rerio because we observe that there are good alignments of the scaffolds in the Haplochromis burtoni. When aligning them with other queries of the family, the alignment of higher quality in the case of GPx5 and GPx6 came from the human query, and not from the GPx3 zebrafish orthologue, so we can deduce that our species has kept this protein. In addition, GPx3 may have been lost, as its alignment is identical to that of GPx5 and GPx6.
However, when looking at literature, we saw that it has been previously described that GPx5 and GPx6 are mammalian-specific duplications of GPx3, and as such should not be present in our fish. As such, this could be consistent with our analysis, as the alignment for these 3 genes in our species is practically the same.
GPx1
Seven distinct scaffolds showed significant hits when the homologous protein from Homo sapiens was aligned to the genome of Haplochromis burtoni. However, the gene located in the JAGKPV010000177.1 scaffold was the only one chosen as the best candidate. The gene coding for GPx1 is located between the 419878 and 521041 genomic coordinates from the scaffold on the positive strand. We discovered, using exonerate, that this protein has two exons and a conserved selenocysteine. Furthermore, SECIS structures were seen in the 3'-UTR region.
GPx2
For GPx2, the scaffold selected for the alignment features is the one in the JAGKPV010000016.1 scaffold, between positions 2764710 and 2865584 on the negative strand, with two exons. We also found a SECIS sequence. It conserves selenocysteine.
GPx3
For D3 we obtained a gene in the JAGKPV010000019.1 scaffold, between 3985595 and 4083241 on the negative strand with four exons and we have predicted a SECIS sequence. It conserves selenocysteine.
GPx4
In this case, we have obtained a gene in the JAGKPV010000041.1 scaffold, between coordinates 1273227 and 1373987 on the negative strand and with 6 exons. It conserves selenocysteine, as the query species, and also contains a SECIS sequence.
GPx5 and GPx6
For these two proteins, we obtained two very similar genes in the same scaffold, JAGKPV010000019.1, located between 3985601 and 4083241 on the positive strand. As mentioned from what we have seen in literature, this duplicated gene is most likely GPx3, due to the fact that we took the queries from the human and that these two genes have recently diverged from GPx3. As such, similar alignments occur.
GPx7 and GPx8
GPx7 and GPx8 are located in JAGKPV010000054.1, between 1366592-1468524 in the negative strand, and JAGKPV010001001.1, between 753-97351 in the negative strand, respectively. Both genes are cysteine homologs, having diverged from GPx4 from a zebrafish and Haplochromis burtoni common ancestor. For both, we found a SECIS sequence.
Msr A is extremely conserved, and its major function is to catalyze the enzymatic reduction of the amino acid methionine-S-sulfoxide using thioredoxin. We obtain only one predicted protein, in the scaffold JAGKPV010000093.1. This protein starts at position 1174138 and ends at position 302160. We note that it is a cysteine homologue, both in zebrafish and our species. Interestingly, we observed a duplication in the JAGKPV010000086.1 scaffold. We found that, despite having lost selenocysteine, it still retains SECI.

SelR1
When the homologous protein from Zebrafish was aligned to the genome of Haplochromis burtoni, four distinct scaffolds exhibited significant hits. However, the gene found in the JAGKPV010000081.1 scaffold was the best candidate, as it presented a good alignment and the greatest possible score in t-coffee. The other gene entry, found in another scaffold, corresponds to the other SelR; they aligned with other scaffolds because the proteins are very similar. In the negative strand, the gene coding for SelR1 is situated between 1128364 and 1229067 places from the scaffold. Using Exonerate, we discovered that this protein has three exons and a conserved selenocysteine. Moreover, we discovered SECIS structures in the 3'-UTR region.
SelR2 SelR3
Sel15 is a thioredoxin-like fold ER-resident protein like SelM.
Following the aforementioned criteria, we detected the scaffold that aligned with the human Sel15 query, because the zebrafish one was misannotated. In this case, we only detected one gene in the JAGKPV010000266.1 scaffold, located between 553559 and 657888 on the positive strand with four exons. This protein maintains selenocysteine, as it happens with the reference species (human). Therefore we could conclude that evolution seeks to maintain this selenocysteine, as it provides important functions.
Again, following the aforementioned criteria, we detected the alignment of our reference species query in the scaffold JAGKPV010000018.1, between 3234933 and 3337138 on the negative strand with 4 exons. We noted that selenocysteine was both conserved in our species and the reference species. Subsequently, we found that the sequence contained two SECIS from which we chose the one located in the 3'UTR region as, detailed in the scheme below.
It catalyzes the conversion of 2-thiouridine to 2-selenouridine at the wobble position in tRNA by transferring selenium from selenophosphate. This protein comes in three different varieties: SelU1, SelU2, and SelU3.
SelU1 SelU2 and SelU3
The function of SelP is not yet well understood, but it is thought that this protein is involved in the supply of selenium to tissues such as the brain and testes, as well as in the creation of some extracellular antioxidant defense.
From the alignment characteristics, we determined that the gene located in the JAGKPV010000034.1 scaffold between 190481-1291306 on the negative strand and with three exons, is the most accurate one, and conserves selenocysteine as the query species. In addition, this gene possesses a copy that aligns worse with our query, located in JAGKPV010000380.1 between 486965-581959 also on the negative strand. But, in this case, we observe that our species loses selenocysteine and therefore becomes a cysteine homologue.
They are members of the pyridine nucleotide-disulfide oxidoreductase family, which is involved in many cell functions including oxidant damage prevention, cell growth and transformation, and ascorbate recycling from its oxidative state. It also reduces other substrates in addition to Trx. TR1, TR2, and TR3, respectively, are found in the cytosol or nucleus, mitochondria, and testis. During development, TR1 and TR2 are critical.
In this scenario we have found different scaffolds aligning with the different members of the TR family. We can also detect duplications and some losses of selenocysteine.
The proteins of Haplochromis burtoni correctly match zebrafish query proteins, as seen in the phylogenetic tree. We found a branch that groups TLR1 and TLR2, which are orthologous to the TLR2 seen in zebrafish. TLR1 was most likely found in the common ancestor, and zebrafish suffered from a particular loss. Moreover, there's another branch that groups the TLR3s, which were most likely a paralog of TLR1 and TLR2 in the common ancestor.
TR1 TR2 TR3
In this family of proteins, we could establish two different proteins, thanks to the characteristics of the alignment of the Haplochromis burtoni genome with the query of zebrafish and human. Actually, we determined the SelW and the SelW2 proteins. These two proteins are encoded for a gene located in the negative strand of the genome of interest. The first one can be found in the genome of Danio rerio and the second one only in humans. This highlights the fact that SelW2 protein is no longer present in the genome of the zebrafish, as this species lost it.
We have seen that SelV protein is lost in our species, as the automated python program has not been able to obtain a blast file from the human query fasta file. As this gene is lost both in zebrafish and Haplochromis burtoni, it is very likely that there has been a loss of gene in the common ancestor.
Regarding the wikipedia page elaboration, we published an article in Catalan about Haplochromis burtoni using different tools and templates. However, later on we had to merge this article with the one on Astatotilapia Burtoni, the most commonly accepted name nowadays to describe the fish especies, since the name Haplochromis is no longer in use. With this, we were able to go deeper into the wikipedia world, getting familiarized with the community and its exemplary goal to spread knowledge into society.
Because it has the greatest score when aligning our species genome to the homologous protein from Zebrafish, JAGKPV010000001.1 was chosen as the scaffold for SelR2. The gene is found between 5475836 and 5576976 on the positive strand. Both our species and the reference species have a cysteine homolog protein. Exonerate predicted 4 exons, while SECISearch3 predicted an SECIS structure.
Four different scaffolds showed significant hits when the homologous protein from Zebrafish was matched to the genome of Haplochromis burtoni. However, the gene contained in the JAGKPV010000081.1 scaffold was the best candidate, with a strong alignment and the highest t-coffee score. The other scaffolds, which match to this SelR3, are produced as a consequence of the homology of the proteins. The gene coding for SelR3 is located between the 841460 and 944785 scaffold coordinates, on the negative strand. Using exonerate, we determined that this protein has six exons , and contains an homologous cysteine. Furthermore, SECIS structures were found in the 3'-UTR region. Remarkably, this protein maintains the SECIS sequence even though it is a cysteine homolog (it does not have a selenocysteine).
Subsequently, when performing the phylogenetic tree, we analysed these gene pairs and observed that the SelR3 in zebrafish is grouped with SelR1 and vice versa. Therefore, we could have made a mistake when evaluating the alignments and that the scaffold we have annotated as SelR1 is that of SelR3 and the other way around. Further phylogenetic studies should be carried out to elucidate this issue.
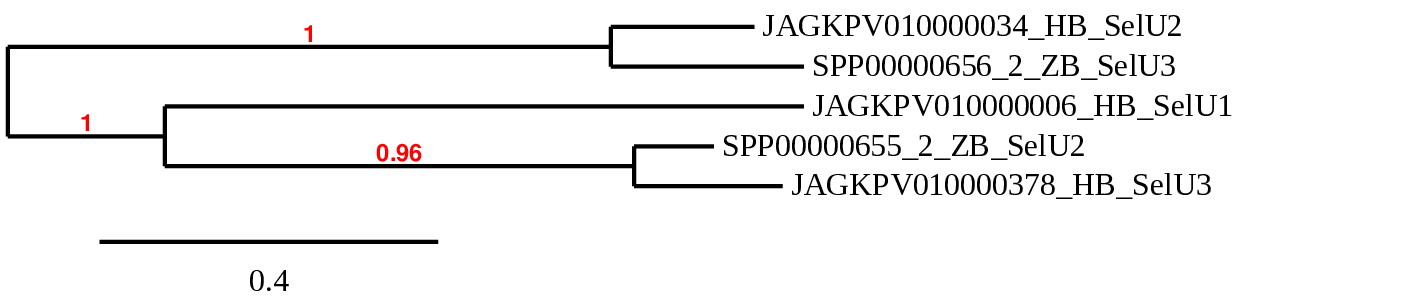
Thanks to the alignment, we were able to determine SelU1 in the JAGKPV010000006.1 scaffold, between 3629785-3731165 in the positive chain and with four exons. A selenocysteine has been detected both by analysing the seblastian file and by studying the presence of a stop codon in the predicted sequence. Both our species and the reference species (Danio rerio) present this selenocysteine, but it is not found in humans, as it is lost and transformed into a cysteine homologue. Therefore, we can conclude that this selenocysteine is lost throughout evolution. The presence of a SECIS sequence has also been detected.
On the other hand, SelU2 can be located in the JAGKPV010000034.1 scaffold, between 1811739-1912297 in the positive strand, and SelU3 in JAGKPV010000378.1 between 124872-225078 in the negative strand, both with 5 exons. These genes are homologues of cysteines that have lost selenocysteine in our species, as is the case in humans and zebrafish (query).
We observed a SECIS sequence in both SelU1, which allows selenocysteine to be established, and SelU3. Interestingly, SelU2 does not maintain this SECIS sequence, so it loses all the necessary characteristics to determine the presence of selenocysteines.
In this case, we could have also inverted the annotation of the genes after assessing the alignments, as observed in the phylogenetic tree.
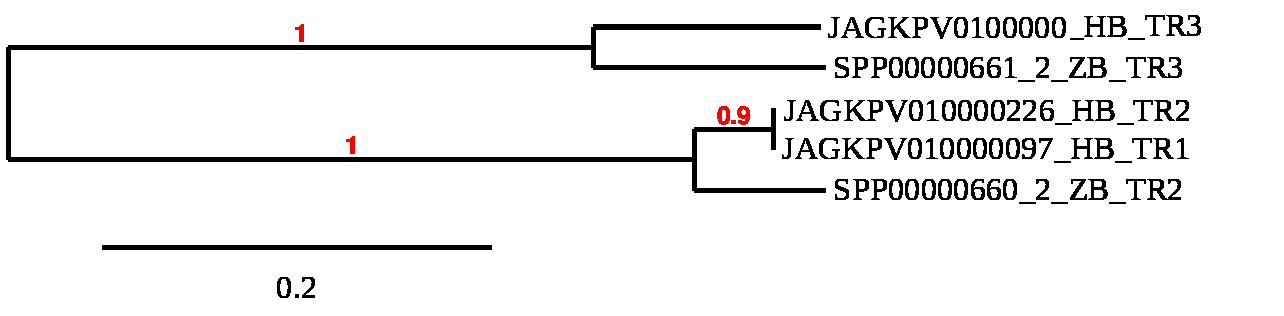
Regarding TR1, thanks to the alignment study in the Haplochromis burtoni genome, we have been able to determine that it is found in the JAGKPV010000097.1 scaffold between the 713972-819571 coordinates in the positive strand and with 13 exons. We can observe that this protein has been lost in zebrafish, but that it is conserved in the common ancestor, since we are able to predict it in our species. It contains a SECIS sequence.
Regarding TR2, thanks to the alignment study in the Haplochromis burtoni genome, we have been able to determine that it is found in the JAGKPV010000226.1 scaffold between positions 229234-346117 in the positive strand, with 16 exons. We can observe that this protein conserves selenocysteine both in our species and in the query species. It also contains a SECIS sequence.
Finally, TR3, through the alignment study on the Haplochromis burtoni genome, we have specified that our gene is in the JAGKPV010000097.1 scaffold between 710912-819571 in the positive strand, and also contains 16 exons. We can observe that this protein conserves selenocysteine both in our species and in the reference species. Contains a SECIS sequence.
The gene determined with the alignment, both SelW and SelW2 are located in the JAGKPV010000052.1 scaffold between 963082-1063622 coordinates of the genome, with 3 exons the first one and with 1 exon the second. As such, it is very likely that both our organism and our reference species only have the SelW gene that is later duplicated throughout evolution ending up with two different proteins in humans.