This is our
Discussion
Important considerations
All the results obtained for each predicted protein were carefully analyzed and discussed, paying special attention to the T-Coffee output and SECIS elements prediction.
The criteria for deciding whether a detected protein in Carettochelys insculpta' genome was a selenoprotein, a cysteine-containing homologue or neither of them was the following:
- Selenoprotein: at least one selenocysteine (Sec, U) was detected in the T-Coffee output and a SECIS element was located at the 3'UTR region of the gene.
- Cysteine-containing homologue: a cysteine (Cys, C) from Carettochelys insculpta’ genome was aligned with a Sec from the Mus musculus or Anolis carolinensis’ genome.
- Others: some proteins could have lost or replaced their Sec position for another amino acid other than Cys. Hence, these proteins are not considered selenoproteins nor Cys-containing homologues.
THIOREDOXIN REDUCTASE (TR)
Based on the scaffolds selected according to the criteria previously described (e-value, coverage; see methodology) a phylogenetic analysis was performed in order to identify those which were more similar to the query protein chosen. From there, the study was carried out with a single region, the closest to the reference protein in the phylogenetic tree.
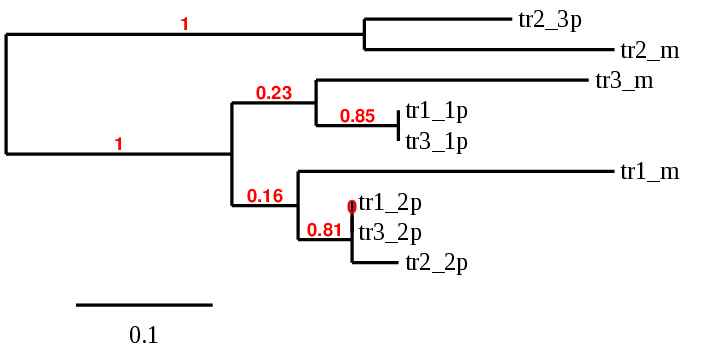
TR1: several hits in 5 scaffolds were found from tblastn in TR 1, being the hits in scaffold ML683785.1 the one we chose for the analysis. Exonerate predicted that the gene was located between positions 14622192 - 14643591, comprising 13 exons in the forward strand. T-coffee alignment between Carettochelys insculpta genome and the protein reference (in this case, obtained from Mus musculus) showed high similarity between the sequences and, from this, we extract that there is a coding sequence for this protein in the turtle genome. Seblastian could predict a SECIS structure at the 3’ end of the genomic sequence. Furthermore, Seblastian could predict a protein in the same region.
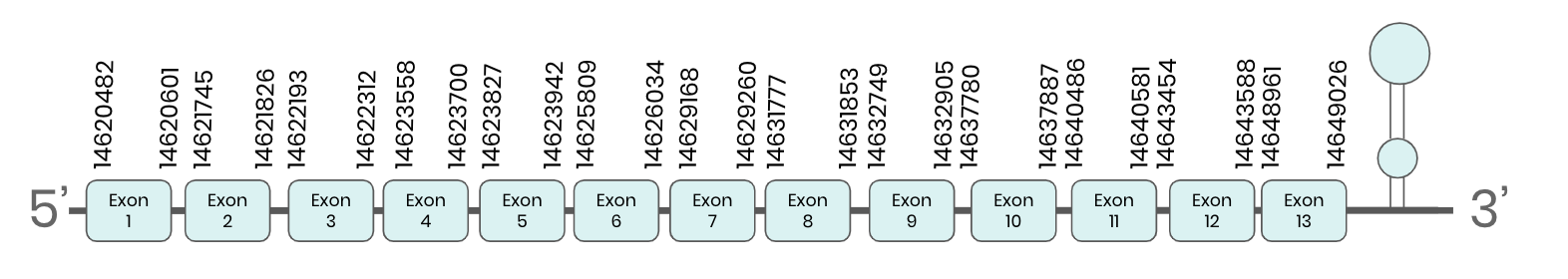
Thus, TR 1 selenoprotein is present in the Carettochelys insculpta genome as a SECIS element and a selenocysteine were found.
TR2: several hits in 5 scaffolds were found from tblastn in TR 2, being the hits in scaffold ML683833.1 the one we chose for the analysis. Exonerate predicted that the gene was located between positions 5662933 - 5714443, comprising 12 exons in the forward strand. T-coffee alignment between Carettochelys insculpta genome and the protein reference (in this case, obtained from Mus musculus) showed high similarity between the sequences and, from this, we extract that there is a coding sequence for this protein in the turtle genome. Seblastian could not predict a SECIS structure at the 3’ end of the genomic sequence. Furthermore Seblastian could not predict a protein in the same region.
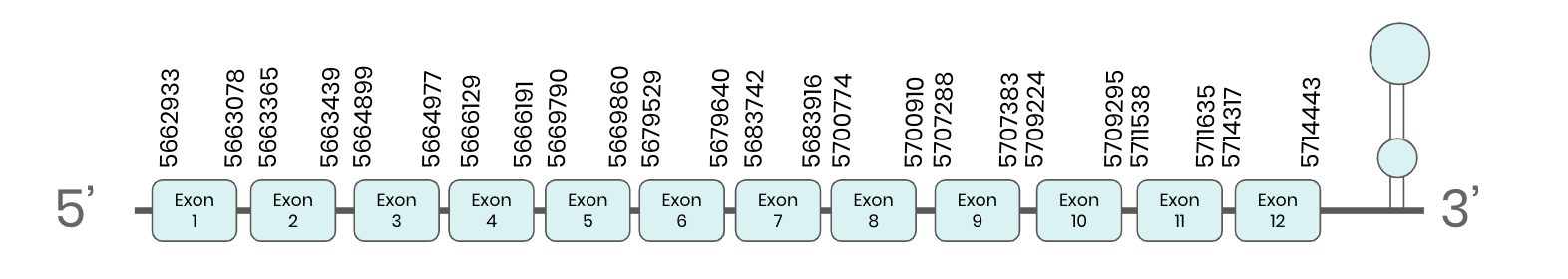
Thus, TR 2 selenoprotein is not present in the Carettochelys insculpta genome because although a selenocysteine element was found, a SECIS was not. Nevertheless, in the reference sequence we could find a selenocysteine as we did in the turtle’s genome. We consider that this protein needs further analysis due to the difficulties in SECIS prediction. Because its sequence is too large, our hypothesis is that the protocol used in this cases affected the sequence of the SECIS element and that this is why Seblastian could not find it.
TR3: several hits in 7 scaffolds were found from tblastn in TR 3, being the hits in scaffold ML683830.1 the one we chose for the analysis. Exonerate predicted that the gene was located between positions 20460135 - 20479549, comprising 16 exons in the reverse strand. T-coffee alignment between Carettochelys insculpta genome and the protein reference (in this case, obtained from Mus musculus) showed high similarity between the sequences and, from this, we extract that there is a coding sequence for this protein in the turtle genome. Seblastian could not predict a SECIS structure at the 3’ end of the genomic sequence. Nevertheless, Seblastian could predict a protein in the same region.
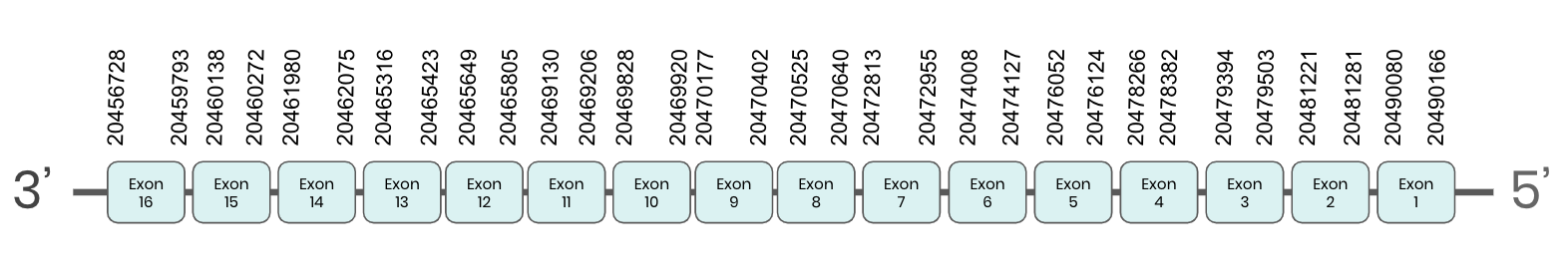
Thus, TR 3 selenoprotein is not present in the Carettochelys insculpta genome because although a selenocysteine element was found, a SECIS was not. Nevertheless, in the reference sequence we could find a selenocysteine as we did in the turtle’s genome. We consider that this protein needs further analysis due to the difficulties in SECIS prediction. Because its sequence is too large, our hypothesis is that the protocol used in these cases affected the sequence of the SECIS element and that this is why Seblastian could not find it.
GLUTATHIONE PEROXIDASE (GPX)
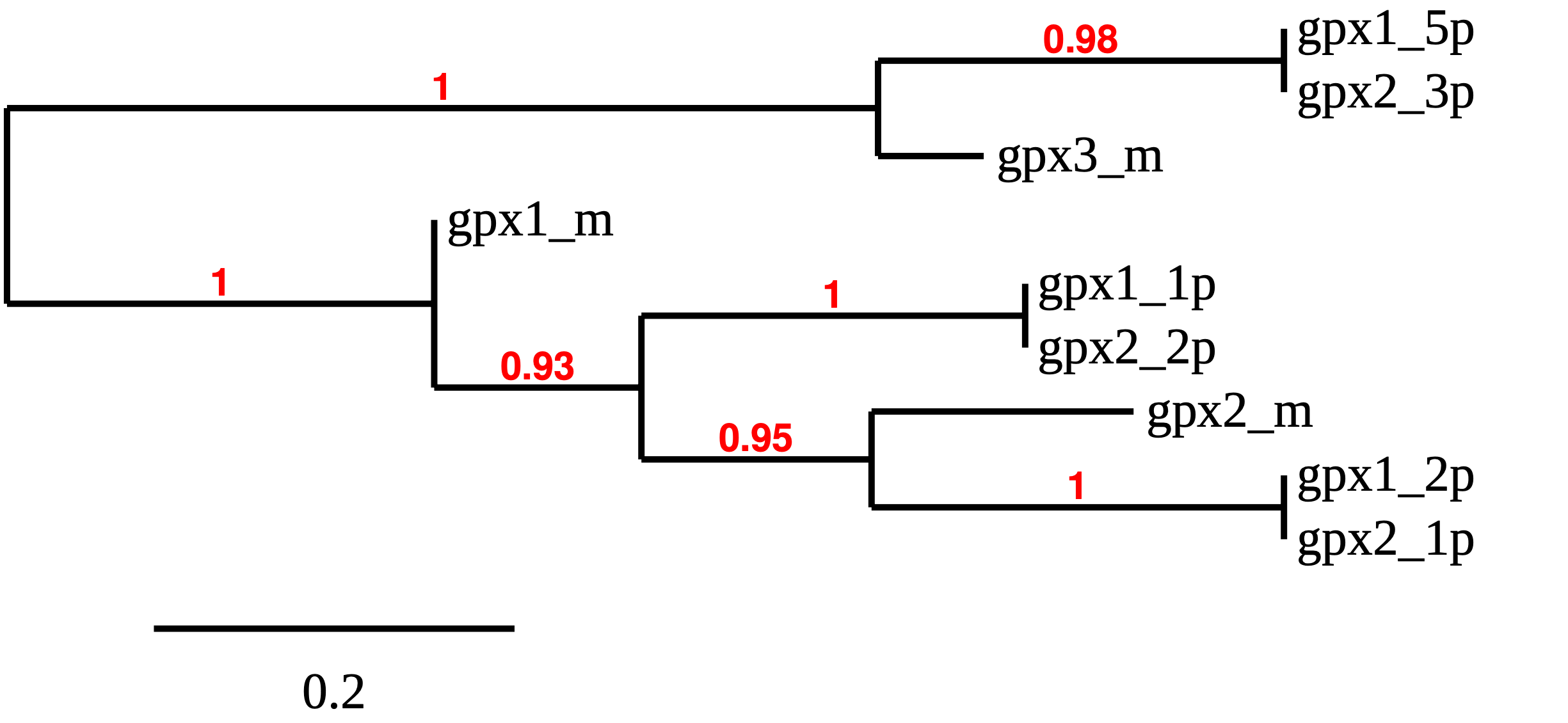
From our tblastn hits, we could see that we have different proteins of this family aligning in the same region of Carettochelys insculpta genome. To be able to solve this puzzle and identify which predicted proteins actually correspond to each of the members of this family, and identify possible duplications and deletions, we had to use a phylogeny (see methods).
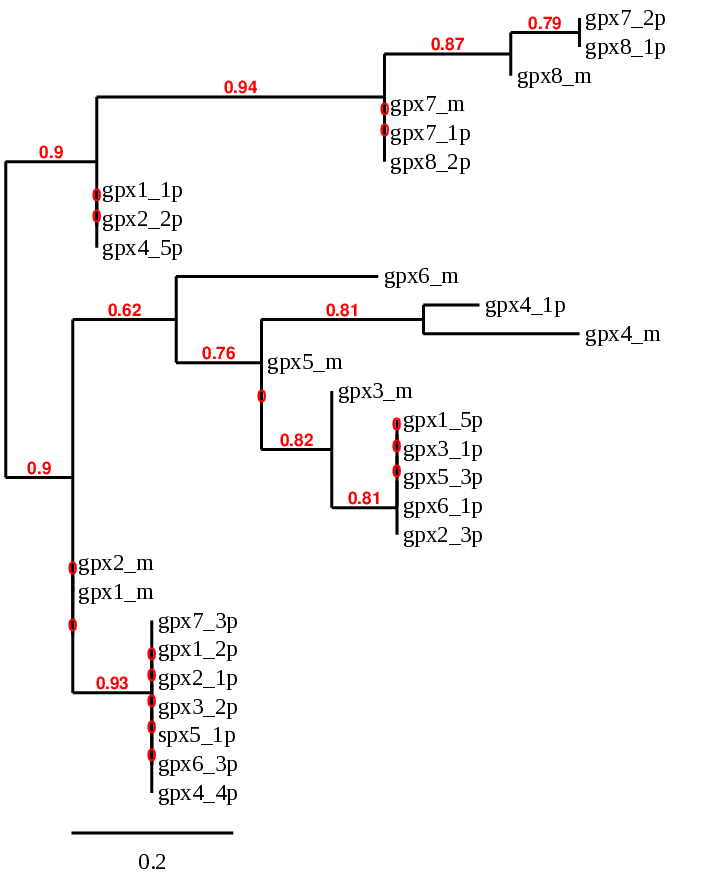
GPX1: several hits in 6 scaffolds were found from tblastn in GPX1, being the hit in scaffold ML683830.1 the one we chose for the analysis. Exonerate predicted that the gene was located between positions 48290797- 48292278, comprising 2 exons in the reverse strand. T-coffee alignment between Carettochelys insculpta genome and the protein reference (in this case, obtained from Mus musculus) showed high similarity between the sequences and, from this, we extract that there is a coding sequence for this protein in the turtle genome. Seblastian could predict a SECIS structure at the 3’ end of the genomic sequence. Nevertheless, Seblastian could not predict a protein in the same region.
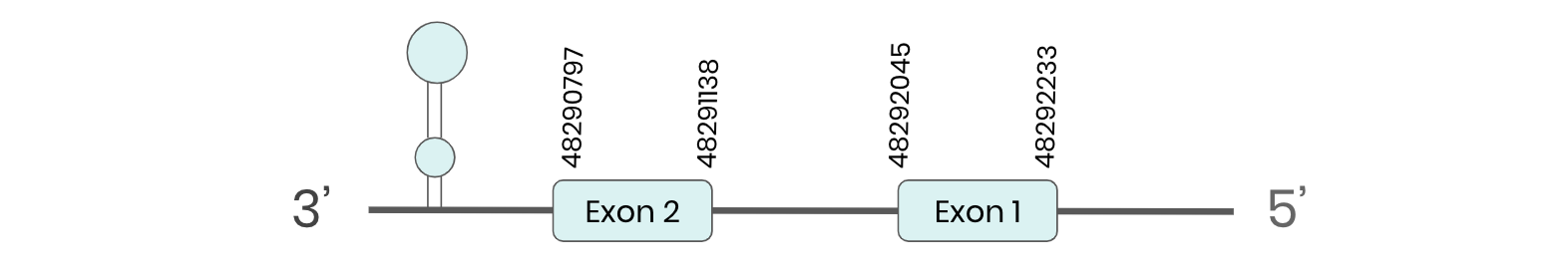
Thus, after looking at the redone tree, we can see that GPX1 is present in the Carettochelys insculpta’s genome as our results predict: a SECIS element and a selenocysteine were found. This result was interesting because, when looking at Anolis carolinensis’ genome, we saw that this protein was not present.
GPX2: several hits in 6 scaffolds were found from tblastn in GPX2, being the hit in scaffold ML683816.1 the one we chose for the analysis. Exonerate predicted that the gene was located between positions 4758110 - 4759914, comprising 2 exons in the reverse strand. T-coffee alignment between Carettochelys insculpta genome and the protein reference (in this case, obtained from Mus musculus) showed high similarity between the sequences and, from this, we extract that there is a coding sequence for this protein in the turtle genome. Seblastian could predict a SECIS structure at the 3’ end of the genomic sequence. Furthermore, Seblastian could predict a protein in the same region.
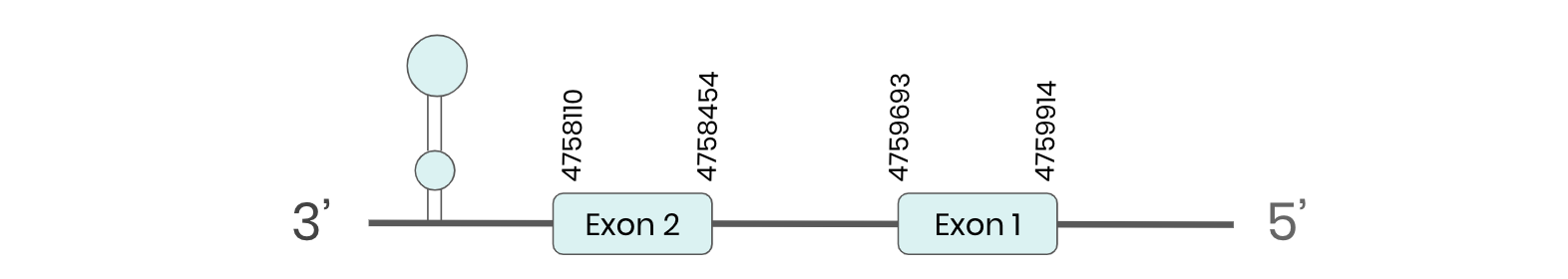
Thus, GPX2 selenoprotein is present in the Carettochelys insculpta genome as a SECIS element and a selenocysteine were found.
GPX3: several hits in 5 scaffolds were found from tblastn in GPX3, being the hit in scaffold ML683799.1 the one we chose for the analysis. Exonerate predicted that the gene was located between positions 30887244-30890286, comprising 5 exons in the forward strand. T-coffee alignment between Carettochelys insculpta genome and the protein reference (in this case, obtained from Mus musculus) showed high similarity between the sequences and, from this, we extract that there is a coding sequence for this protein in the turtle genome. Seblastian could not predict a SECIS structure at the 3’ end of the genomic sequence. Furthermore, Seblastian could not predict a protein in the same region.
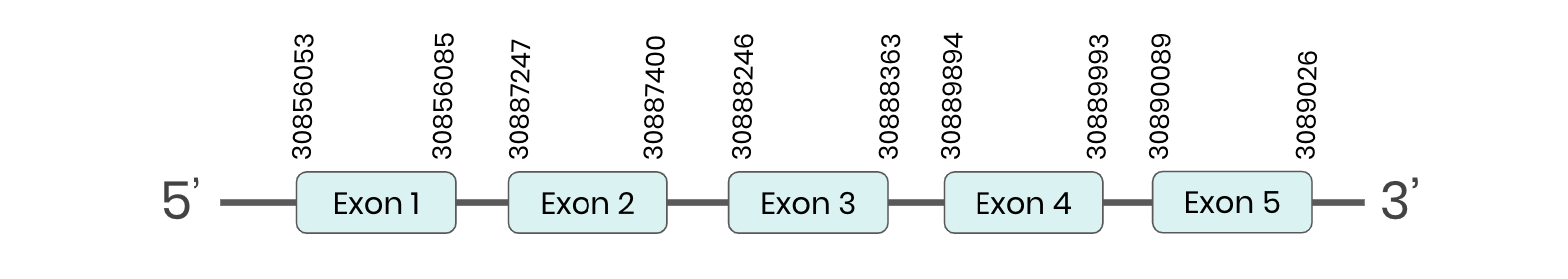
Thus, even though the GPX3 gene is present in the Carettochelys insculpta genome, we cannot consider it a selenoprotein because it doesn’t meet the established criteria: although a selenocysteine was found, a SECIS element was not. Nevertheless, in the reference sequence we could find a selenocysteine as we did in the turtle’s genome. We consider that this protein needs further analysis due to the difficulties in SECIS prediction. Because its sequence is too large, our hypothesis is that the protocol used in this cases affected the sequence of the SECIS element and that this is why Seblastian could not find it.
GPX4: several hits in 6 scaffolds were found from tblastn in GPX4, being the hit in scaffold ML683883.1 the one we chose for the analysis. Exonerate predicted that the gene was located between positions 4779344 - 4780898, comprising 7 exons in the reverse strand. T-coffee alignment between Carettochelys insculpta genome and the protein reference (in this case, obtained from Mus musculus) showed high similarity between the sequences and, from this, we extract that there is a coding sequence for this protein in the turtle genome. Seblastian could predict a SECIS structure at the 3’ end of the genomic sequence. Furthermore, Seblastian could predict a protein in the same region.
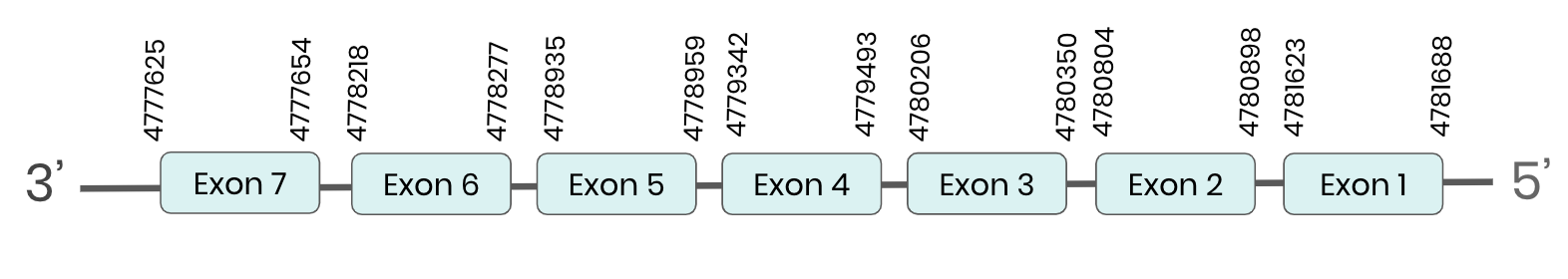
Thus, GPX 4 selenoprotein is present in the Carettochelys insculpta genome as a SECIS element and a selenocysteine were found.
GPX5: several hits in 5 scaffolds were found from tblastn in GPX5, being the hit in scaffold ML683799.1 the one we chose for the analysis. The region was located between positions 30887244-30890280.
GPX6: several hits in 5 scaffolds were found from tblastn in GPX6, being the hit in scaffold ML683799.1 the one we chose for the analysis. The region was located between positions 30887244- 30890286. As we can see, both queries align in the same region of the same scaffold, and this is also the case for GPX 3 (scaffold: ML683799.1; positions: 30887244-30890280). Furthermore, after looking again at the phylogenetic tree, we can hypothesize that GPX 5 and GPX 6 are not present in Carettochelys insculpta genome and the queries are aligning with the region of GPX 3.
This makes sense when looking at the Anolis carolinensis genome because these genes are not present in it, probably meaning a deletion throughout the evolution of reptiles.
GPX7: several hits in 6 scaffolds were found from tblastn in GPX7, being the hit in scaffold ML683911.1 the one we chose for the analysis. Exonerate predicted that the gene was located between positions 7519367-7540018, comprising 3 exons in the forward strand. T-coffee alignment between Carettochelys insculpta genome and the protein reference (in this case, obtained from Mus musculus) showed high similarity between the sequences and, from this, we extract that there is a coding sequence for this protein in the turtle genome. Seblastian could not predict a SECIS structure at the 3’ end of the genomic sequence. Furthermore, Seblastian could not predict a protein in the same region.
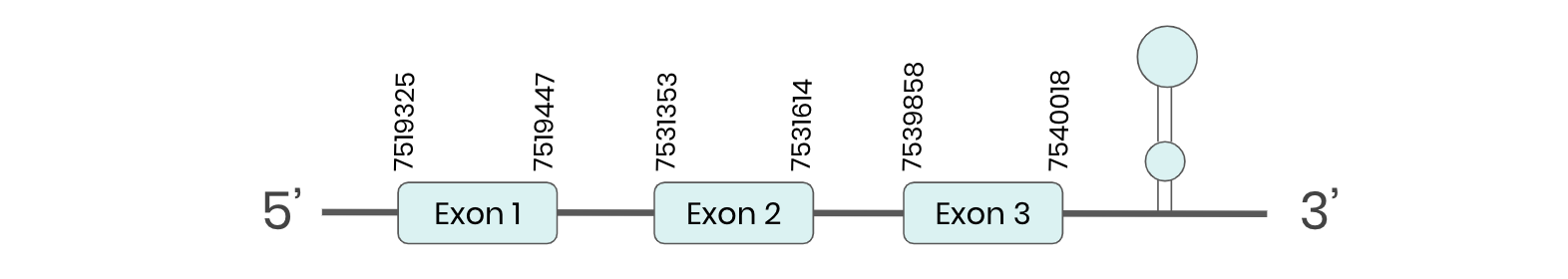
Thus, even though the GPX7 gene is present in the Carettochelys insculpta genome, we cannot consider it a selenoprotein because it doesn’t meet the established criteria: neither the SECIS element nor the selenocysteine were present.
GPX8: several hits in 4 scaffolds were found from tblastn in GPX8, being the hit in scaffold ML683798.1 the one we chose for the analysis. Exonerate predicted that the gene was located between positions 29770397- 29775548, comprising 3 exons in the reverse strand. T-coffee alignment between Carettochelys insculpta genome and the protein reference (in this case, obtained from Mus musculus) showed high similarity between the sequences and, from this, we extract that there is a coding sequence for this protein in the turtle genome. Seblastian could not predict a SECIS structure at the 3’ end of the genomic sequence. Furthermore, Seblastian could not predict a protein in the same region.
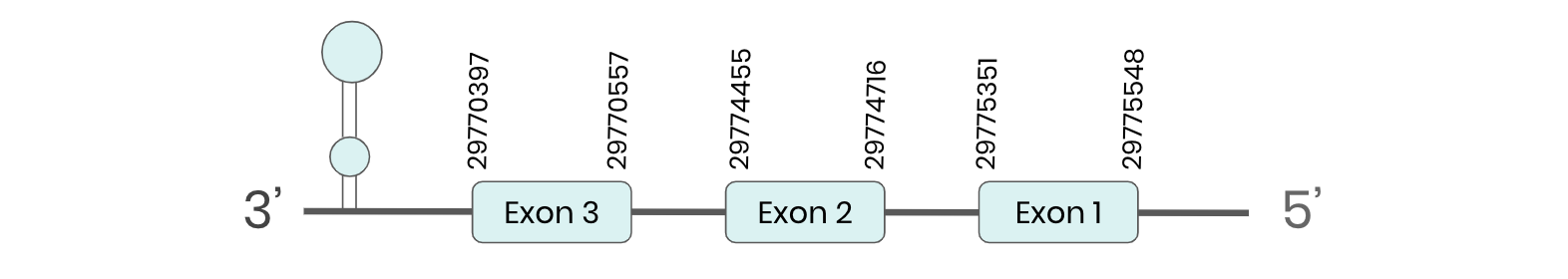
Thus, even though the GPX8 gene is present in the Carettochelys insculpta genome, we cannot consider it a selenoprotein because it doesn’t meet the established criteria: neither the SECIS element nor the selenocysteine were present.
IODOTHYRONINE DEIODINASES (DI)
Although the protocol followed for the analysis consisted in taking together all the hits found in the best scaffolds, the fact that we manually checked our TBLASTn hits allowed us to identify situations in which the hits found in a given scaffold were analyzed separately because they aligned overlapping regions of the query in non-overlapping regions of the scaffold, indicating two different regions of interest in the same scaffold. This was the case of this protein family.
DI1: several hits in 2 scaffolds were found from tblastn in DI1, being the hits in scaffold ML683816.1 (positions for the first region 102620543-102621208, and positions 86731875- 8673124 for the second region) the ones that passes our filters.
DI2: several hits in 2 scaffolds were found from tblastn in DI2, being the hits in scaffold ML683816.1 (positions for the first region 86731294 - 86746134 and positions 102620645-102621199 for the second region) the ones that passed our filters.
Furthermore, as both of the splitted sequences of both proteins (DI1 and DI2) aligned in very similar positions of the same scaffold (ML683816.1), a phylogenetic analysis was performed in order to shed light on what was going on in this case.
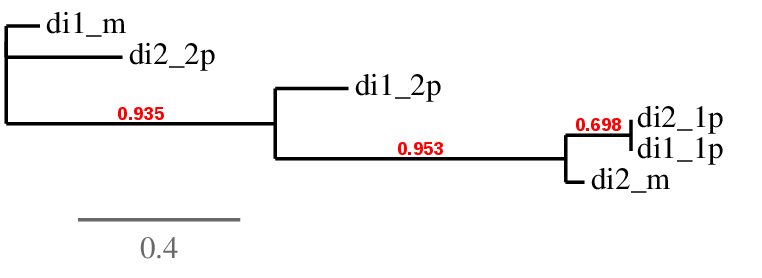
Taking into account the tree and the data analysis, we hypothesize that the predicted DI2_1 is actually the same as DI1_2 and correspond to DI2 query. On the other hand, from the phylogenetic tree we deduce DI1_1 is the prediction for DI1 query even though it is further away than expected.
Therefore we continue the analysis:
DI1: several hits in 2 scaffolds were found from tblastn in DI1, being the hits in contig ML683816.1 the one we chose for the analysis. Exonerate predicted that the gene was located between positions 102620543-102621208 comprising one exon in the forward strand. T-coffee prediction of the alignment between Carettochelys insculpta genome and the protein reference (in this case, obtained from Mus musculus) showed a coding sequence for this protein in the turtle genome. Seblastian could predict a SECIS structure at the 3’ end of the genomic sequence. Nevertheless, Seblastian could not predict a protein in the same scaffold.
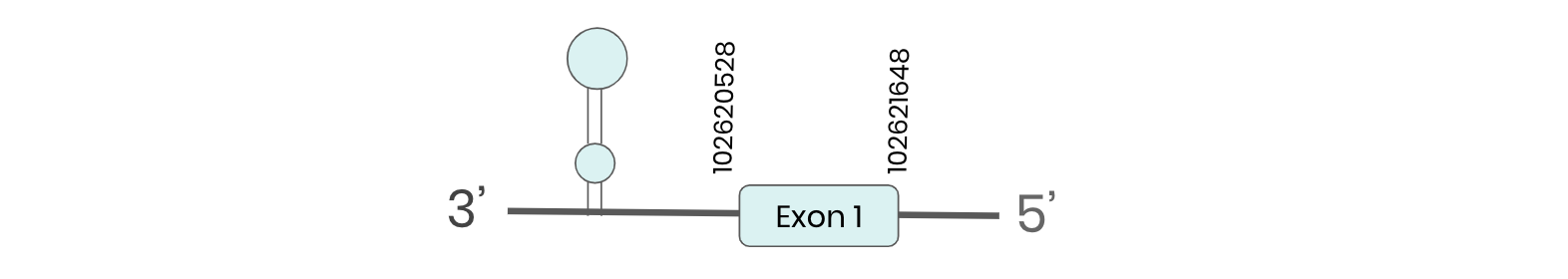
Thus, DI1 selenoprotein is not present in the Carettochelys insculpta genome because although a SECIS element was found in the first region of interest, a selenocysteine was not in any of the two analysed regions.
For DI2: exonerate predicted that the gene was located between positions 86731294- 86746134 comprised 2 exons in the reverse strand. T-coffee alignment between Carettochelys insculpta predicted protein and the protein reference (in this case, obtained from Mus musculus) showed high similarity between the sequences and, from this, we extract that there is a coding sequence for this protein in the turtle genome. Seblastian could predict a SECIS structure at the 3’ end of the genomic sequence. Furthermore, Seblastian could predict a protein in the same region.
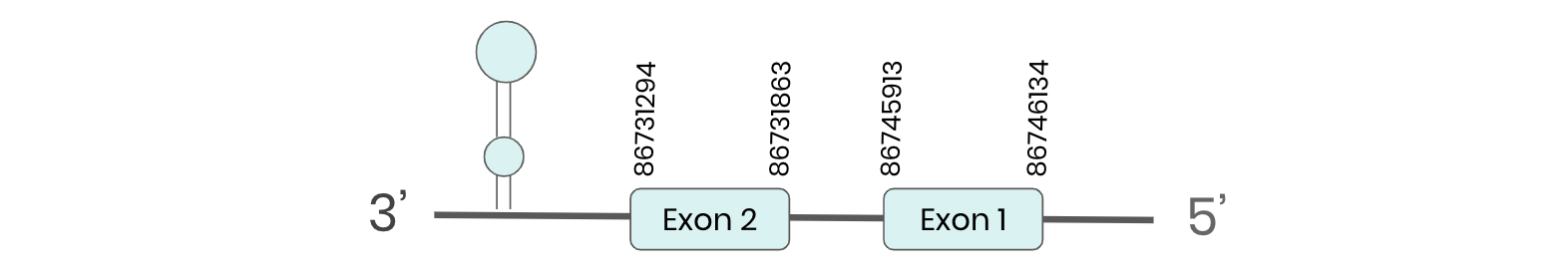
Altogether, the predicted proteins cannot be considered selenoproteins, because it they do not meet the established criteria mentioned above: although SECIS elements were found in both regions, a selenocysteine was not found in any of them.
These results fall within expectations, given that the sequence of the reference protein does not present a selenocysteine either. For this reason, according to the criteria used, it would not be considered a selenoprotein.
Selenoprotein P
SEL P: based on the scaffolds selected according to the criteria previously described (e-value, coverage; see methodology) a phylogenetic analysis was performed in order to identify those which were more similar to the query protein chosen. From there, the study was carried out with a single region, the closest to the reference protein in the phylogenetic tree.
Several hits in 2 scaffolds were found from tblastn in SEL P, being the hits in scaffold ML683798.1 the one we chose for the analysis. Exonerate predicted that the gene was located between positions 34197594-34204279, comprising 3 exons in the forward strand. T-coffee alignment between Carettochelys insculpta genome and the protein reference (in this case, obtained from Anolis carolinensis) showed high similarity between the sequences and, from this, we extract that there is a coding sequence for this protein in the turtle genome. Seblastian could predict a SECIS structure at the 3’ end of the genomic sequence. Furthermore, Seblastian could predict a protein in the same region.
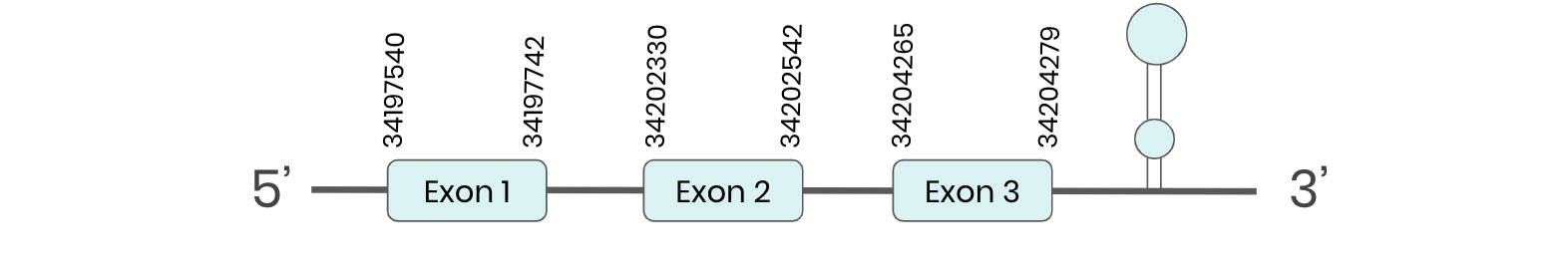
Thus, SEL P selenoprotein is present in the Carettochelys insculpta genome as a SECIS element and a selenocysteine were found.
Selenoprotein W
SEL W2: several hits in 2 scaffolds were found from tblastn in SEL W2, being the hits in scaffold ML683865.1 the one we chose for the analysis as it is the only one that passed our filters. Exonerate predicted that the gene was located between positions 12864592 - 12866698, comprising 3 exons in the forward strand. T-coffee alignment between Carettochelys insculpta genome and the protein reference (in this case, obtained from Mus musculus) showed high similarity between the sequences and, from this, we extract that there is a coding sequence for this protein in the turtle genome. Seblastian could predict a SECIS structure at the 3’ end of the genomic sequence. Nevertheless, Seblastian could not predict a protein in the same region.
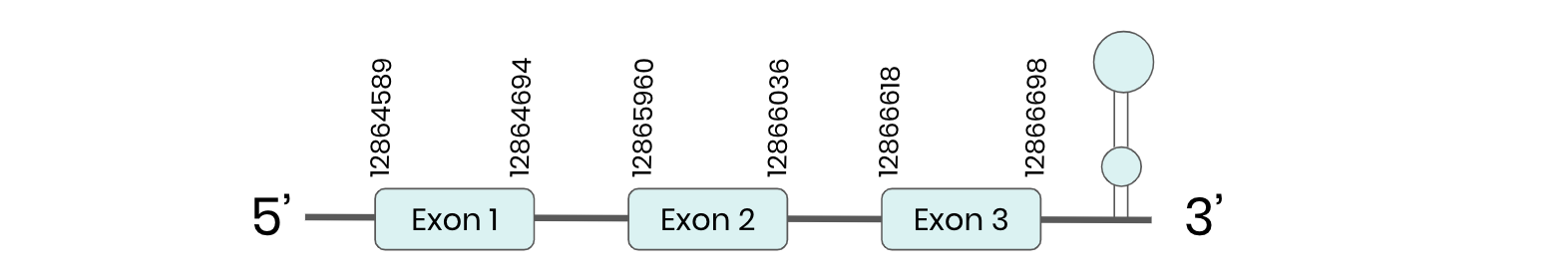
Thus, even though the SEL W2 gene is present in the Carettochelys insculpta genome, we cannot consider it a selenoprotein because it doesn’t meet the established criteria: although a SECIS element was found, a selenocysteine was not.
This results falls within expectations, given that the sequence of reference protein does not present a selenocysteine either. For this reason, according to the criteria used, it would not be considered a selenoprotein.
Selenoprotein S
SEL S: the protein was first analyzed in Mus musculus and as it had a very low coverage we analyzed it afterwards in Anolis carolinensis expecting a better result but again it did not pass the filters.
Our hypothesis is that this protein is not present in Carettochelys insculpta’s genome or it is very different from the reference protein. And due to these reasons, it did not pass the filter, with a percentage of coverage lower than 60%.
Selenoprotein 15
SEL15: Several hits in the ML683911.1 scaffold were found from tblastn in SEL 15. Exonerate predicted that the gene was located between positions 26894675-26917169, comprising 4 exons in the reverse strand. T-coffee alignment between Carettochelys insculpta genome and the protein reference (in this case, obtained from Mus musculus) showed high similarity between the sequences and, from this, we extract that there is a coding sequence for this protein in the turtle genome. Seblastian could predict a SECIS structure at the 3’ end of the genomic sequence. Furthermore, Seblastian could predict a protein in the same region.
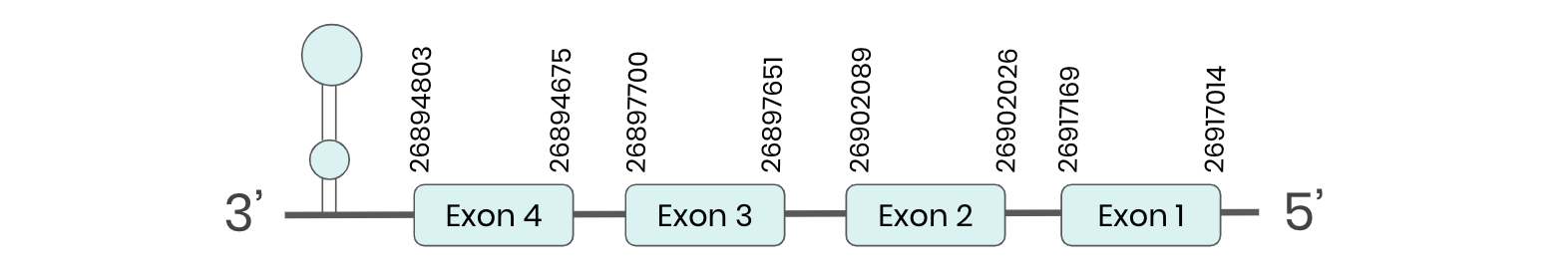
Thus, SEL 15 selenoprotein is present in the Carettochelys insculpta genome as a SECIS element and a selenocysteine were found.
Selenoprotein M
SEL M: several hits in the ML683833.1 scaffold were found from tblastn in SEL M. Exonerate predicted that the gene was located between positions 17903382-1704095, comprising 3 exons in the reverse strand. T-coffee alignment between Carettochelys insculpta genome and the protein reference (in this case, obtained from Anolis carolinensis) showed high similarity between the sequences and, from this, we extract that there is a coding sequence for this protein in the turtle genome. Seblastian could predict a SECIS structure at the 3’ end of the genomic sequence. Furthermore, Seblastian could predict a protein in the same region.
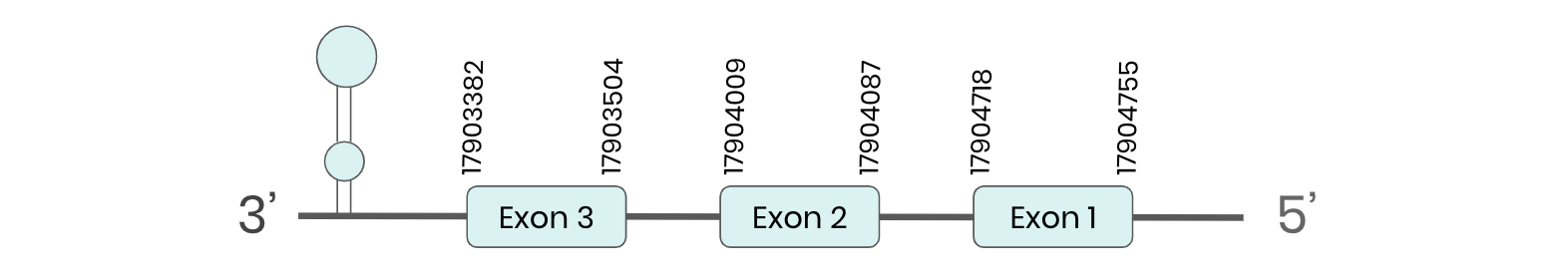
Thus, even though the SEL M gene is present in the Carettochelys insculpta genome, we cannot consider it a selenoprotein because it doesn’t meet the established criteria: although a SECIS element was found, a selenocysteine was not.
Since in the query used as a reference a U (Sec) is present and Seblastian could predict a SECIS element, our hypothesis is that this selenoprotein has been lost in the turtle recently in evolutionary terms.
Selenoprotein R
MSRB 1: several hits in 1 scaffold were found from tblastn in MSRB 1, being the hits in scaffold ML683829.1 the one we chose for the analysis. Exonerate predicted that the gene was located between positions 4053677-4054112, comprising 2 exons in the forward strand. T-coffee alignment between Carettochelys insculpta genome and the protein reference (in this case, obtained from Anolis carolinensis) showed high similarity between the sequences and, from this, we extract that there is a coding sequence for this protein in the turtle genome. Seblastian could predict a SECIS structure at the 3’ end of the genomic sequence. Furthermore, Seblastian could predict a protein in the same region.
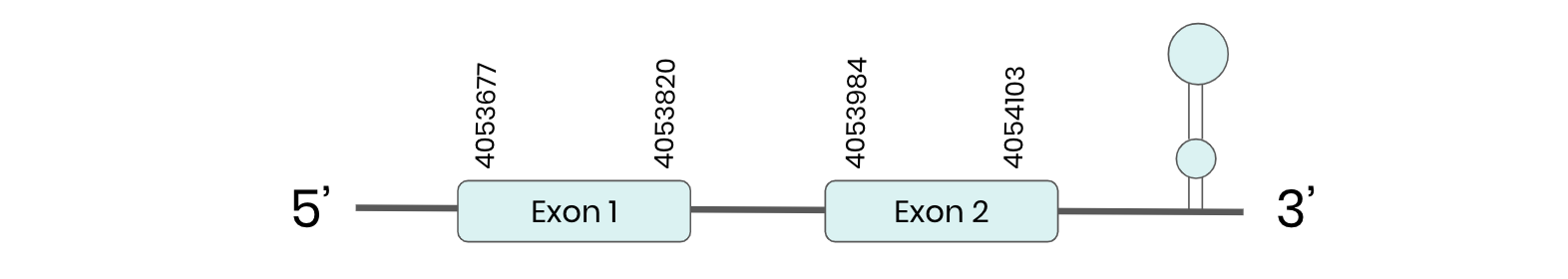
Thus, MSRB 1 selenoprotein is present in the Carettochelys insculpta genome as a SECIS element and a selenocysteine were found.
MSRB 2: the protein was directly analyzed in Anolis carolinensis but it did not pass the filters.
Our hypothesis is that this protein is not present in Carettochelys insculpta’s genome or it is very different from the reference protein. And due to these reasons, it did not pass the filter, with a percentage of coverage lower than 60%.
MSRB 3: several hits in 1 scaffold were found from tblastn in MSRB3, being the hits in scaffold ML683785.1 the one we chose for the analysis. Exonerate predicted that the gene was located between positions 55146461-55232218, comprising 6 exons in the reverse strand. T-coffee alignment between Carettochelys insculpta genome and the protein reference (in this case, obtained from Anolis carolinensis) showed high similarity between the sequences and, from this, we extract that there is a coding sequence for this protein in the turtle genome. Seblastian could not predict a SECIS structure at the 3’ end of the genomic sequence. Furthermore, Seblastian could not predict a protein in the same region.
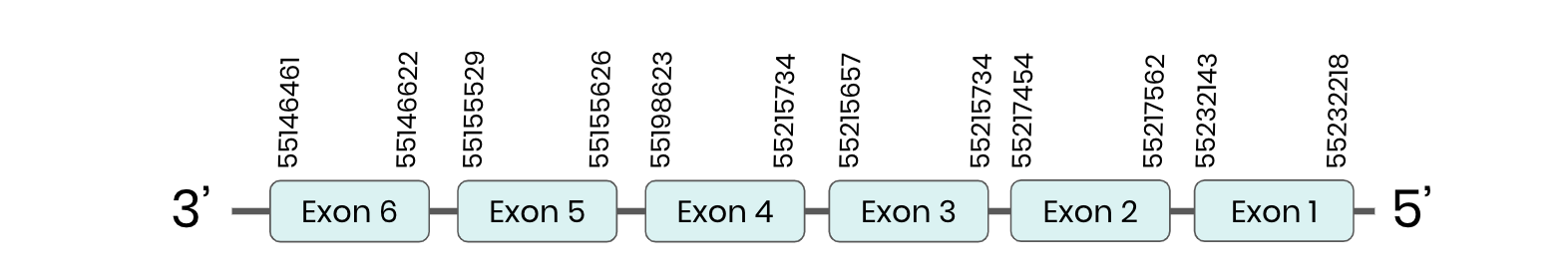
Thus, even though the MSRB 3 gene is present in the Carettochelys insculpta genome, we cannot consider it a selenoprotein because it doesn’t meet the established criteria: neither the SECIS element nor the selenocysteine were present.
Methionine sulfoxide reductase A
MSRA: several hits in 4 scaffolds were found from tblastn in MSRA, being the hits in scaffold ML683834.1 the one we chose for the analysis. Exonerate predicted that the gene was located between positions 6139053-6268719, comprising 4 exons in the forward strand. T-coffee alignment between Carettochelys insculpta genome and the protein reference (in this case, obtained from Mus musculus) showed high similarity between the sequences and, from this, we extract that there is a coding sequence for this protein in the turtle genome.
In order to run Seblastian properly we needed to split the scaffold in two because the hits were separated more than 100.000 positions amongst each other and the programme did not accept sequences of such length .
Seblastian could not predict a SECIS structure at any of the 3’ end of the genomic sequences. Furthermore, Seblastian could not predict any protein in the same region.
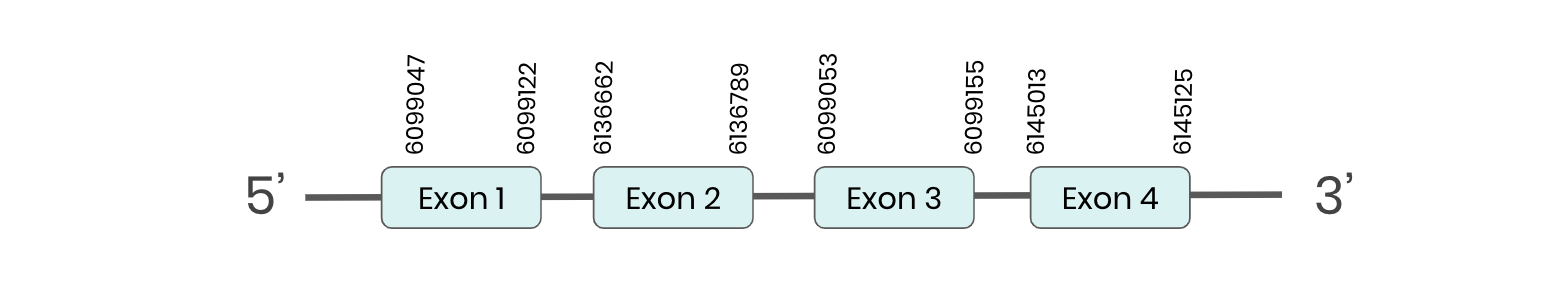
Thus, even though the MSRA gene is present in the Carettochelys insculpta genome, we cannot consider it a selenoprotein because it doesn’t meet the established criteria: neither the SECIS element nor the selenocysteine were present.
Selenoprotein U
Based on the scaffolds selected according to the criteria previously described (e-value, coverage; see methodology) a phylogenetic analysis was performed in order to identify those which were more similar to the query protein chosen. From there, the study was carried out with a single region, the closest to the reference protein in the phylogenetic tree.
SEL U1: several hits in the ML683777.1 scaffold were found from tblastn in SEL U1. Exonerate predicted that the gene was located between positions 58477660-58487608, comprising 5 exons in the reverse strand. T-coffee alignment between Carettochelys insculpta genome and the protein reference (in this case, obtained from Anolis carolinensis) showed high similarity between the sequences and, from this, we extract that there is a coding sequence for this protein in the turtle genome. Seblastian could predict a SECIS structure at the 3’ end of the genomic sequence. Nevertheless, Seblastian could not predict a protein in the same region.
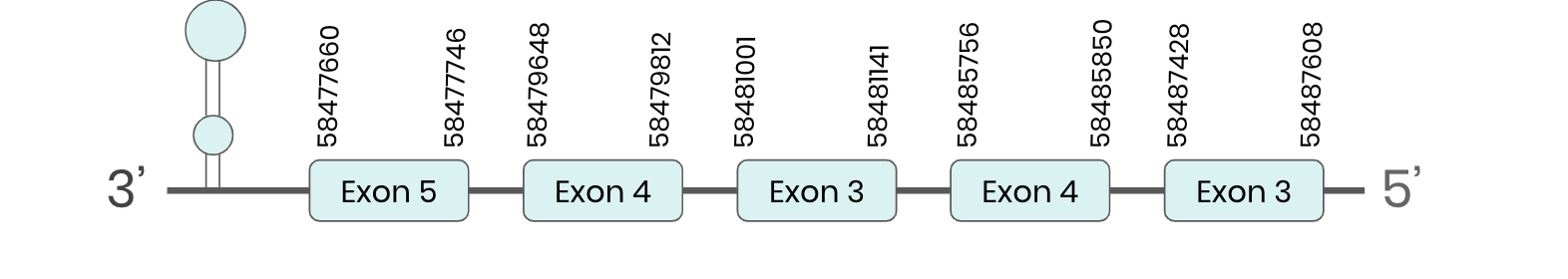
Thus, even though the SEL U1 gene is present in the Carettochelys insculpta genome, we cannot consider it a selenoprotein because it doesn’t meet the established criteria: although a SECIS element was found, a selenocysteine was not.
Since in the query used as a reference a U (Sec) is present and, in the same position, a C (Cys) can be found in the turtle’s genome, our hypothesis is that this protein suffered a conversion from Sec to Cys recently in evolutionary terms. Because of this, Sel U1 is a cysteine-containing homologue protein.
SEL U2: several hits in the ML683929.1 scaffold were found from tblastn in SEL U2. Exonerate predicted that the gene was located between positions 484030-489152, comprising 6 exons in the reverse strand. T-coffee alignment between Carettochelys insculpta genome and the protein reference (in this case, obtained from Anolis carolinensis) showed high similarity between the sequences and, from this, we extract that there is a coding sequence for this protein in the turtle genome. Seblastian could not predict a SECIS structure at the 3’ end of the genomic sequence. Furthermore, Seblastian could not predict a protein in the same region.
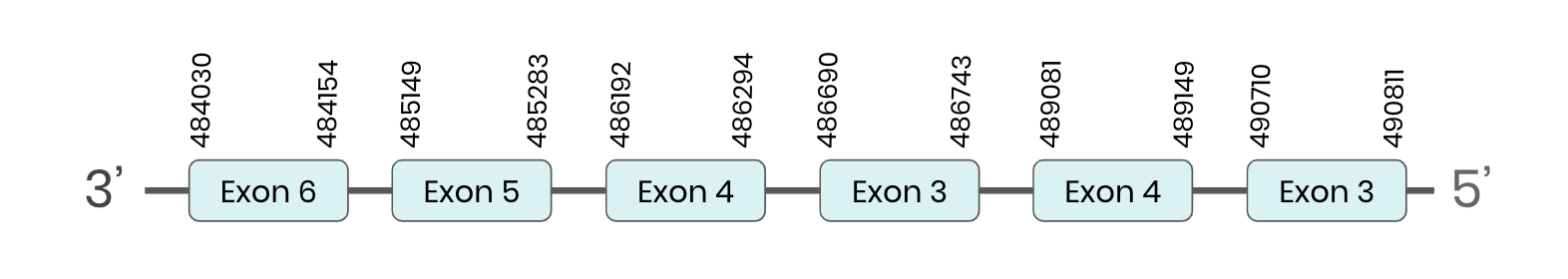
Thus, even though the SEL U2 gene is present in the Carettochelys insculpta genome, we cannot consider it a selenoprotein because it doesn’t meet the established criteria: neither the SECIS element nor the selenocysteine were present.
Selenoprotein T
SEL T: several hits in 5 scaffolds were found from tblastn in SEL T, being the hits in scaffold ML683805.1 the one we chose for the analysis. Exonerate predicted that the gene was located between positions 24996193 - 25003335, comprising 5 exons in the forward strand. T-coffee alignment between Carettochelys insculpta genome and the protein reference (in this case, obtained from Mus musculus) showed high similarity between the sequences and, from this, we extract that there is a coding sequence for this protein in the turtle genome. Seblastian could predict a SECIS structure at the 3’ end of the genomic sequence. Nevertheless, Seblastian could not predict a protein in the same region.
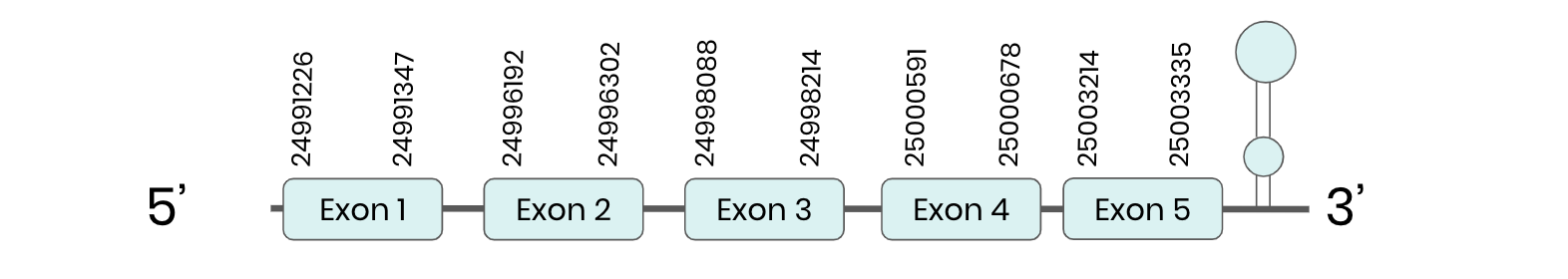
Thus, SEL T selenoprotein is present in the Carettochelys insculpta genome as a SECIS element and a selenocysteine were found.
Selenoprotein H
SEL H: several hits in the ML683816.1 scaffold were found from tblastn in SEL H. Exonerate predicted that the gene was located between positions 5901356-5902068, comprising 2 exons in the forward strand. T-coffee alignment between Carettochelys insculpta genome and the protein reference (in this case, obtained from Anolis carolinensis) showed high similarity between the sequences and, from this, we extract that there is a coding sequence for this protein in the turtle genome. Seblastian could predict a SECIS structure at the 3’ end of the genomic sequence. Furthermore, Seblastian could predict a protein in the same region.
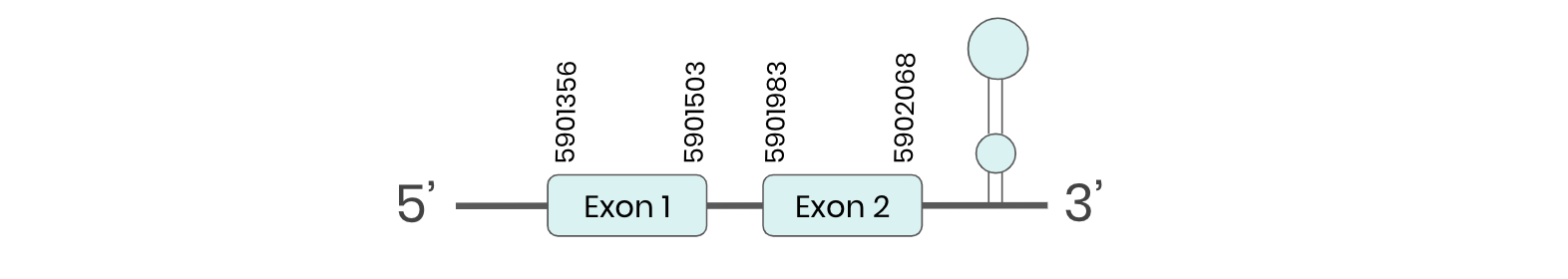
Thus, SEL H selenoprotein is present in the Carettochelys insculpta genome as a SECIS element and a selenocysteine were found.
Selenoprotein I
SEL I: several hits in 4 scaffolds were found from tblastn in SEL I, being the hits in scaffold ML683834.1 the one we chose for the analysis. Exonerate predicted that the gene was located between positions 8137936 - 8151418, comprising 10 exons in the forward strand. T-coffee alignment between Carettochelys insculpta genome and the protein reference (in this case, obtained from Mus musculus) showed high similarity between the sequences and, from this, we extract that there is a coding sequence for this protein in the turtle genome. Seblastian could predict a SECIS structure at the 3’ end of the genomic sequence. Furthermore, Seblastian could predict a protein in the same region.
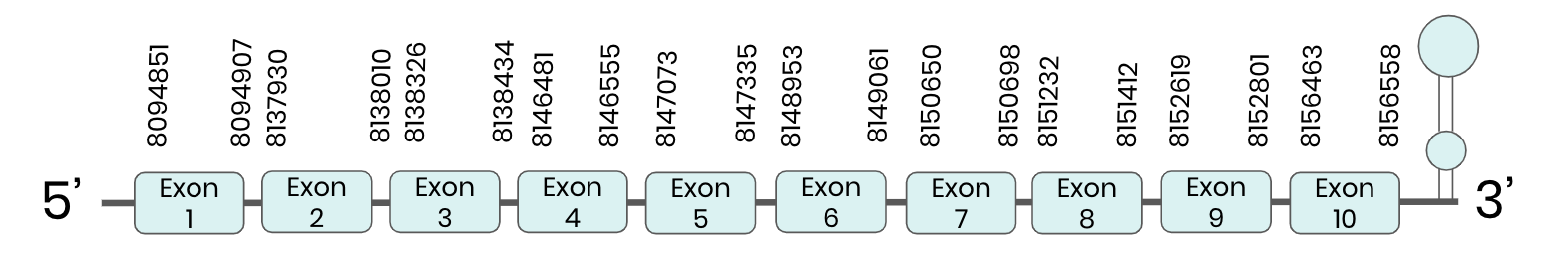
Thus, SEL I selenoprotein is present in the Carettochelys insculpta genome as a SECIS element and a selenocysteine were found.
Selenoprotein K
SEL K: the protein was first analyzed in Mus musculus and as it had a very low coverage we analyzed it afterwards in Anolis carolinensis expecting a better result but again it did not pass the filters.
Our hypothesis is that this protein is not present in Carettochelys insculpta’s genome or it is very different from the reference protein. And due to this reason, it did not pass the filter, with a percentage of coverage lower than 60%.
Selenoprotein O
SEL O: several hits in 2 scaffolds were found from tblastn in SEL O, being the hits in scaffold ML683802.1 the one we chose for the analysis. Exonerate predicted that the gene was located between positions 32545077-32560228, comprising 9 exons in the forward strand. T-coffee alignment between Carettochelys insculpta genome and the protein reference (in this case, obtained from Anolis carolinensis) showed high similarity between the sequences and, from this, we extract that there is a coding sequence for this protein in the turtle genome. Seblastian could not predict a SECIS structure at the 3’ end of the genomic sequence. Furthermore, Seblastian could not predict a protein in the same region.
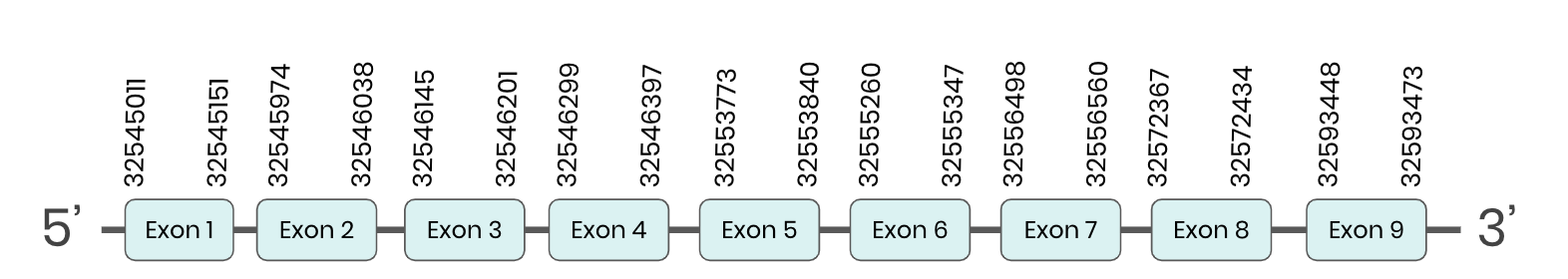
Thus, even though the SEL O gene is present in the Carettochelys insculpta genome, we cannot consider it a selenoprotein because it doesn’t meet the established criteria: neither the SECIS element nor the selenocysteine were present.
Selenoprotein N
In the case of SEL N, we performed a phylogenetic analysis because, from tblastn we got 2 two scaffolds aligning with the same region of the reference protein. Both accomplished the criteria to be analyzed. With this analysis, we were able to hypothesize that there was a duplication in the Carettochelys insculpta’s genome. The following data analysis supported this idea:
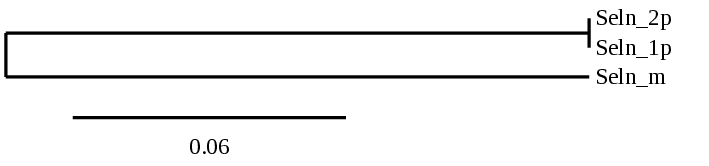
Several hits in 2 scaffolds were found from tblastn in SEL N, being the hits in scaffolds ML683823.1 and ML683824.1 the ones we chose for the analysis. Exonerate predicted that the genes were located between positions 7077804-7097351 and 7082400-7101945, respectively. Both are comprising 11 exons in the reverse strand. T-coffee alignments between Carettochelys insculpta genome and the protein reference (in this case, obtained from Mus musculus) showed high similarity between the sequences and, from this, we extract that there are two coding sequences for these proteins in the turtle genome. Seblastian could predict SECIS structures at the 3’ end of the genomic sequences. Nevertheless, Seblastian could not predict any protein in the same region.
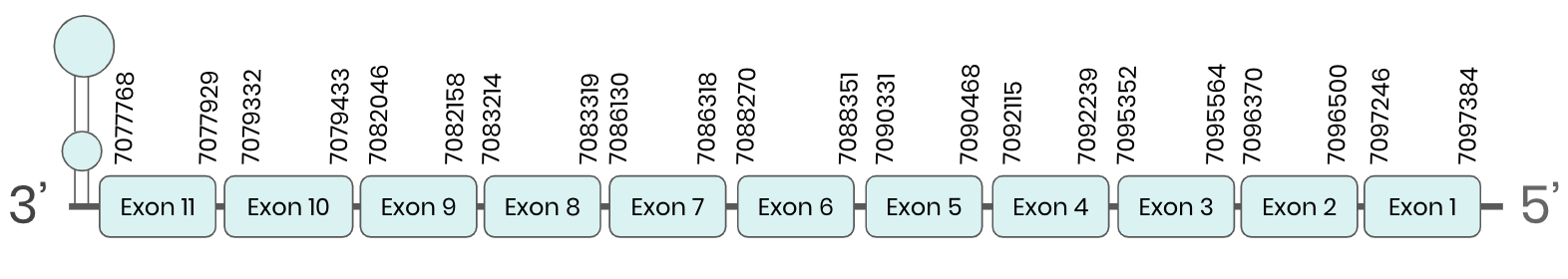
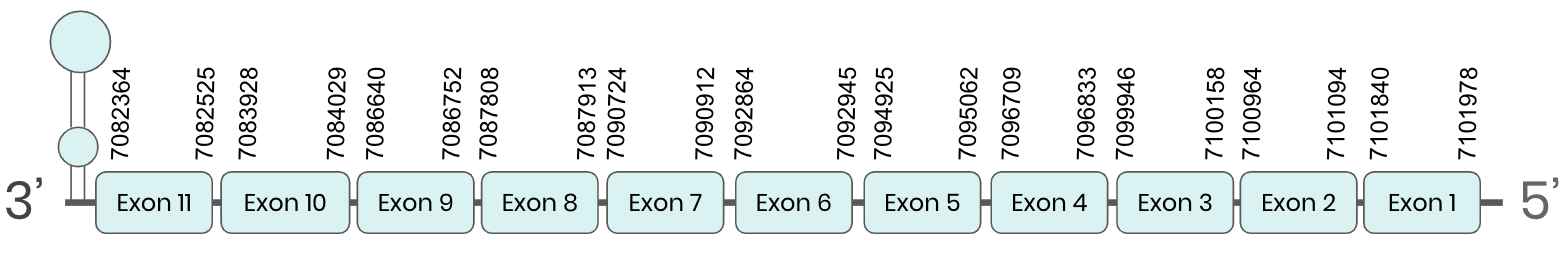
Thus, SEL N selenoprotein is present in the Carettochelys insculpta genome as a SECIS element and a selenocysteine were found. Furthermore, from the analysis we have done, we were able to conclude that there was a duplication in the turtle’s genome.
Translation machinery
eEFsec: several hits in the ML683830.1 scaffold were found from tblastn in eEFsec. Exonerate predicted that the gene was located between positions 22337455-22338099, comprising 1 exon in the forward strand. T-coffee alignment between Carettochelys insculpta genome and the protein reference (in this case, obtained from Anolis carolinensis) showed high similarity between the sequences and, from this, we extract that there is a coding sequence for this protein in the turtle genome. Seblastian could not predict a protein in the same region.
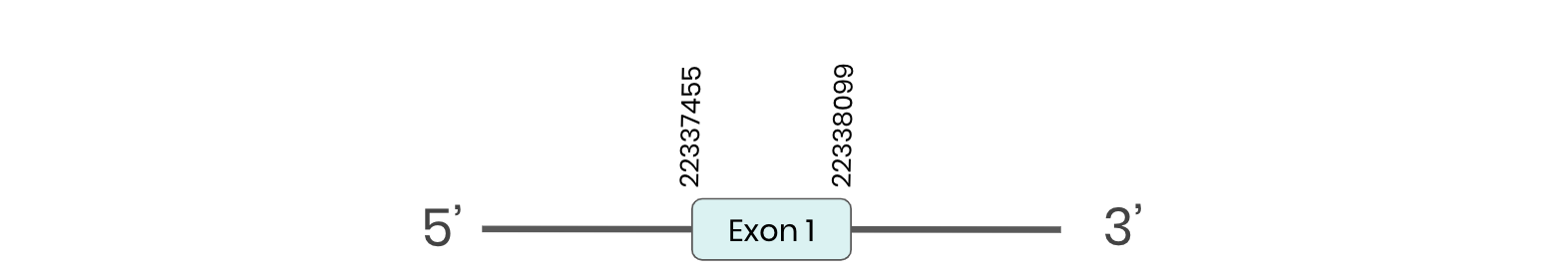
In conclusion, eEFsec was found in the Carettochelys insculpta genome.
Sbp2: several hits in 2 scaffolds were found from tblastn in sbp2, being the hits in scaffold ML683808.1 the one we chose for the analysis. Exonerate predicted that the gene was located between positions 24553077-24583094, comprising 16 exons in the reverse strand. T-coffee alignment between Carettochelys insculpta genome and the protein reference (in this case, obtained from Anolis carolinensis) showed high similarity between the sequences and, from this, we extract that there is a coding sequence for this protein in the turtle genome. Seblastian could not predict a protein in the same region.
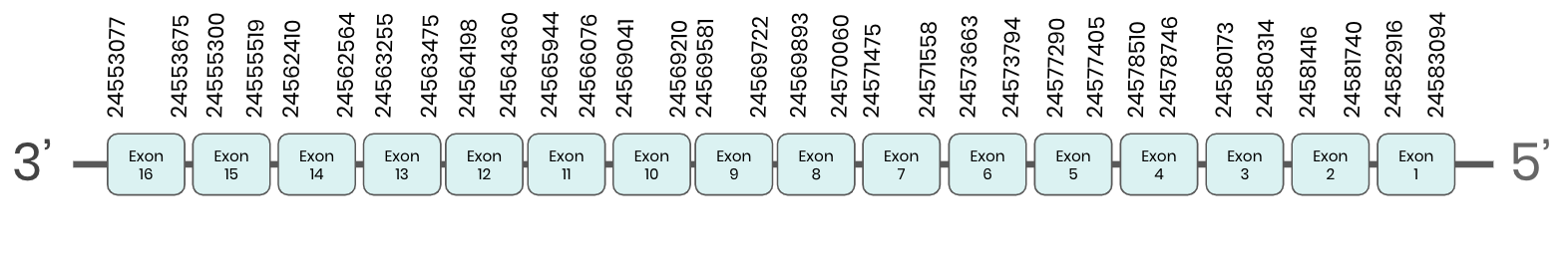
In conclusion, SBP 2 was found in the Carettochelys insculpta genome.
Pstk: several hits in the ML683777.1 scaffold were found from tblastn in PSTK. Exonerate predicted that the gene was located between positions 20969139-20977362, comprising 6 exons in the forward strand. T-coffee alignment between Carettochelys insculpta genome and the protein reference (in this case, obtained from Anolis carolinensis) showed high similarity between the sequences and, from this, we extract that there is a coding sequence for this protein in the turtle genome. Seblastian could not predict a protein in the same region.
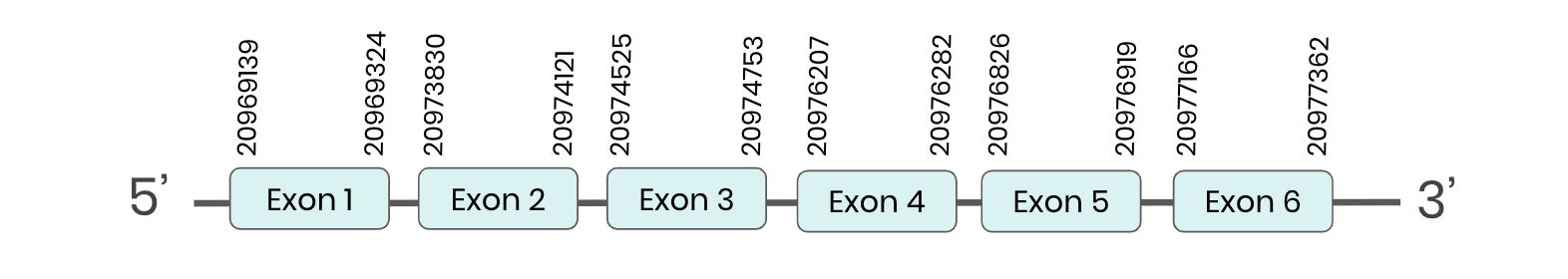
In conclusion, PSTK was found in the Carettochelys insculpta genome.
Secp43: the protein was directly analyzed in Anolis carolinensis but it did not pass the filters.
Our hypothesis is that this protein is not present in Carettochelys insculpta’s genome or it is very different from the reference protein. And due to these reasons, it did not pass the filter, with a percentage of coverage lower than 60%.
SecS: several hits in the ML683766.1 scaffold were found from tblastn in secS. Exonerate predicted that the gene was located between positions 32663167-32696188, comprising 11 exons in the reverse strand. T-coffee alignment between Carettochelys insculpta genome and the protein reference (in this case, obtained from Anolis carolinensis) showed high similarity between the sequences and, from this, we extract that there is a coding sequence for this protein in the turtle genome. Seblastian could not predict a protein in the same region.
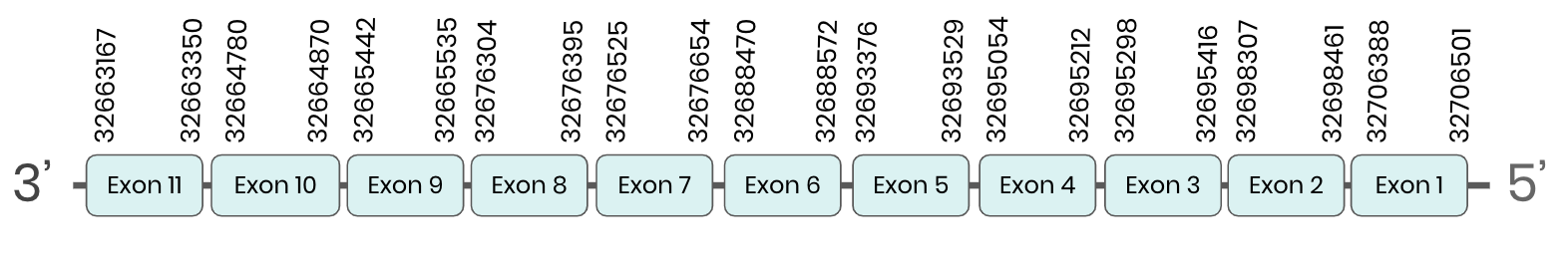
In conclusion, secS was found in the Carettochelys insculpta genome.
SELENOPHOSPHATE SYNTHETASE (SPS)
Based on the scaffolds selected according to the criteria previously described (e-value, coverage; see methodology) a phylogenetic analysis was performed in order to identify those which were more similar to the query protein chosen. From there, the study was carried out with a single scaffold, the closest to the reference protein in the phylogenetic tree.
SPS1: several hits in 2 scaffolds were found from tblastn in SPS 1, being the hits in scaffold ML683785.1 the one we chose for the analysis. Exonerate predicted that the gene was located between positions 106782848-106809201, comprising 8 exons in the forward strand. T-coffee alignment between Carettochelys insculpta genome and the protein reference (in this case, obtained from Mus musculus) showed high similarity between the sequences and, from this, we extract that there is a coding sequence for this protein in the turtle genome. Seblastian could not predict a protein in the same region.
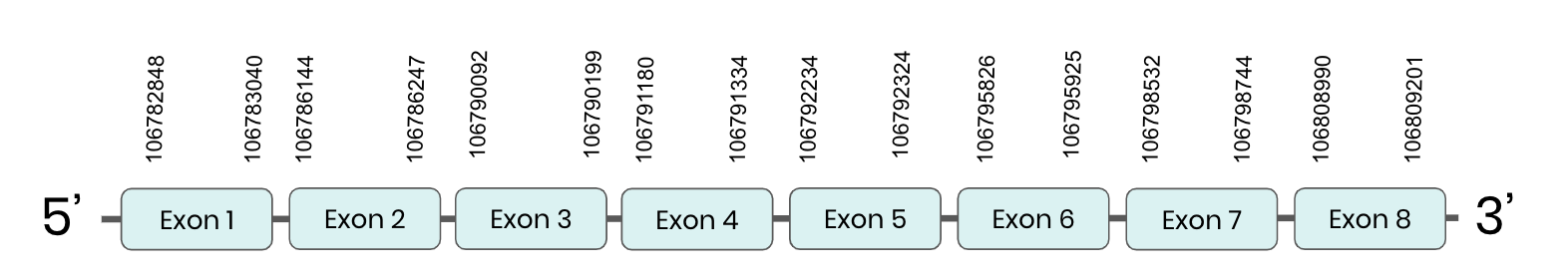
In conclusion, SPS 1 was found in the Carettochelys insculpta genome.
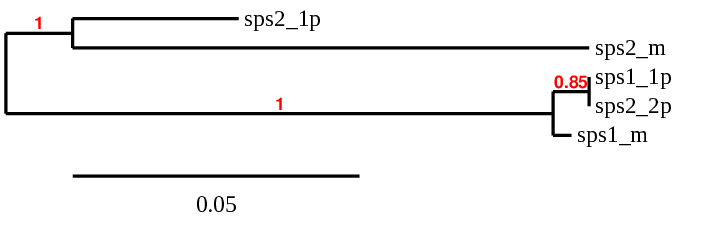
SPS2: several hits in 2 scaffolds were found from tblastn in SPS 2, being the hits in scaffold ML683863.1 the one we chose for the analysis. Exonerate predicted that the gene was located between positions 6968-13735, comprising 7 exons in the forward strand. T-coffee alignment between Carettochelys insculpta genome and the protein reference (in this case, obtained from Mus musculus) showed high similarity between the sequences and, from this, we extract that there is a coding sequence for this protein in the turtle genome. Seblastian could predict a protein in the same region.
In conclusion, SPS 2 was found in the Carettochelys insculpta genome.