Fig 1. Selenocystein's codon in the genetic code. 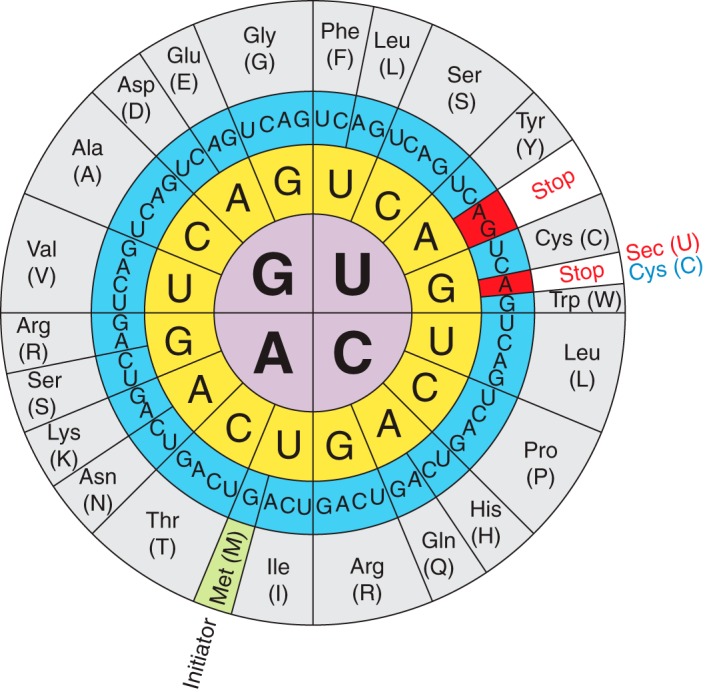
At least one selenocysteine (Sec or U), a Se-containing aminoacid, is found in selenoproteins. These proteins can be found in each of life's tree domains (eukarya, archaea, and bacteria). Sec is considered to be the 21st aminoacid of the genetic code. It is not found in a standard way in the genetic code, as it is encoded by the codon, which normally acts as a stop codon. As a result, Sec synthesis involves the use of specialized translation machinery.
Approximately 50% of mammalian selenoproteins have known functions.
Fig 2. Comparison between the structure of the amino acids selenocysteine and cysteine. 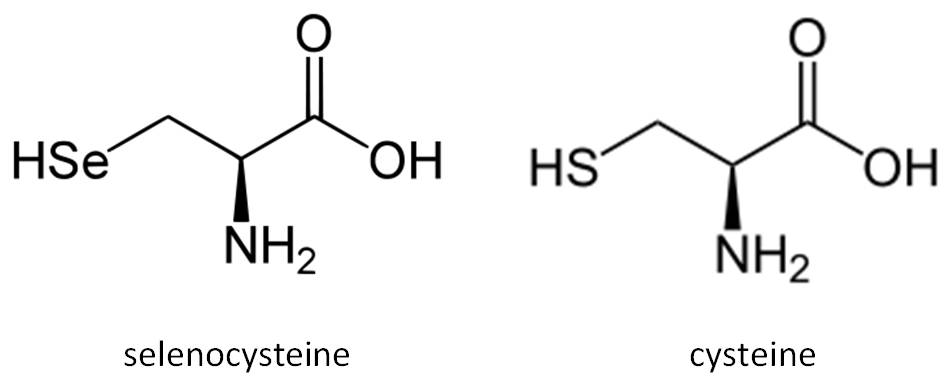
Many selenoproteins that have been functionally described have a role in redox control. Housekeeping and stress-related selenioproteins are the two main kinds of selenioproteins. Stress-related selenoproteins are not needed for survival and often show lower expression under Se-deficient situations. Housekeeping selenoproteins participate in tasks vital to cell viability. The selenoproteome is a collection of selenoproteins found in an organism. In the case of humans there are 25 selenoprotein genes. Several groups of terrestrial animals reduced their reliance on selenium by replacing selenoproteins with Cys homologs or losing selenoproteins, according to comparative investigations of organisms with substantial selenoproteomes and those missing selenoproteins. Cys mutants of selenoproteins can keep some of their functions. Aquatic creatures have huge selenoproteomes, showing that the environment plays an important role in selenoproteome evolution. The substitutions in mammals were always from Sec to Cys, never the other way around. Although several selenoprotein genes (GPx6, SelV, GPx5, the latter of which was quickly transformed to a Cys variant) first emerged in mammals, they evolved through gene duplication.
Sec tRNA [Ser] Sec is the only known amino acid in eukaryotes whose biosynthesis occurs after the precursor amino acid (serine) has already been linked to the tRNA. This tRNA differs from other tRNAs due to some sequence variations and is subjected to post-transcriptional modifications. Trsp is the gene that codes for tRNA[Ser]Sec. Except from fish, every organism has a single copy of this gene.
The first step in the byosinthesis of Sec is the aminoacylation of tRNA[Ser]Sec by seryl-tRNA synthetase (SerS). Because of it's modifications the tRNA's structure, serine recognizes it. Sec synthase (SecS), which incorporates selenophosphate, the active form of selenium, into the aminoacid backbone and forms Sec-tRNA, must then convert this seryl-tRNA[Ser]Sec to selenocysteyl-tRNA[Ser]Sec. Prior to this step, phosphoseryl-tRNA kinase must first form a phosphoseryl-tRNA (PTSK).
The SPS2 enzyme is also involved in the biosynthesis of cysteine (Cys). Although Cys is an essential amino acid in mammals, its synthesis on tRNA[Ser]Sec is a newly discovered de novo pathway. SPS2 is a selenoprotein that may function as a selenoprotein synthesis autoregulator.
Sec is broken down by the enzyme Sec lyase (SCL), which catalyzes the conversion of Sec to L-Ala and selenium. During the degradation of selenoproteins, SCL is thought to recycle selenium from Sec.
UGA codons in selenoprotein mRNAs control the incorporation of Sec into proteins. The SECIS element is needed for this mRNA's unique recognition.
When a ribosome encounters a UGA codon, the Sec machinery interacts with the translation machinery to boost UGA codon coding potential.
Sec-tRNA[Ser]Sec translates UGA to Sec in response to the SECIS element. The Sec-specific translation elongation factor (eEFSec) as well as the SECIS binding protein 2 are required for this process (SBP2).
SBP2 is associated with ribosomes and contains an RNA-binding domain that interacts with eEFSEc, which recruits Sec-tRNA[Ser]Sec, and binds to SECIS elements.
Fig 3. Schematic representation of selenoproteins' synthesis.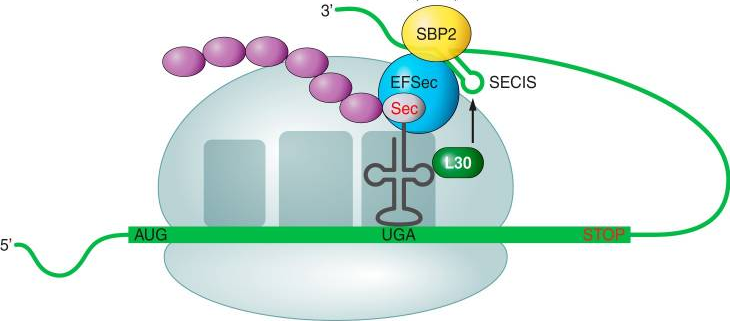
Ribosomal protein L30 (which is part of the basal Sec insertion machinery), nucleolin, and eIF4a3 are other SECIS-binding proteins. These proteins are regulatory proteins that control selenoprotein synthesis. In the 3'-UTR of selenoprotein mRNAs, SECIS elements are cis-acting stem-loop RNA structures. SECIS elements are found just after the coding region in bacteria, but they can also be noticed much further in eukarya and archaea. Eukaryotic SECIS elements are conformed by two helixes separated by an internal loop, a GA Quartet (SECIS core) structure, and an apical loop or bulge. The GA Quartet is responsible for interacting with SBP2. In terms of the presence of an apical bulge, eukaryotic SECIS can be divided into two groups.
| Kingdom | Phylum | Class | Order |
|---|---|---|---|
| Animalia | Chordata | Actinopterygii | Cichliformes |
| Family | Genus | Specie |
|---|---|---|
| Cichlidae | Haplochromis | H. burtoni |
Haplochromis burtoni, more commonly known as Astatotilapia burtoni (most accepted name today) is a species of fish in the family Cichlidae. It was discovered by the british zoologist Albert Günther in 1894. H. burtoni is commonly used as a model organism for the study of the social behavior and physiological systems of this family of fish. Thus, it is a very good animal model for delving into the mechanisms of development and embryogenesis of cichlids. In addition, this species of fish is considered an example of enviable hormonal and neuronal plasticity.

Fig 4. H.burtoni
The species Haplochromis burtoni is original form Lake Tanganyika and its surrounding waterways, including parts of Burundi, Rwanda, Tanzania, and Zambia. It also inhabits associated rivers like Lake Kivu. Its natural habitats are rivers, intermittent rivers, swamps, freshwater lakes, freshwater marshes, intermittent freshwater marshes, and inland deltas.
Fig 5. Distribution of subspecies of Haplochromis burtoni.
Organisms of Haplochromis burtoni are approximately 12 cm long, although males can usually reach a maximum size of 15 cm, while females up to 7 cm. In addition, its approximate weight is 100 g.
Males of Haplochromis burtoni have two reversible phenotypes, which means they can easily switch between being territorial and non-territorial depending on the social environment in which they are found. Males in dominant environments are territorial, possess bright coloration, aggressive behavior while defending territory and an active role in sexual reproduction with females. On the other hand, subordinate and non-territorial males have a coloration similar to that of females (sand-colored, thus disappearing), have no initiative to chase females and are reproductively suppressed due to the regression of the gonads. It should be noted that these transitions between different social roles cause various changes in the brain and reproductive system, so that social transformation affects them both behaviorally and physically.
It is suggested that the stress hormone, cortisol, may play a direct role in social status, as the hormone may change the biological priorities of the cichlid system. Under chronic stress, the animal may experience reproductive regression (as shown in the change from the territorial male to the non-territorial type) as a result of the body’s efforts to combat stress.

Fig 6. H. burtoni
Dominant H. burtoni males will have courtship displays to attract females. This display begins with the male trying to get the female's attention by showing her his side while shaking his body. During mating, the male releases sperm and fertilizes the eggs in the female's mouth, which will raise the eggs in her mouth, following a behavior called maternal mouthbrooding.
H. burtoni incorporates multiple sensory systems, including the chemosensory, visual and especially the acoustic, to be able to interact socially with other organisms of the same species. Acoustic information has been shown to play an important role in the sexual reproduction of the species. Specifically, dominant males were observed to emit auditory signals to attract females, suggesting that sounds associated with body quivers are intentionally produced for mating.