INTRODUCTION
Selenium
Selenium (Se) is an essential micronutrient for all living creatures. Mainly, it can be found in the amino acids selenocysteine and selenomethionine. However, selenium may be toxic at high concentrations. Nevertheless, its got many important physiological functions [1]. It is a powerful antioxidant since it acts as a cofactor for the reduction of antioxidant enzymes such as glutathione peroxidases and thioredoxin reductases [2]. Its function is attributed to its presence in proteins as the 21st naturally occuring amino acid, selenocysteine (Sec). Thus, it has a major role in neutralizing free radicals, stimulating the immune system and in the functioning of the thyroid gland [1].
Having done this brief introduction to selenium, we wanted to present the main focus of our project: selenoproteins.
Selenocysteine
The amino acid selenocysteine (Sec) is known as the 21st amino acid. It is a cysteine analog that contains selenium instead of sulfur (a much more reactive element) in the radical chain. Sec is encoded by UGA codons which, usually act as STOP codons.
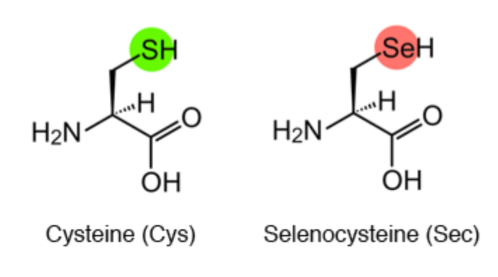
Figure 1. Synthesis mechanism of 21st amino acid Selenocysteine. Image extracted from: http://www.spring8.or.jp/en/news_publications/press_release/2013/130405/
Selenoproteins
Selenoproteins are proteins containing the amino acid selenocysteine. Members of this protein family have different functions, but their synthesis depends on a common set of cofactors and on dietary selenium [3]. They can be found in all three domains of life: bacteria, archaea and eukaryota [4].
Cys-containing homologous
Sec differs from cysteine by a single atom (Se versus S), conferring a lower pKa (5.2 versus 8.3) and higher reactivity. Certain selenoproteins that have cysteine instead of Sec at their active sites showed up to 100-fold lower catalytic efficiency [5]; however, the expected evolutionary selection towards the cysteine containing proteins due to their higher efficacy has been compensated by availability of Se, which has produced a complex distribution of Sec- and Cys-containing homologs throughout nature [6].
Selenoprotein synthesis
In order to synthesize a selenoprotein, the inclusion of the Sec is needed, requiring a specific sequence located in the 3'-UTR region of the mRNA in eukaryotes named SECIS (SElenoCysteine Insertion Sequence), which is a conserved structure [7]. Eukaryotic SECIS elements consist in two helixes separated by an internal loop, a GA quartet structure which is the SECIS core, and an apical loop or bulge [8].
This stem loop structure allows the binging of SBP2 (SECIS binding protein 2), which is a key step in the coding of the selenocisteine [8].
Furthermore, other elements participate in the process [see Figure 2].
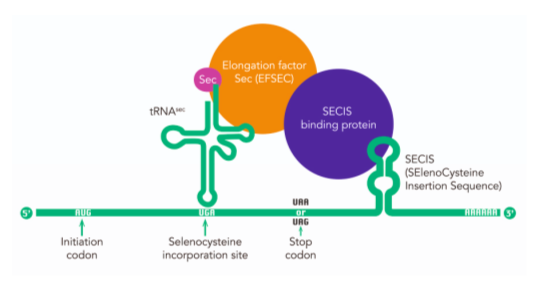
Figure 2. Rare, but essential. Image extracted from: https://researchfeatures.com/2017/06/19/amino-acid-selenocysteine/
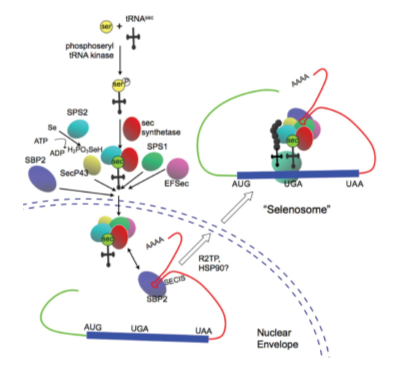
Figure 3. Regulation and function of selenoproteins in human disease. Image extracted from: http://www.biochemj.org/content/422/1/11
As it has been previously mentioned, the UGA codon has the ability to perform two functions: codifying selenocysteines and STOP codons [4]. This fact remained ignored over the years, so it could have led to a lot of misinterpretations.
In fact, the first comprehensive analysis of all the selenoprotein families in vertebrates and mammals was published in 2012. However, there is still a long way to completely understand all the ins and outs of this proteic family [4].
Deepening in the understanding of selenoprotein synthesis, the introduction of the Sec requires a complex machinery and a variety of regulatory mechanisms and genes [9].
To accomplish the decoding of UGA as a SeCys residue, the presence of conserved cis (SECIS elements and the in-frame UGA codon) and trans (SPS1, SPS2, SecS, PSTK, eEFsec, SBP2 and the Sec-tRNA[Ser]Sec)- acting factors is required. All these factors are globally known as the selenoprotein translation machinery [10].
A main step is that cell machinery must recognise UGA not as a stop codon but as a new aminoacid. In order to do that, the tRNA needs to suffer some modifications [10,14].
First of all, it is aminoacylated with serine, then it will be phosphorylated to form phosposeryl-tRNA by PSTK and converted to selenocysteyl-tRNA by SecS, responsible of binding the atom to the serine, completing the synthesis of the tRNA. Also SPS are needed to prepare the Selenium to be incorporated. Selenocystein is then incorporated [11, 12, 14].
So when a ribosome reads the UGA codon, usually meaning the end of the translation, SECIS binds to SBP2, which allows the union of eEFsec, which in turn recruits the Sec-tRNA, enabling the incorporation of Sec, and allowing the translation to continue [10,14].
Other additional proteins needed in the process are L30, Secp43 or Nucleolin.
Philogeny
The distribution of selenoproteins among the evolutionary tree is still incomplete due to their complexity [13]. In fact, there are several families whose function is not assigned yet, and even new proteins encoded with the UGA codon, from which we know nothing about [4]. Nevertheless, there are some things that are well known, for instance, that there is a vast amount of selenoproteins (50 families described to this date) and a lot of variety amongst them too [14].
It is also important to mention that at larger evolutionary distances some selenoproteins are lost and a few evolve and duplicate. Furthermore, some species have lost Sec, replacing them by Cys-containing homologues, which do not always have the same function [4].
On the other hand, in closer related species selenoproteomes are more similar to each other [13].
Moreover, the selenoproteins are irregularly distributed among different species. For example, higher plants and yeast do not present any selenoprotein, but some green algae and vertebrates have over 20 selenoproteins within their genome, while red algae, insects and nematodes code for less than 5. Furthermore, some trends in selenoprotein distribution can be observed, such as larger selenoproteomes in aquatic organisms if compared to terrestrial organisms. Besides, generally speaking, mammalian species tend to reduce their amount of selenoproteins [4]. In figure 4 the evolution of the vertebrate selenoproteome can be observed, which makes the phylogeny and distribution fo the selenoproteins more clear.
Throughout evolution, selenoproteins have been modified by natural selection. 45 selenoproteins have been found in total in sequenced vertebrates, 28 of which were present in mammals, although none of them have all these proteins. Bony fishes are the ones that have the largest selenoproteome and organisms such as frogs and some mammals have the smallest one (Figure 4). Therefore, 21 selenoproteins were found in all vertebrates: GPx1-4, TR1, TR3, Dio1, Dio2, Dio3, SelH, SelI, SelK, SelM, SelN, SelO, SelP, MsrB1, SelS, SelT1, SelW1, Sep15. The rest or selenoproteins may be detected in some lineages, whereas others were lost or replaced their Sec with cysteine [4]. In conclusion, there are 28 proteins in the ancestral vertebrate selenoproteome and 25 in the ancestral mammalian selenoproteome as it is highlighted in Figure 4.
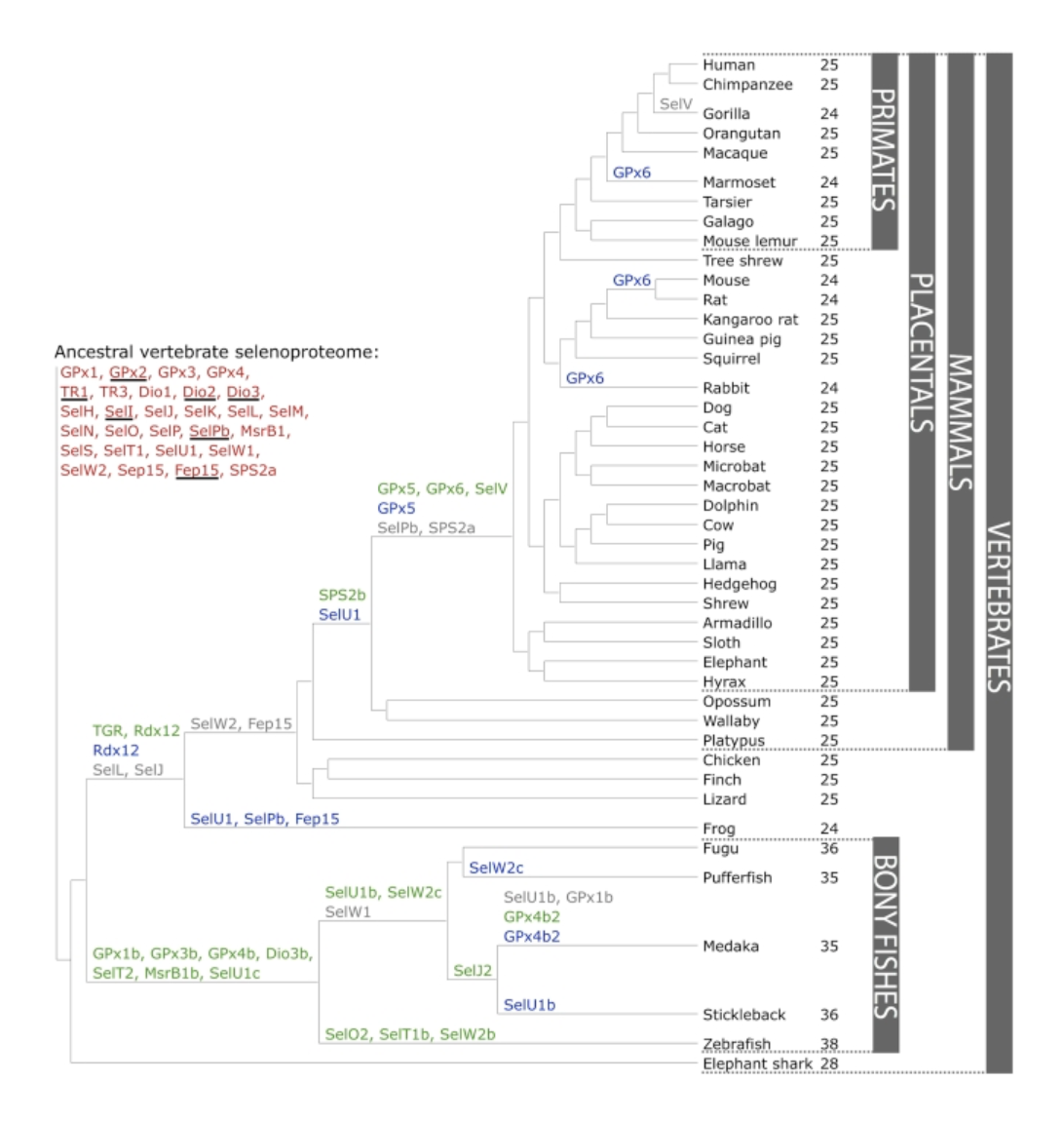
Figure 4. Evolution of the vertebrate selenoproteome. The ancestral vertebrate selenoproteins are written in red in the top left part of the Figure. Their changes throughout the phylogenetic tree are also represented. Besides, the ancestral selenoproteins only found in vertebrates are underlined. The duplication of a selenoprotein is shown in green, while the loss is shown in grey. The cysteine homologous proteins (change from selenocysteine into cysteine) is shown in blue (with the exception of SelW2c in pufferfish, which is with arginine). There were no Cys to Sec conversions found. In the right part, the numbers denote the number of selenoproteins predicted within each specie. Extracted from: https://www.ncbi.nlm.nih.gov/pubmed/22479358
Nowadays a lot of ongoing projects are focused on genotyping organisms to expand the number of genomes with the distribution of their selenoproteins. This annotation would allow us to have a better understanding of their phylogeny, and to better appreciate their function.
Selenoprotein families
Selenoproteins are classified depending on their function and their structure. The set of selenoproteins in the human genome consists of 21 known selenoprotein families, as it is shown in SelenoDB. In the following list we mention the main families of selenoproteins, containing the ones that conform the human proteosome and other ones that we considered important to mention that are present in other vertebrates but not in humans (SecS, SELENOL, PSTK, SECp43).
This protein is a thioredoxin-like located in the endoplasmic reticulum that regulates the correct folding of glycoproteins. Its loss causes cataracts [14,15] |
||
This protein of selenoprotein synthesis machinery takes on the role of elongation factor specific to selenocysteines [14]. |
||
GPx2 GPx3 GPx4 GPx5 GPx6 GPx7 GPx8 |
SPP00000005 SPP00000006 SPP00000007 SPP00000008 SPP00000009 SPP00000010 SPP00000011 |
It is composed by 8 enzymes, which preserve a catalytic cysteine at the N-terminal extremity, ancested cysteine peroxidica (CysP-S-). They reduce hydroperoxidases and peroxynitrites, helping to the maintenance of the homeostasis with redox reactions. [14, 16, 17] |
DIO2 DIO3 |
SPP00000002 SPP00000003 |
They all are transmembrane proteins which regulate the activation or inactivation of the thyroid hormone. We find mainly three types [14]. |
It is an antioxidant enzyme found in all domains of life that catalyzes the reduction of methionine-S-sulfoxide (MSO) to methionine in proteins and free amino acids [14]. |
||
This protein binds to the SECIS element which has been specifically stimulated by a Sec-specific elongation factor [14]. |
||
SPS2 |
SPP00000033 |
Its function is to generate the selenophosphate necessary to incorporate selenium into selenocysteine. We find two paralogical forms of the protein [14]. |
Transcription factor. It protects neurons against damage caused by UVB by inhibiting apoptosis by enhancing mitochondrial activity. They preserve a Cys-x-x-Sec motif [14]. |
||
Involved in the maintenance of vesicular membranes, regulation of the lipid metabolism and folding of proteins [14]. |
||
Selenoprotein K is required for Ca2+ influx in immune cells and plays a role in T-cell proliferation and in T-cell and neutrophil migration. Also, it is involved in endoplasmic reticulum-associated degradation of soluble glycosylated proteins. This family is grouped by structural similarity although they are distal between them [14]. |
||
they have a thioredoxin-like domain and a surface with an accessible redox motif. This selenoprotein is located in the endoplasmic reticulum and codify for an N-peptide that is cleaved once translocated into the endoplasmic reticulum [14]. |
||
Its precise function is unknown. It is found in many tissues during embryonic development and it is thought to play a relevant role in myogenesis: during embryogenesis and adulthood [14]. |
||
It is located in the mitochondria where it controls redox reactions. It is the longest selenoprotein in mammals that has orthologues [14]. |
||
It is related to the extracellular transport of the selenium or its antioxidant action. Mostly expressed in the liver, which is secreted in plasma, although we find it present in many tissues [14]. |
||
SelR2 SelR3 |
This involves a family of enzymes which contain zinc. They are all involved in the conversion of methionine sulfoxide to methionine [14]. |
|
This family is grouped by structural similarity although they are distal between them [14]. |
||
It gives protection to dopaminergic neurons in Parkinson's and also controls the homestase of glucose in beta-pancreatic cells [14, 18]. |
||
SelU2 SelU3 |
SPP00000027 SPP00000028 |
Unknown function. It is hypothesized that it may have a redox function [4]. |
It is believed to be a duplication of selenoprotein SelW. Preserves a Cys-x-x-Sec motif [14, 18], suggesting a redox function for this gene. This protein is specifically expressed in the testis. Alternatively spliced transcript variants have been found for this gene. |
||
SelW2 |
SPP00000031 |
It is expressed in multiple tissues and we can find different homologues in vertebrate organisms. They preserve a Cys-x-x-Sec motif [14, 18],suggesting a redox function for this gene. Studies in mouse show that this selenoprotein is involved in muscle growth and differentiation, and in the protection of neurons from oxidative stress during neuronal development. |
TR2 TR3 |
SPP00000035 SPP00000036 |
These proteins are oxyreductases of the pyridine nucleotide. They reduce thioredoxins among other substrates while playing an important role in the metabolism of selenium and the protection against oxidative stress [14]. |
*The different colors mean the differences that are in these proteins: in orange, selenoproteins; in green, Cys-containing homologs; in red, selenium molecular machinery; and in blue, other amino acid containing homologous.
Through the evolution, diverse selenoproteins were lost across vertebrates when the terrestrial environment was colonized. Thus, it supports the idea that mammals decreased the utilization of selenocysteine compared with fishes [19]. Due to it, there are several selenoprotein families that are found only in fishes such as selenoprotein L (SelL), which is contained in the thioredoxin superfamily with the common sequence UxxU (U is a selenocysteine and x any amino acids). The same happens with SElJ [4] and SECp43 [20].
Moreover, the selenoprotein SelW2 was also lost and, currently, it can be only found in fish and frog. Perhaps, that is why SelW2 is present in humans as Cystein-containing homolog [4].
Urocitellus Parryii
The Arctic ground squirrel (Urocitellus parryii, also noun as Spermophilus parryii) is a hibernating rodent native to Holarctic ecozone which is abundant, colonial and non cyclical small mammals. [21] This squirrel belongs to the group of species that are threatened with extinction, included on the IUCN Red List (Arkive 2013) on the category of least concern and with its conservation status that is "secure".
Description

Adult Arctic ground squirrels average about 39 cm in length from head to end of tail with a weigh around 750 g females and 100 g heavier the males. It is important to clarify that the exact body mass is tricky to determine since they undergo drastic seasonal changes.
U. parryii is a grayish tan and gray in the upper backside and rusty to tan on the belly side. This squirrel has a short face with a rusty-tan-colored nose and white markings around its eyes. It also has a dark tail and four toes on their feet. It walks on all four legs but can sit up on two legs as well. They change their coat from summer to winter, in summer including red/yellow colorations along the cheeks and sides of the animal. In fall, these red patches are replaced with silvery fur [22].
Classification
Kingdom: Animalia
Phylum: Chordata
Class: Mammalia
Order: Rodentia
Family: Sciuridae
Genus: Urocitellus
Species: U. parryyi
Image extracted from: http://www.olsvik.info/Diverse/Reiser/Denali_2014.html
Distribution and habitat
The Arctic ground squirrel can be found in regions of Northern Canada ranging from the Arctic Circle to northern British Columbia, and down to the southern border of the Northwest Territories, as well as Alaska and Siberia [23].
It is native to the North American Arctic tundra in open meadows, or above the treeline, in river valleys and meadow-steppe places [24]. This squirrel lives in sandy soil forming colonies with complex system of shallow burrows [25].
Behaviour and diet:
The diurnal Arctic ground squirrel lives on the tundra and is prey to the Arctic fox, the red fox, wolverine, lynx, the grizzly bear, and eagles. Moreover, it hibernates from October until March. During hibernation, its brain and body temperature decrease and its heart rate drops to ~1BPM. Peripheral, colonic, and blood temperatures become subzero [24].
Although being omnivorous, in the summer it feeds mainly on plants, seeds and fruits to increase body fat for its winter hibernation. They also eat insects and, less often, mice, snowshoe hares and caribou [26].
These squirrels reproduce only once a year and, besides, they can communicate between themselves through vocal and physical means [22].
Furthermore, we extended the entrance of U. parryii in Vikipedia, as you can see in this link.