The importance of
Selenoproteins
Selenium and selenoproteins
Selenium (Se) is an essential trace element that is incorporated in the small but vital family of proteins, namely the selenoproteins, as the selenocysteine amino acid residue. Selenocysteine (Sec, U) is considered the 21st amino acid and it is analogous to cysteine (Cys, C). The difference between them is found in the side chain: while in Cys we can find an atom of sulfur (S), in Sec we find selenium (Se) [1]. The most remarkable trait of selenoprotein us the recoding of a UGA codon, normally decoded as a stop signal [2, 3].

Selenoproteins were discovered in 1989 and, since then, they represent a big line of research. After a couple of decades, despite many advances in the field and the discovery of many essential and regulatory components, the precise mechanism of UGA-selenocysteine recoding remains elusive and more complex than anticipated, with many layers of control [4].
Se is mostly involved in redox systems and it has antioxidant protection capability [5]. The set of selenoproteins in an organism is known as selenoproteome.
Biosynthesis of selenoproteins
Selenium is incorporated from the diet. It is reduced several times by processes that are not well known yet.
The first step of biosynthesis of Sec is the charge of a Ser to a special tRNA: tRNASec. This is done by the the seryl-tRNA synthetase. The Ser-tRNASec is next converted into Sec-tRNASec by an enzime called selenocysteine synthase (either directly or via prhosphoseryl intermediated produced by the phosphoseryl-tRNa kinase, using monoselenophosphate as the substrate) [6]. Once the Sec-tRNASec is formed, it can be incorporated creating a selenoprotein.
On the other hand, how the cellular protein translation machinery uses the UGA codon either as a selenocysteine insertion signal for selenoprotein mRNAs or as a stop signal for all other cellular mRNAs remains an unresolved and fascinating issue. Foremost, every selenoprotein mRNA harbors a stem-loop-stem-loop RNA structure of +/- 100 nucleotides, so it is called SECIS element. These RNA motifs play a pivotal role in the mechanism of UGA recoding. Initially believed to be a recruitment platform of recoding factors, the SECIS actually controls the translation of selenoprotein mRNAs by governing selenocysteine insertion efficiency.

There are also necessary in this process: the Sec-specific elongation factor (EF Sec), SECIS-binding protein 2 (SBP2), ribosomal protein L30, 43-kDa RNA binding (SECp43) and selenophosphate synthetase 2 (SPS2) [7]. Nevertheless, the SECIS element and the Sec-tRNA[Ser]Sec are the central components dictating selenocysteine insertion together with these associated proteins [8].
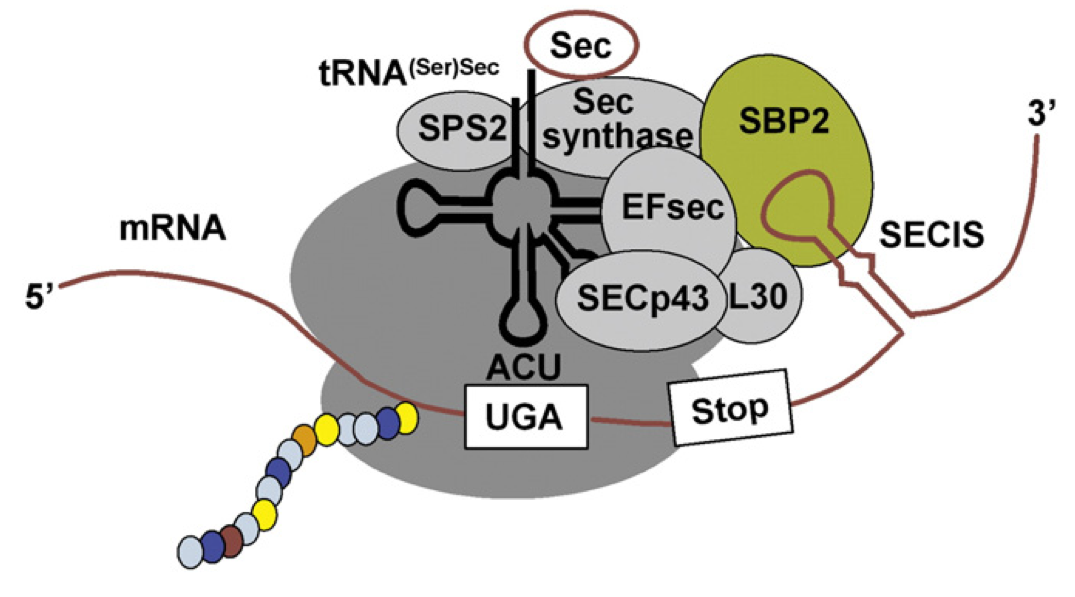
This is an schematic representation of selenoprotein synthesis. As explained before, in order for UGA to encode for selenocysteine insertion and not translational termination, the selenoprotein mRNAs require a downstream stem loop structure, the SECIS.
The SECIS element binds SBP-2, which in turn interacts with the selenocysteine-specific elongation factor, EFsec. EFsec also binds the selenocysteine tRNA (Sec-tRNASec) and promotes selenocysteine incorporation in the elongating protein by the ribosome at the UGA codon.
An additional SECIS binding protein, L30, can displace SBP-2 and anchor the loaded SECIS complex to the ribosome. This protein has been shown either to interact directly with or facilitate the interaction between many components needed for selenoprotein synthesis [9, 10].
Evolution of selenoproteins
Selenoproteins exist in archaea, bacteria and eukaryotes, but not all species have them. For example, yeast and plants do not have selenoproteins but cysteine-containing homologues [11]. The predicted ancestral vertebrate selenoproteome is composed by 28 proteins, but several selenoproteins were lost after the terrestrial environment was colonized, providing us with the idea that tetrapods reduced their utilizations of Sec compared with fiches [12].
They were found 45 families of selenoproteins, 28 of them were present in mammals although nowadays there are known more than 50. 21 of them were found in every vertebrate and the others, only in some lineages. This difference shows a dynamic process by which we obtain new selenoproteins generated by duplication or by losing or changing Sec by Cys [13].
Comparing the sequence of vertebrate selenoproteins in order to make a phylogenetic study, it was observed that the most conserved selenoprotein was selenoprotein T or SelT (conserved even at the nucleotide level) [14].
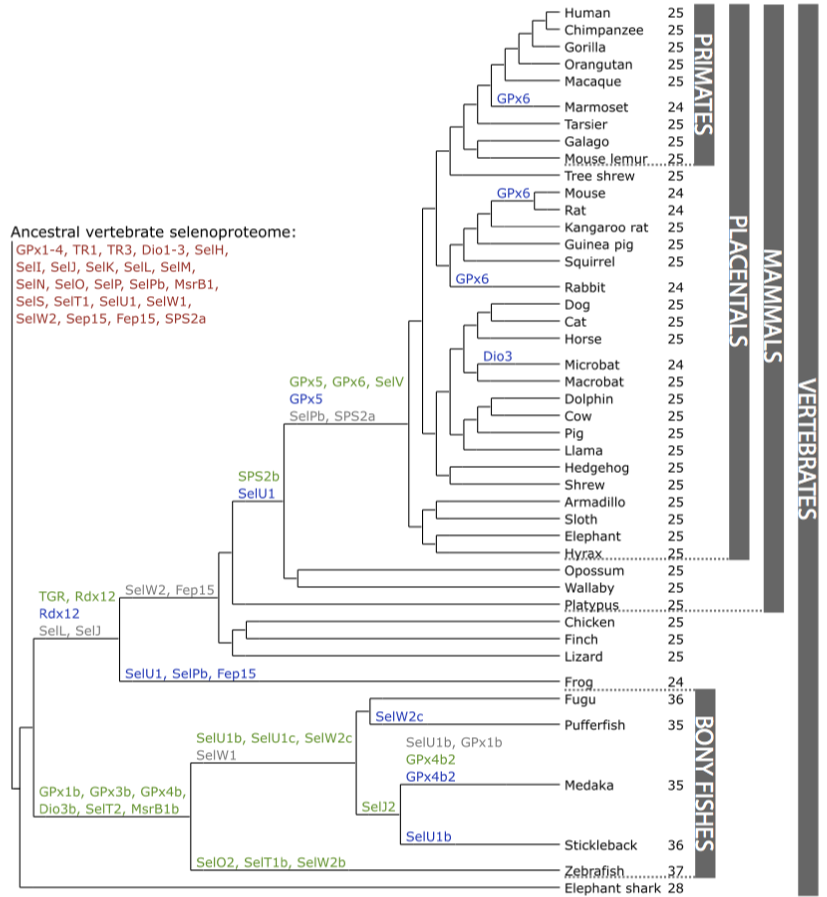
Families of selenoproteins
The function of more than half selenoproteins is still unknown, but there are some families that have been widely studied. The following information only describes the mammal selenoprotein families found in other reptiles.
| Protein | Function |
|---|---|
| Thioredoxin reductase (TR) | These selenoproteins are the only enzymes capable of reducing oxidized Trx (Thioredoxin). TrxRs also participate in many cellular signaling pathways. Cellular proliferation, cell viability and apoptosis are other cell functions in which TrxRs are involved [15]. |
| Glutathione peroxidase (GPx) | The incorporation of selenium in the form of selenocysteine was first reported in proteins of the GPx family. This family is the largest selenoprotein family in vertebrates (8 in mammals), containing 4 different proteins in lizards [15, 16, 17, 18]. |
| Iodothyronine deiodinase (DIO) | This family consists of proteins involved in the regulation of activation/inactivation of thyroid hormones. These oxidoreductases are classified according to their location. |
| Selenoprotein P (SEL P) | SEL P contains multiple selenocysteine residues. As mentioned previously in the GPx section, both GPx3 and SEL P play an important role in selenium transport and distribution [16, 18, 19]. |
| Selenoprotein W (SEL W) | SEL W is catalogued as a stress-related protein as its expression depends on selenium levels. Along with SEL T and SEL H, these proteins belong to the Rdx super-family [18, 20]. |
| Selenoprotein S (SEL S) | It plays an important role in the removing of misfolded proteins from this organelle to the cytosol for degradation [18, 20]. |
| Selenoprotein 15 (SEL 15) | This protein has a thiol-disulfide isomerase activity and is possibly involved in disulfide bond formation. SEL 15 is highly expressed in prostate, liver, kidney and testis [21]. |
| Selenoprotein M (SEL M) | This protein presents functional and structural homology with SEL 15, with a thiol-disulfide activity and a signal peptide at the N-terminal end. Thus, it has also been proposed as a protein with an important role in disulfide bond formation in the ER. Redox-active motifs observed in SEL M structure indicate that this protein has a redox function [21]. |
| Selenoprotein R (MSRB) | Methionine-R-Sulfoxide Reductase family includes three proteins, one of them containing selenocysteine (MSRB1) and two of them being homologues with cysteine (MSRB2 and 3) [22, 23]. |
| Methionine sulfoxide reductase A (MSRA) | Methionine sulfoxide reductase A (MSRA) is an antioxidant enzyme found in all domains of life that catalyzes the reduction of methionine-S-sulfoxide (MSO) to methionine in proteins and free amino acids. |
| Selenoprotein U (SEL U) | The function and structure of this family remain unknown [18]. |
| Selenoprotein T (SEL T) | This thioredoxin-like protein is predominantly located in ER and Golgi, and is ubiquitously expressed during embryonic development and in adult tissues[21, 22]. |
| Selenoprotein H (SEL H) | SEL H binds specifically to sequences that contain heat shock and stress response elements. Furthermore, SEL H possesses an intrinsic glutathione peroxidase activity and can increase glutathione levels. In response to stress, this proteins may up-regulate other selenoproteins involved in the regulation of oxidative stress such as GPx [18, 22]. |
| Selenoprotein I (SEL I) | This protein is only found in vertebrates, indicating a recent appearance in the phylogenetic line. It is a transmembrane protein containing seven transmembrane domains and three conserved aspartic residues, which are required for SEL I catalytic activity [22, 23]. |
| Selenoprotein K (SEL K) | This protein is implicated in ER-associated degradation of misfolded proteins, and has been implicated in inflammation and the immune response [22]. |
| Selenoprotein O (SEL O) | This protein is one of the less characterized selenoproteins. It has one single Sec residue located at C-terminus and analysis on its sequence revealed a mitochondrial targeting peptide. Its function remains still unknown [23]. |
| Selenoprotein N (SEL N) | SEL N is an ER-resident transmembrane glycoprotein highly expressed during embryonic development. It is ubiquitously expressed in all tissues, but in adult skeletal muscle tissue is believed to have an important role in the maintaining of satellite cells and reparation of muscles after injury or stress [23].Although the biologic function of SEL N remains unknown, it is clear that it plays a role in muscle tissue [20, 18]. |
It is necessary to briefly describe the proteins involved in the synthesis of Sec transference RNA and its attachment to the growing selenoprotein chain in order to achieve an optimal understanding of how selenoproteins are synthesized. These proteins, formally known as translation machinery, do not include selenocysteine residues in their peptidic chain: therefore, they are not considered as selenoproteins.
| Protein | Function |
|---|---|
| Selenocysteine-specific elongation factor (eEFSec) | eEFsec is a GTP-binding protein that delivers the Sec-tRNAsec to the ribosome. It is also implicated in Sec-biosynthesis [18, 24]. |
| SECIS-Binding protein 2 (SBP2) | SBP2 is ubiquitously expressed in all body tissues in multiple isoforms formed by alternative splicing [17, 18]. |
| Phosphoseryl t-RNA kinase (PSTK) | PSTK is a kinase which phosphorylates the seryl-tRNAsec complex in the presence of ATP and Mg2+. This pSer-tRNAsecintermediate serves as a substrate for the Sec synthetase (SecS) [22, 25] |
| tRNA Sec 1 associated protein 1 (SECP43) | This protein has two ribonucleoprotein-binding domains (RNP) which constitute an RNA-recognition motif (RRM), and is usually located in the nuclear compartment. Its expression regulates the levels of GPx1 and methylated Sec tRNA[Ser]Sec. SECp43 also plays a role in the formation or stabilization of the EFSec-SBP2-Sec tRNA[Ser]Sec complex [18]. |
| Selenocysteine synthase (SECS) | SecS protein is a member of the pyridoxal phosphate-dependent transferase superfamily. This protein mediates Sec incorporation into selenoproteins by dephosphorylating O-phosphoseryl tRNA[Ser]Sec and transfers monoselenophosphate to the tRNA. SecS solely depletion has no described effect [18, 22, 26]. |
| Selenophosphate synthetase (SPS) | SPS proteins, also known as a SEPHS, are involved in the synthesis of seleno-phosphate molecules. Therefore, besides the fact that these selenoproteins allow the synthesis of Sec-tRNA, it is believed that they act as regulators of selenium bioavailability and prevent excessive incorporation of this element [16, 19]. |