DISCUSSION
As we have previously mentioned, the purpose of this study was to characterize and annotate every selenoprotein and machinery genes present in Urocitellus parryii’s genome. TheUrocitellus parryii’s genome was compared with the Human selenoproteins. The search was supplemented with the analysis of SECIS elements via SEBLASTIAN and SECISearch3. That way, orthologous proteins were studied and the results are here commented one by one, taking into account our results and showing a representation of the gene, its exons and where exactly the selenoprotein (if found) is.
Human genome presents 21 families of selenoproteins, containing in total 38 proteins which were obtained from SelenoDB.1 and have been considered our queries. As it was expected, the human selenogenome is well annotated and we do not expect a lot of differences because selenoproteins are well conserved among vertebrates.
In some of our protein predictions, the first amino acid encountered was not a methionine, so it led us to a deeper interpretation of the prediction that will be discussed within each protein section. In general, this could have been due to a) query’s poor annotation, which is not likely to happen since we used as reference genome the human genome from SelenoDB.1; b) Urocitellus parryii’s poor genome annotation.
Moreover, in some cases we lacked the initial part of the protein in the main scaffold studied. As it has already been explained in ‘Methods’, for some proteins we ran several different scaffolds since the tblastn results were accurate enough in all of them. Although the T-coffee results of these scaffolds allowed us to choose the most suitable one, we also examined the others in case the lacking part of the protein was coded in another scaffold. Nevertheless, we realised that in the Seblastian output, besides from the SECIS prediction, an alignment between Urocitellus parryii and other organisms was also performed. This allowed us to reconstruct the whole protein, which will be attached in a text document within each protein section.
Furthermore, some criteria were born in mind in order to choose whether the predicted protein was a selenoprotein, a cys-containing homolog or non of them. For selenoproteins, it was indispensable that a TGA (UGA) codon was aligned with a selenocysteine or with a cysteine in the query and that the sequence after this particular codon offers a perfect alignment with the query. Moreover, to determine whether it was a selenoprotein or not it was also necessary to find the SECIS predicted element in the 3’ UTR region. In the case of cysteine-containing homologs, a cysteine in Urocitellus parryii’s genome must be found aligned with a cysteine or with a selenocysteine in the query; as in the previous case, the alignment following this cysteine should be suitable. It was also taken into account the fact that, in some cases, SECIS elements could also be found in cys-containing homologs. As they were initially selenoproteins, they might have been converted into cysteines but maintained the SECIS element. Eventually, proteins that lost their selenocysteine but replaced it with any other amino acid but cysteine were classified as others.
Eventually, phylogenetic trees will also be attached to see the distance between our final predicted protein and its human orthologous through evolution. It is relevant to mention that specifically in the case of the GPx, other phylogenetic trees were performed, inputting the query protein and all the predictions done for each protein, which helped us to choose which was the most suitable scaffold for each protein. However, we considered it was not worth it to include these GPx trees since they were just used as another tool to discern between scaffolds within this large family of proteins.
Selenoproteins
Iodothyronine deiodinases (DIO)Iodothyronine deiodinases have an essential role in regulating the activation of the thyroid hormone. This family consists in two proteins: DIO1 and DIO2.
DIO1Regarding the significant results of the tblastn (e-value smaller than 0.001), the scaffold QVIC01000022.1 showed 3 hits with a relatively high percentage of identity, between 82 and 84%. The T-coffee results show a 1000 score and no gaps were found in the alignment. We finally chose this as the scaffold where our predicted protein would be located. However, the N-terminal part of the protein is not present in this scaffold. We expect it to be in scaffold QVIC01006785.1, which presents a lower percentage of identity than the first one mentioned above but an alignment of 118 nucleotides. We finally run this scaffold and we found the N-terminal part of the protein, so the final predicted protein is found attached in this document.
The predicted gene, which is composed by 3 exons, is located between 5209759-5218191 positions in the reverse strand and contains a total amount of 8432 nucleotides (nt). A selenoprotein (Sec) was found in the first exon (counting from 5’) in the same position as the Sec in Human’s query. This fits with the expected results since DIO is a selenoprotein [see figure below].
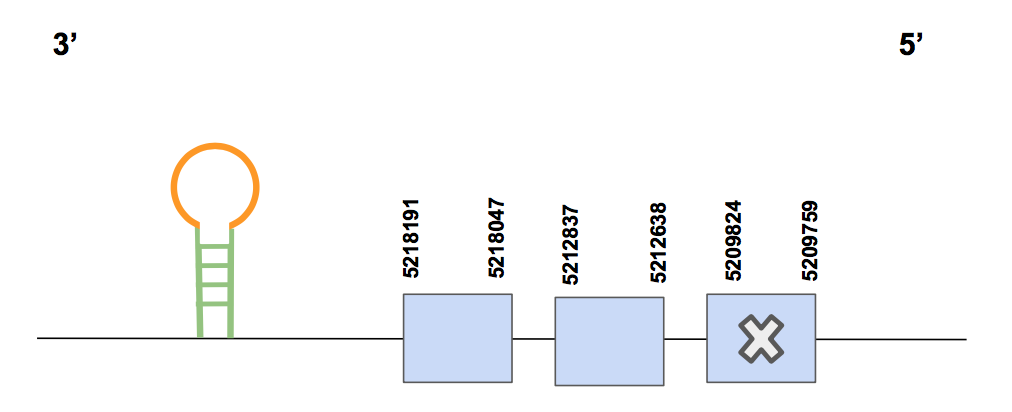
Seblastian predicted two suitable SECIS elements, both found in the 3’ UTR region, in positions 5208400-5208469 and 5206793-5206872, being of grade A and B respectively. Thus, we conclude that DIO1 is a selenoprotein found in Urocitellus parryii encoded by one gene divided in two scaffolds.
DIO22 significant hits of 87-93% of identity were found for the scaffold QVIC01000009.1 when running the tblastn. Regarding the results of the T-coffee, we can conclude that DIO2 was completely identified, since the alignment contains the methionine in N-terminal end, just small changes of amino acids and 0 gaps, which results in a score of 1000. The predicted gene is located in the forward strand between 7764302-7772754 positions and contains a total of 8452 nt, having two exons.
A Sec is found in the second exon (counting from 5’) where it matches with the Sec of Human’s query, as it is expected because DIO2 is a selenoprotein. However, another Sec is found in the exon 2 (counting from 5’), which does not match neither with a Sec nor with a cysteine within the human query. For more graphic information see figure below.
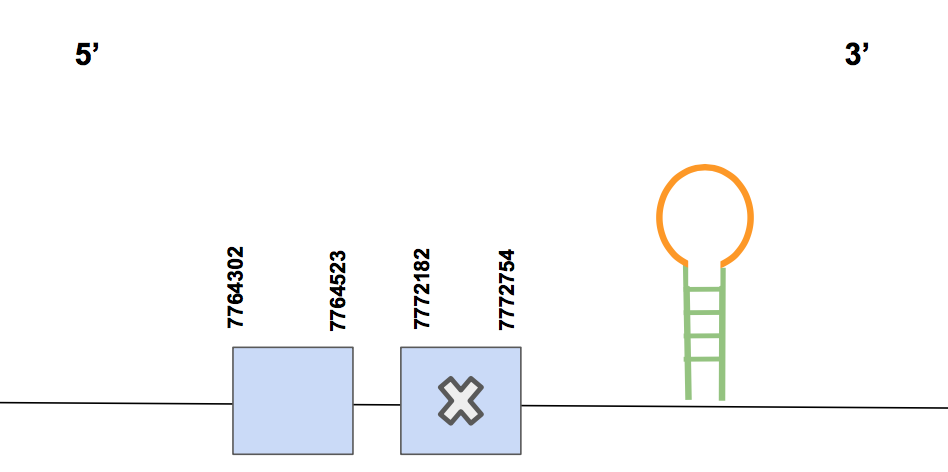
SECISearch3 predicted 3 possible SECIS, one of them being in the reverse strand, but our protein is coded by the forward strand, so we can discard this prediction. The other two predictions are located in the forward strand and only one of them located in 3’ UTR (positions 7777403-7777475). Thus, this third prediction is selected as it fulfils the conditions. Moreover, it has an A grade. Eventually, we establish that DIO2 was identified in Urocitellus parryii with its respective SECIS element.
DIO3In this case, scaffold QVIC01000918.1 showed one significant hit with an identity percentage of 96%. Analyzing the T-coffee alignment, it seems that this protein has been successfully characterized from the starting point (since the methionine is included) to the end, except for the last amino acid. It is worth mentioning that the last amino acid is lost in the T-coffee when comparing the predicted squirrel sequence with the human query. Nevertheless, in Seblastian it can be appreciated that comparing with Marmota marmota this amino acid does not exist, so we hypothesised that maybe humans have an extra amino acid that our squirrel may not have. Apart from that, the rest of the T-coffee alignment is almost perfect.
This protein is located in the reverse strand, between the positions 177323 and 178147 (824 nt), and it has only one exon in which the Sec is located. This Sec matches with the Sec of the query, as expected [see figure below].
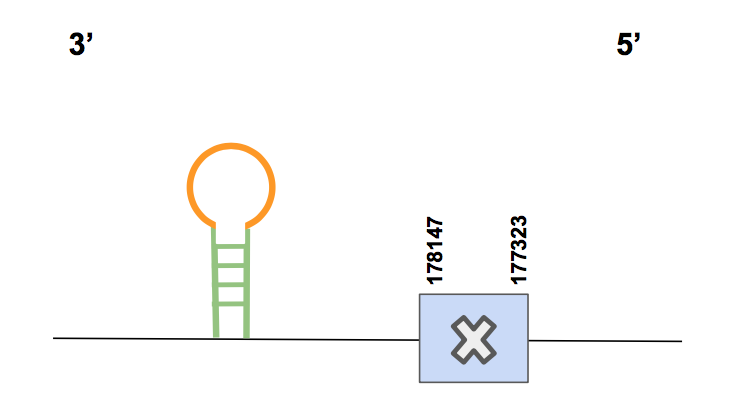
A SECIS of grade A was predicted by Seblastian in positions 7777403-7777475, so it is located in 3’UTR and this prediction is valid. Finally, regarding the Sec’s match, the T-coffee and Seblastian alignment, we may claim that DIO3 is present in Urocitellus parryii.
A phylogenetic tree was performed in order to double-check if our predicted proteins are accurate enough.
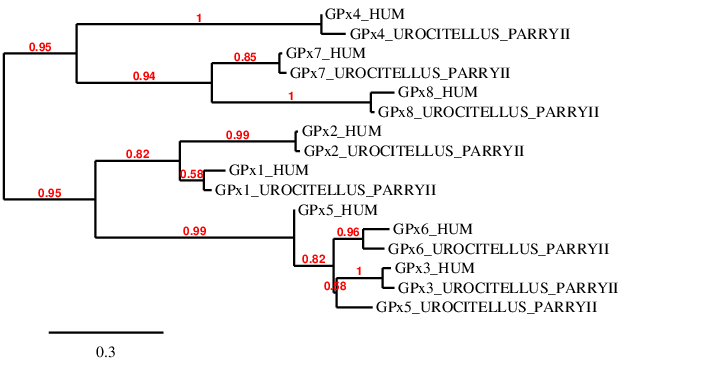
This is the biggest family of selenoproteins found in vertebrates. It contains 8 enzymes that contribute to the maintenance of homeostasis through redox reactions and have an important role in the detoxification of hydroperoxides and hydrogen peroxide signalling.
It is worth mentioning that as these 8 proteins belong to the same family, they might have common domains. This fact made it more difficult to discern in which scaffold was located each predicted protein, since at first sight some of them could have been suitable for the prediction of more than one GPx.
GPx1Taking into account the significant scaffold results from the tblastn (e-value "<" 0.001), two scaffolds were first selected to examine their T-coffee results, whose output would make it clear to decide in which scaffold we expect to find our protein. Besides, in this case we graphed a phylogenetic tree between GPx1 and GPx2 since both had the same two scaffolds selected. This helped to select just one scaffold for each protein. Scaffold QVIC01000027.1 was selected for GPx4. It presented 2 hits in the tblastn output, with a really high percentage of identity (88-90%). Moreover, the T-coffee had a relatively accurate alignment, with just one gap. In this case, we have been able to fully predict the protein. However, the first amino acid does not correspond to methionine, while it does within the query of the human genome. As it has been introduced, this could be because of a poor genome’s annotation of Urocitellus parryii.
The predicted gene is located between 8616764-8617598 positions in the forward strand, containing a total number of 834 nt. It includes 2 exons. A Sec was located in the first exon (counting from 5’), matching with the Sec in the query. This makes sense since we actually expected the GPx1 to be a selenoprotein [see figure below].
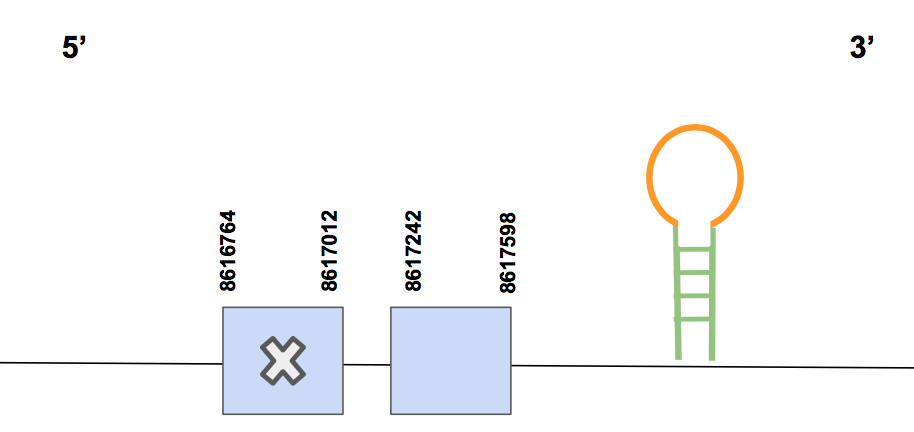
Seblastian found just one predicted SECIS element in 3’UTR, specifically in positions 8617645-8617719 (grade A). Hence, taking into account the information given by the T-coffe alignment, the alignment obtained from the Seblastian output, the phylogenetic tree and the matching between the Sec from Urocitellus parryii and our query sequence and the prediction of a SECIS element, we conclude that GPx1 is a selenoprotein present in the genome of Urocitellus parryii, encoded by a gene located in scaffold QVIC01000027.1.
GPx2Among all the significant results in the tblasnt output, two scaffolds were chosen in order to obtain their T-coffee results. As it has already been explained, a phylogenetic tree with the GPx1-2 from the query and their possible predicted proteins (using the two selected scaffolds: QVIC01000027.1 and QVIC01000037.1) was done. The most suitable one for GPx2 was QVIC01000037.1, were we expect our predicted protein to be located. In the tblastn output this scaffold showed 2 hits with a high identity value (88-93%). The T-coffee results offer a 1000 score, with no gaps and just a few amino acid changes (these changes were between similar amino acids): the alignment is almost perfect and besides our predicted protein does begin with methionine.
The predicted gene is found between the positions: 4264834-4267622, in the reverse strand. The total amount of nucleotides is 2788 and it contains 2 exons. A Sec was found in the first exon (counting from 5’), matching with the Sec of the human sequence. This suits with our expectations since we already predicted the GPX2 to be a selenoprotein [see figure below].
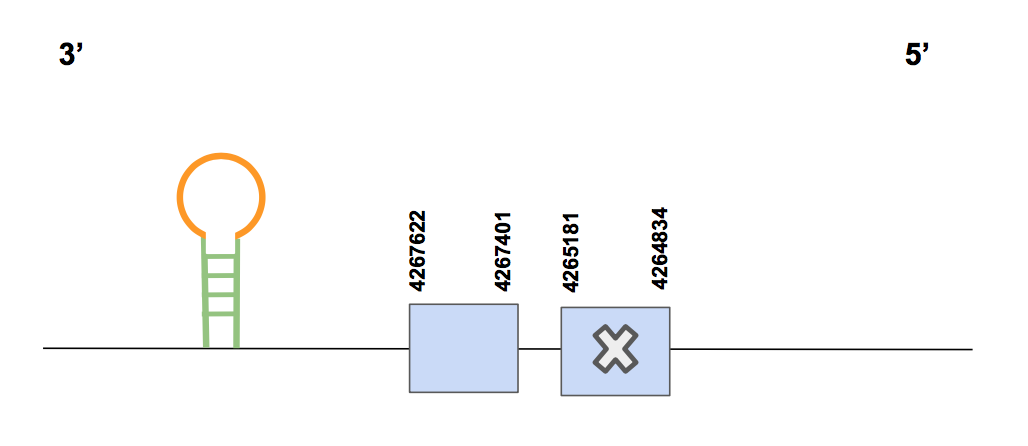
One SECIS prediction was done by Seblastian, which was found in 3’UTR, between the positions 4264551-4264615 (grade A). Eventually, bearing in mind all the results from the alignments, the matching of Sec’s and the presence of a SECIS element in 3’UTR, we can conclude that GPx2 is a selenoprotein found in Urocitellus parryii encoded by one gene in the scaffold QVIC01000037.1.
GPx3The choice of the more suitable scaffold among the more significant results from the tblastn output had to be taken bearing in mind that these scaffolds appeared in more proteins from the same family. So again, the conclusion of which was the scaffold where the GPx3 predicted protein would be allocated was done considering the results from the phylogenetic trees and the T-coffee alignments. The QVIC01000223.1 was the scaffold were we finally expected to find our protein. In the tblastn output this scaffold presented 4 hits, with a relatively low percentage of identity. The T-coffee revealed a score of 1000. Our predicted protein begins with a methionine and there is a gap at the end of the sequence of 5 amino acids, which lead us to question if this small part of the protein is encoded in another scaffold that we have not taken into account, although might also be due to a shorter protein in Urocitellus parryii if compared to humans.
The predicted gene can be found between 3047444-3054798 positions, specifically in the forward strand. It is composed by a total of 7354 nucleotides and 5 exons can be found. A Sec alligned with a Sec from the query was found in exon 3. As it was already expected, we corroborate that GPX3 is a selenoprotein [see figure below].
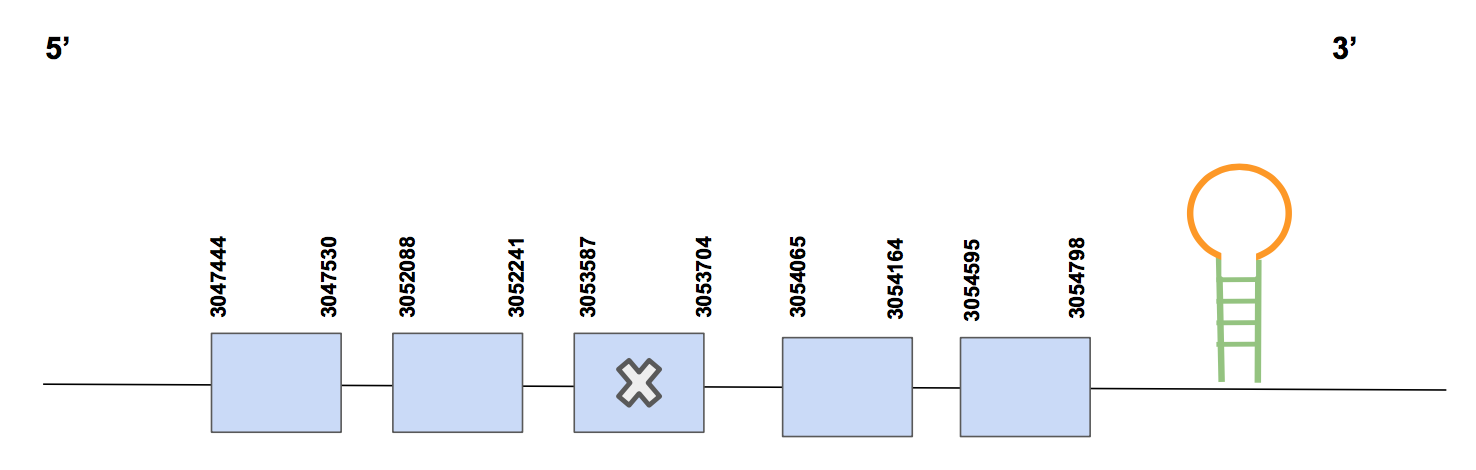
In this case, Seblastian was not able to predict SECIS elements, so SECISearch3 was used and two SECIS were predicted, both of them found in 3’UTR and in positions: 3055323-3055405 and 3061844-3061888 and of grade A and B, respectively. We hence determine that GPx3 is a selenoprotein found in Urocitellus parryii, as the alignment results from T-coffee and phylogeny show.
GPx4Among the significant scaffolds from the output of tblastn, QVIC01000612.1 was chosen, following the same choosing criteria as explained before. This scaffold presented 3 hits in the tblastn results, with a general high percentage of identity. The T-coffee resulted in a 996 score, and a gap of 2 amino acids almost at the beginning of the sequence. According to the T-coffee alignment, our predicted protein does not start with methionine; however, if we focus on the Seblastian alignment (Urocitellus parryii with Ictidomys tridecemlineatus), the initial methionine is present. Moreover, we checked whether the U. parryii sequence matched with the one from T-coffee and it did from one amino acid onwards, but not in the first part. It revealed that 5 amino acids on our predicted protein were missing in the T-coffee alignment and that the initial methionine was also missing. Hence, our final predicted protein does begin with methionine, as the final prediction text document shows: GPx4.finalprediction.txt.
The predicted gene is formed by 7 exons and is located in the reverse strand. It can be found between the following positions: 420695-422546 and the total amount of nucleotides is 1851. A Sec in U. parryii predicted GPx4 protein is aligned with a Sec in the query, specifically in exon 4. This result is in accord with our expectations, since we already anticipated GPx4 to be a selenoprotein. For more information about the exon and selenoprotein distribution, see figure below.
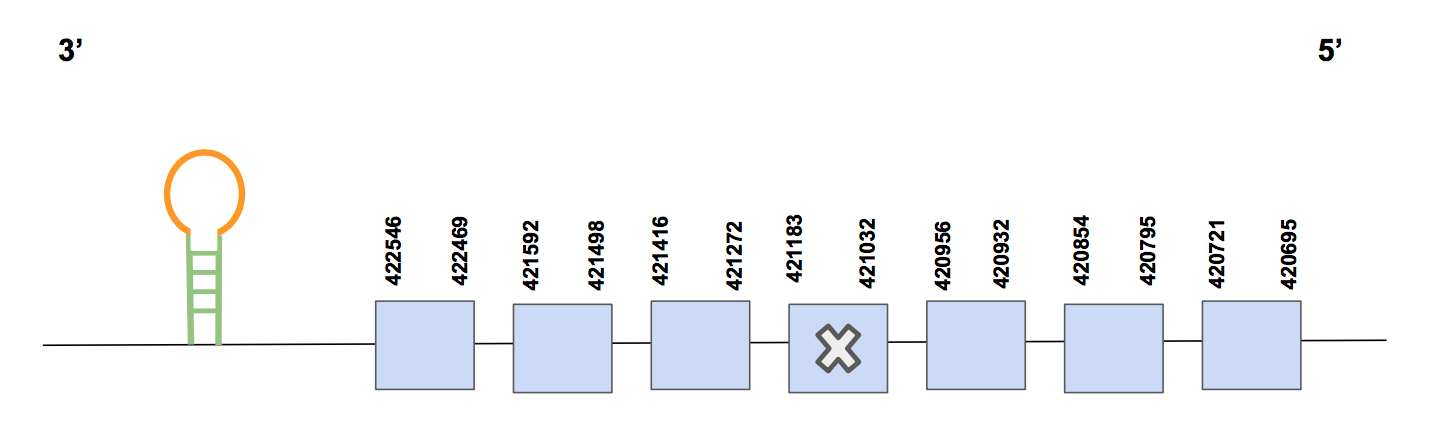
Eventually, the SECIS prediction was performed using the programme Seblastian, and found one suitable prediction; the SECIS element was located in 3’UTR, concretely between positions: 420566-420637 (grade A). Therefore, considering all the predictions and alignments developed, we prove that GPx4 is a selenoprotein that can be found in U. parryii encoded by a gene located in scaffold QVIC01000612.1.
GPx5In the case of this protein, the significant tblastn results offered 3 possible scaffolds where our predicted protein could be found. However, following the same methodology as in the other GPx proteins, the scaffold QVIC01000637.1 was the one chosen to do our prediction. Nevertheless, the exonerate output resulted in two different genes: gene 1 in the forward strand and gene 2 in the reverse strand. In order to choose which gene was the most suitable one to do our prediction, we examined the T-coffee results; the scores did not give conclusive results, since both had high scores. In gene 1 there was one Sec in the U. parryii genome aligned with a cysteine in the human query, meanwhile there was no Sec in gene 2. Furthermore, the gene 1 alignment was shorter and with more amino acid changes. Taking all of this into account and expecting GPx5 to be a cysteine- containing homolog, the gene 2 from the scaffold QVIC01000637.1 was chosen to do our prediction. The T-coffee alignment showed no gaps and although there were some amino acids changes, they were mostly between similar amino acids. Moreover, our predicted protein begins with methionine.
The predicted gene is encoded by 5 exons and located in the reverse strand. It can be found between positions 354859-363524 and it presents a total of 8665 nucleotides. As it has already been mentioned, no Secs were found, which actually makes sense since we expected our protein to be a Cysteine-containing homolog (several cys-cys matches between U. parryii predicted protein and the query were found).
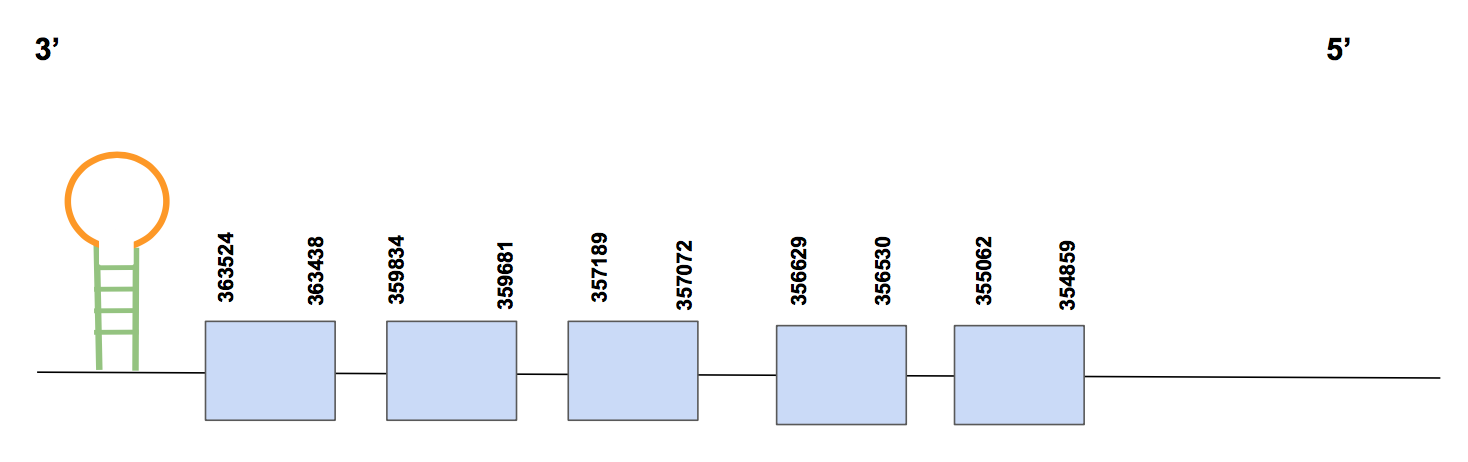
Finally, a SECIS prediction was obtained from Seblastian, located in 3’UTR and between positions 411199-411274 (grade A). We hence determine that GPx5 is a cystein-containing homolog present in U. parryii, according to the results of the alignments and supported by the maintenance of the SECIS structure.
GPx6The significant tblastn results for this protein gave a high amount of different scaffolds, among which we chose to run three of them and then decide from these three which one would fit best our prediction. Again, we discarded two of them following the same procedure (alignment results and phylogenetic tree). Finally, the scaffold QVIC01000637.1 was the one chosen. As it has already been explained, GPx5 was predicted in gene 2 of this same scaffold. Analysing both genes again, we concluded that gene 1 from this scaffold suited GPx6 prediction. The T-coffee results showed a score of 999 and in the alignment there was just one gap in the last position. However some amino acids changes were found and, most importantly, the first part of the protein was lacking.
We analysed the T-coffee alignments of the other two scaffolds that we decided to run (QVIC01000223.1 and QVIC01000027.1) but this first part was in none of them. Nevertheless, the Seblastian alignment allowed us to reconstruct this first part. In this case, Seblastian run the alignment between U. parryii and Ictidomys tridecemlineatus. It is worth mentioning that if the initial part of the protein of the human’s query and the initial part of the protein of Ictidomys tridecemlineatus are compared, they are slightly different. Meanwhile, the alignment of this first part between U. parryii and I. tridecemlineatus is almost perfect (no gaps, just one amino acid change and both beginning with methionine). Hence, we can conclude that the T-coffee failed to predict this first part of U. parryii’s GPx6 because it is quite different from the human’s GPx6. However, we managed to reconstruct the whole U. parryii’s GPx6, which can be found in the attached document: GPx6.finalprediction.txt.
The predicted gene is formed by 4 exons in the forward strand. The gene is concretely located between 406209-411025 positions, containing a total amount of 4816 nt. A Sec aligned with a Sec from the human’s protein is found in exon 1 (counting from 5’), which corresponds with what we expected: GPx6 is a selenocysteine.
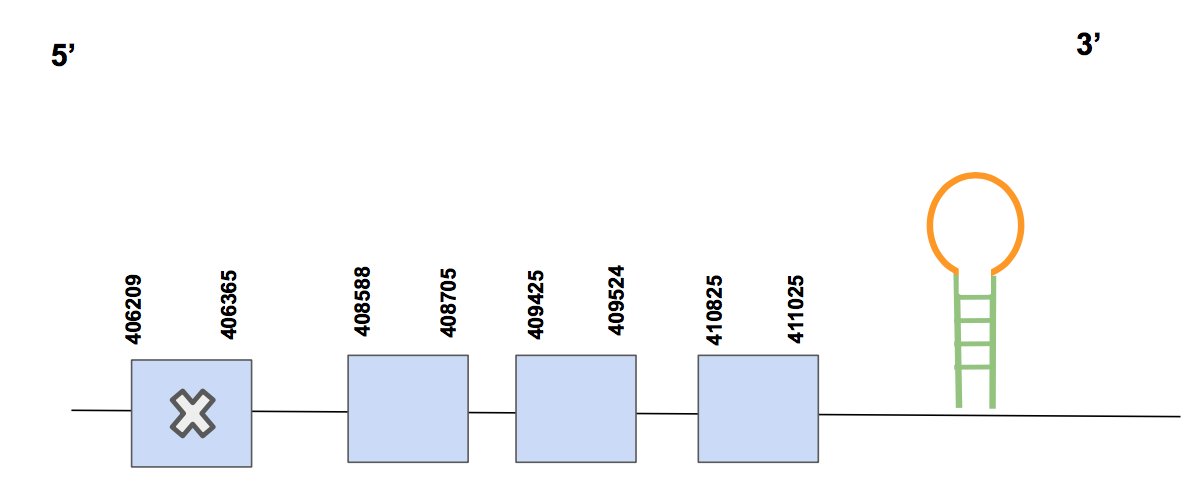
We then used Seblastian, which predicted one SECIS element found in 3’UTR and between positions 411199-411274 (grade A). We can now conclude that GPx6 is a selenoprotein found in U. parryii, more precisely in scaffold QVIC01000637.1 in a gene located in the forward strand.
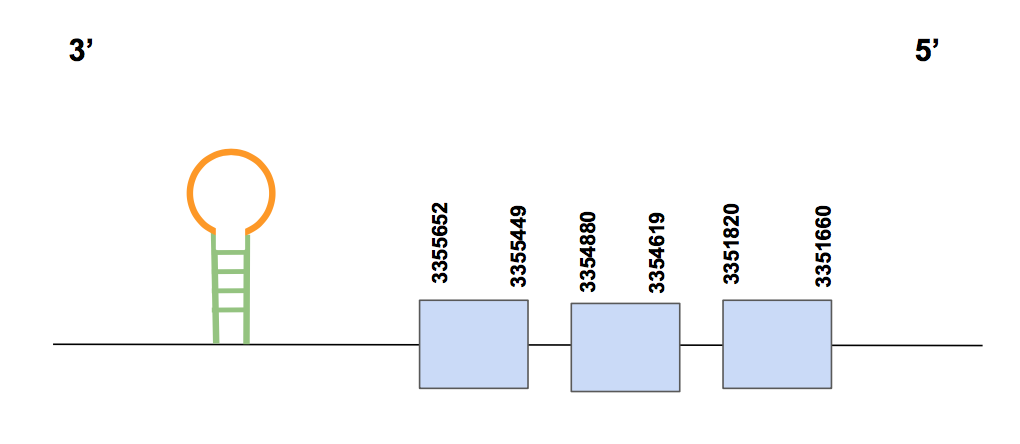
GPx7Among the significant results from the tblastn output, one scaffold was chosen: QVIC01000022.1, which is the one in which our protein prediction has been based on. Three hits were found within this scaffold, with a percentage of identity between 75-96%. The T-coffee revealed a score of 999, with no gaps and almost no amino acid changes. The protein has been fully identified; however, the first amino acid is not a methionine, but a valine, which could be do to an annotation mistake within the U. parryii genome.
The predicted gene is formed by 3 exons and is found in the forward strand. Its exact location is between positions 406209-411025 and is formed by 6497 nt. No Sec is found neither in the U. parryii sequence nor in the human query, which matches with our expectations since we already predicted GPx7 to be a cysteine-containing homolog. Specifically, three pairs of aligned cysteines can be found in the T-coffee output.
In this case, no SECIS structure was found neither with Seblastian nor with SECISearch3. Hence, we conclude GPx7 is a cysteine-containing homolog found in U. parryii, explicitly in a gene present in scaffold QVIC01000022.1.
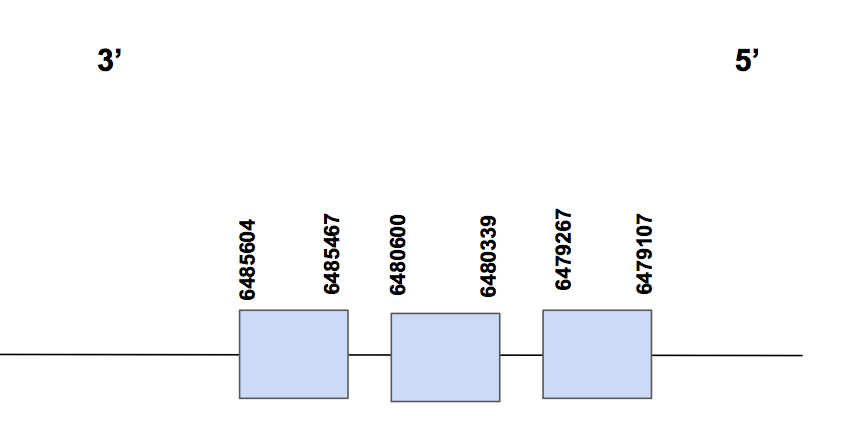
GPx8The scaffold QVIC01000042.1 was chosen from the significant results of the tblastn output (e-value "<" 0.001) in order to do our prediction. Three hits were found and the percentage of identity was up to 93%. The T-coffee revealed a score of 1000, with no gaps and almost no amino acid changes, and the few changes are between similar amino acids. Moreover, the predicted protein begins with methionine and we have been able to fully predict it.
3 exons form the predicted gene, which can be found in the reverse strand. It is located between 6485604-6479107 positions and include a total amount of 6497 nt. No Sec was found in none of the sequences, which is what we expected: GPx8 is a cysteine-containing homolog. Actually, 4 pairs of cys-cys alignments between U. parryii and the human sequence can be found in the T-coffee output.
Seblastian was not able to predict a SECIS element in this case, but one suitable prediction was obtained using SECISearch3. This was located in 3’UTR and more specifically between positions 3332751-3332821 (grade B). Thus, taking into account the information provided by the alignments and by the fact that a SECIS element is maintained, we conclude GPx8 is a cys-containing homolog which is present in U. parryii.
A phylogenetic tree was performed in order to double-check if our predicted proteins are accurate enough.
Methionine sulfoxide reductase A is an antioxidant enzyme found in all domains of life that catalyzes the reduction of methionine-S-sulfoxide (MSO) to methionine in proteins and free amino acids.
MsrAThe human query SPP00000012 showed six significant hits with a percentage up to 100% for the scaffold QVIC01000204.1. Once we checked the results of the T-coffee, although there are several amino acid changes and 2 short gaps (of 2 aa each one), it is a relatively good alignment that starts with a methionine. Besides, the Sec is not found but several Cysteines match with the Cysteines of the query, which is consistent with this protein since it is a Cysteine-containing homolog.
The predicted protein has six exons and is in the reverse strand, being located between 1052141-1383186 (331045 nt) as it can be seen in the figure below.
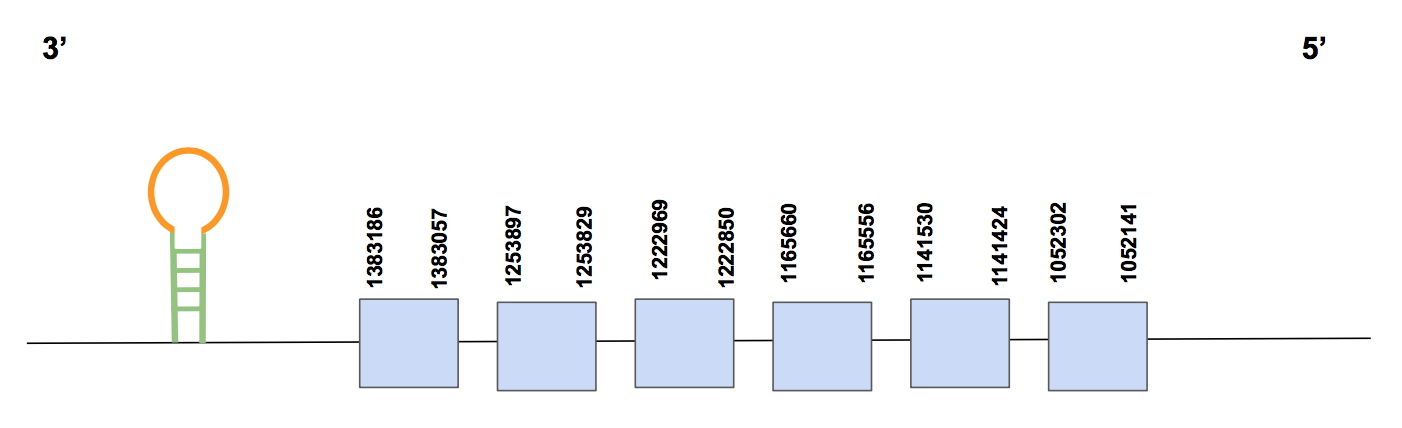
SECISearch3 predicted 4 possibles SECIS, two of them are not valid because they are in the forward strand. Among the other two options, only one is located in the 3’UTR (positions 1031491-1031564). Therefore, we state that Urocitellus parryii contains the protein MsrA, since it is supported by the T-coffe alignment, the Cysteine matches are found and it has still the SECIS structure. This could be explained because it is easier to have an aa mutated (from Sec to Cys) than to loss a complex structure such as SECIS.
15 kDa selenoprotein (Sel 15)15 kDa selenoprotein (Sel 15) is a thioredoxin-like protein that ensures the correct folding of glycoproteins.
Sel15SPP00000013 shows 3 significant hits for the scaffold QVIC01000289.1, whose identity is between 77-96%. The T-coffee alignment score is 1000 but, most importantly, there are just a few amino acid changes and 0 gaps. Furthermore, the Sec found in the third exon (counting from 5’) of the predicted protein matches with the query, consistently with the expectations since Sel15 is a selenoprotein.
The protein has 4 different exons and is in the reverse strand and has 34115 nt in total (positions: 383681-417796) [see figure below].
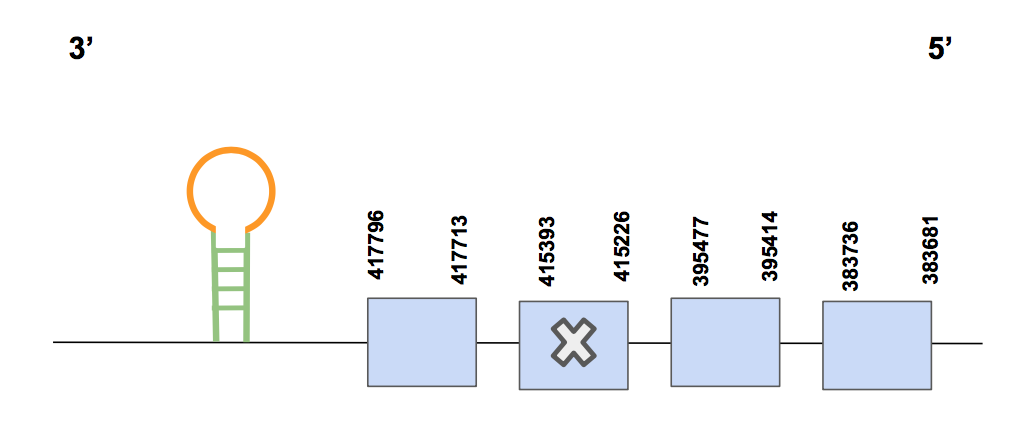
In this case, Seblastian was not able to predict any SECIS element, but SECISearch3 predicted one SECIS of grade A located in 3’UTR (382968-383045 positions) as we expected. Therefore, the Sel15 of U. Parryii was completely and successfully annotated.
Selenoprotein H (SelH)Selenoprotein H has a major role in protecting neurons against damage caused by UVB by inhibiting apoptosis and regulating mitochondrial activity. It is worth mentioning that it preserves a Cys-x-x-Sec motif, so we expect it to be a selenoprotein.
SelHScaffold QVIC01000337.1 presents 2 significant hits with identity percentage of 92 and 100%. With regard to the T-coffee results, it is observed that the alignment is proper since it is complete (from the first amino acid (Methionine) until the end) and it does not have any gap and almost no amino acids changes are found. In this case, a Sec match is found in exon 2.
The predicted protein has 3 exons, located between 1268971 - 1269554 (583 nt), being in the reverse strand [see figure below].
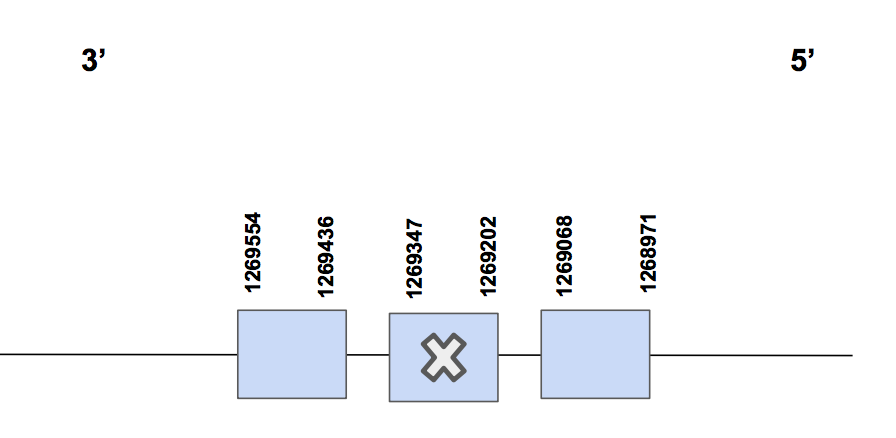
Unfortunately, not SECIS was found neither with Seblastian nor SECISearch3, so we can not conclude that the protein SelH is present in Urocitellus parryii, since the selenocysteine is found but not the SECIS, which is needed for the inclusion of the Sec.
Selenoprotein I (SelI)Selenoprotein I is involved in the maintenance of vesicular membranes and lipid metabolism, as well as regulation of protein folding.
SelIFor the prediction of this protein we used two scaffolds with significant hits to make the prediction: QVIC01000001.1 i QVIC01009008.1. The second one, as the T-coffee alignment shows, seems to have a well conserved small fragment of the protein near to the C-terminal extrem. Nevertheless, this result was not consistent because it does not contain the Sec and is too small. On the other hand, the scaffold QVIC01000001.1 is divided in two genes, but it is due to an annotation error of the exonerate since these regions are a little overlapped from position 52540 to 50000. This problem was illustrated in order to clarify it: the orange region (gene 2) is contained in region blue (gene 2) as it can be seen in the figure below.
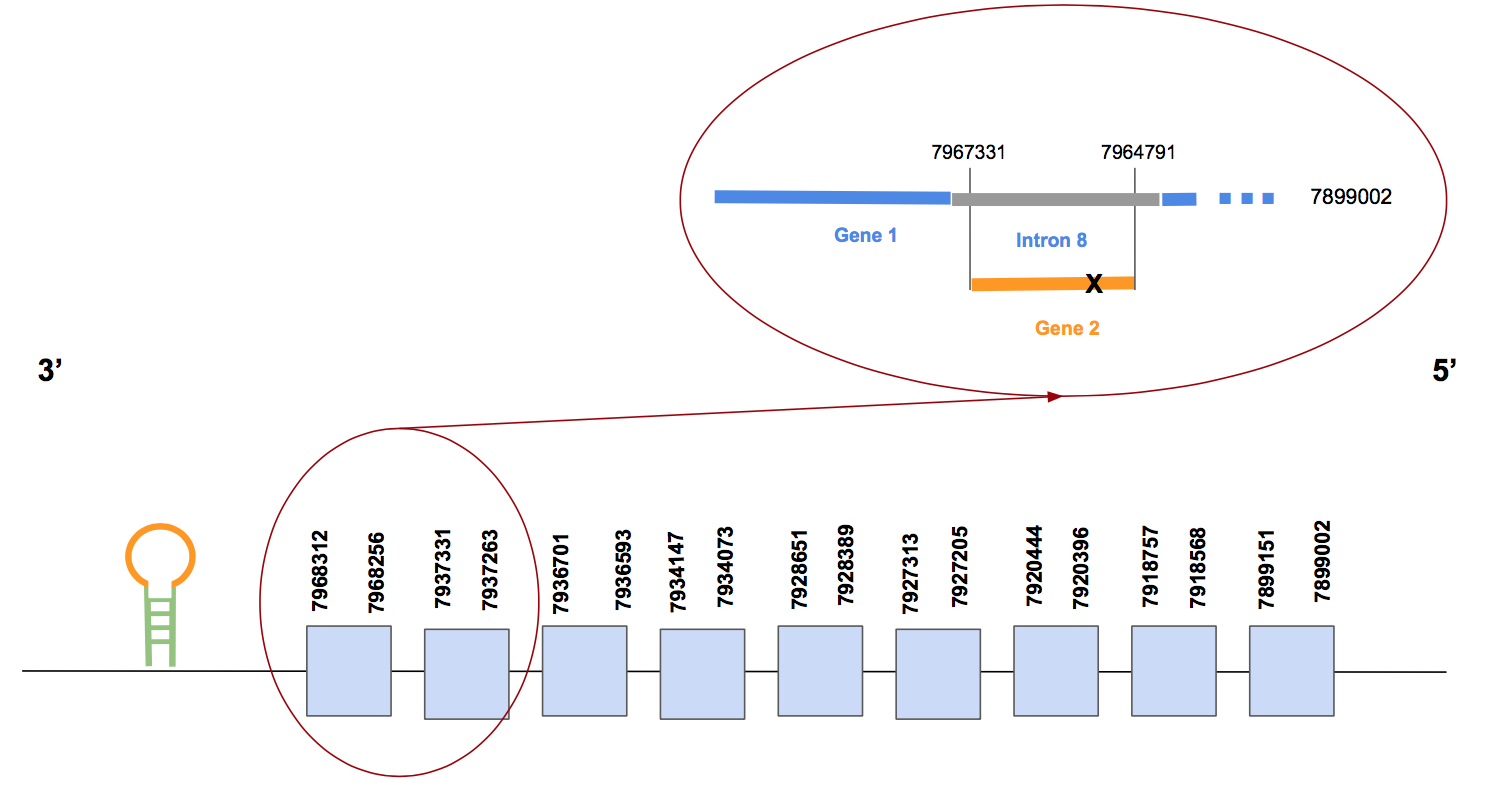
This fact affects our results, because it seems that the human protein is found in the scaffold named QVIC01000001.1 but splitted in both genes. As a consequence, the T-coffee shows that the gene 1 has the longest alignment, highly conserved including the methionine, but the C-terminal end is lost. On the other hand, the T-coffee of the gene two contains only the C-terminal part of the protein, containing the Sec in its first exon located in the forward strand. Thus, we should consider both genes united as the selenoprotein, but to make the interpretation easier we are going to focus only in the gene 1 because we have the longer alignment, although the Sec is not there.
The gene 1 of the scaffold QVIC01000001.1 has 9 exons and it is in the reverse strand. The exact location is from 7899002 to 7968312, it means 69310 nt totals [see the figure above again].
Finally, by using Seblastian, a SECIS structure was found in 3’UTR, located between 7913438 and 7913519. Therefore, we can affirm that the selenoprotein SelI is in U. Parryii genome, although it is divided in two different genes of the same scaffold as a result of an annotation error of the exonerate. The final prediction is attached here: SelI.finalprediction.txt
Selenoprotein K (SelK)Selenoprotein K is required for Ca2+ influx in immune cells and plays a role in T-cell proliferation and in T-cell and neutrophil migration. Also, it is involved in endoplasmic reticulum-associated degradation of soluble glycosylated proteins.
SelKFor this query, the scaffolds QVIC01000061.1. and QVIC01000157.1 were selected since both showed significant hits whose identity percentage was high, but, regarding the T-coffee alignment, we considered that QVIC01000061.1. was the best one. It has a score value of 1000, no gaps and small aa changes, including the Methionine as the first amino acid and the last aa of the human’s query sequence.
The predicted sequence is in the reverse strand, from 1576934 to 1582758 (5824 nt in total). It has four exons and the Sec may be detected in the first exon (considering 5’ → 3’ direction) [see figure below].
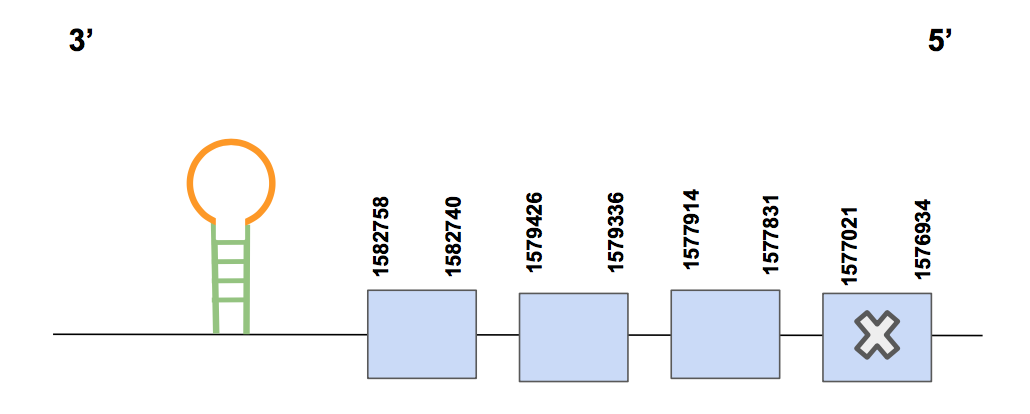
The Seblastian prediction indicates that there is a SECIS structure of grade A in the reverse strand located between positions 1527839-1527839 (3’UTR). Moreover, the Seblastian alignment support our hypothesis that SelK has been completely retrieved and it can be found in U.parryii’s genome.
Selenoprotein M (SelM)Selenoprotein M has a thioredoxin-like domain and a surface with an accessible redox motif. This selenoprotein is located in the endoplasmic reticulum and codify for an N-peptide that is cleaved once translocated into the endoplasmic reticulum.
SelMOnly one significant hit with 69% of identity percentage was reported by the tblastn, corresponding to the scaffold QVIC01000404.1. The T-coffee score is quite high, approximately 994, it has small aa changes and 0 gaps. Besides, one Sec match is found in the exon 4 (5’ → 3’ direction), which makes sense since we already expected SelM to be a selenoprotein.
The predicted protein has 5 exons and it is located in the forward strand between positions 833700 - 835916 (2216 nt) [see figure below].
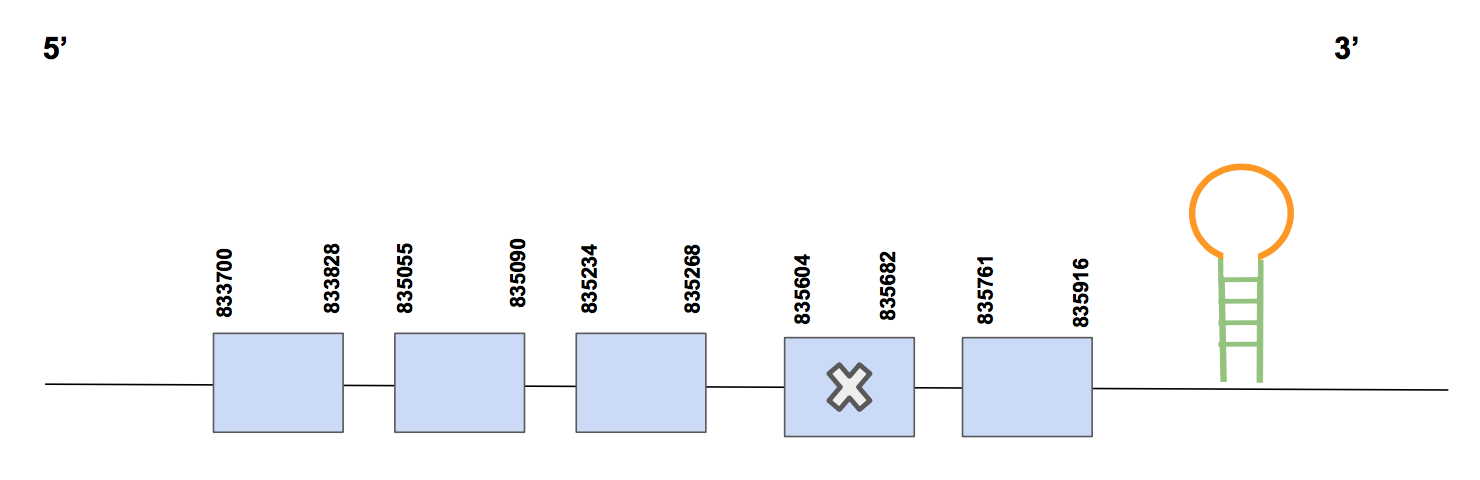
Eventually, Seblastian was not able to predict any SECIS, but SECISearch3 predicted one SECIS element of grade A in the forward strand located in 3’UTR positions 835960-836032. Thus, SelM is reported to be a selenoprotein present un U.parryii’s genome encoded by 5 exons.
Selenoprotein N (SelN)Selenoprotein N may be found in several tissues during embryonic development and it is thought to play a relevant role in myogenesis, although its exact function remains still unclear.
SelNRegarding this query, 9 hits were found for the same scaffold named as QVIC01000093.1, whose identity percentage is up to 90%. The T-coffee alignment shows several aa changes, although the majority are homologous changes. Furthermore, two small gaps were found, but, despite this, the alignment is still considered as a suitable one since it has the starting methionine and it covers the sequence until the last aa.
The predicted protein is in the reverse strand, between positions 5212494 - 5225573 (13079 nt). Two selenocysteines were found in the squirrel genome, one of them at the beginning of the sequence, specifically in exon 1. This one matches with the Human’s Sec. However, there is another Sec in the 3rd exon. This one does not match with another Sec in the human’s sequence. Nevertheless, if we look 5 positions onwards from this point (from where the second Sec is found in the U.parryii sequence), we find a second Sec in the human’s sequence. Despite this, we can consider both selenocysteines as valid since one matches and the other is almost a match [see figure below].
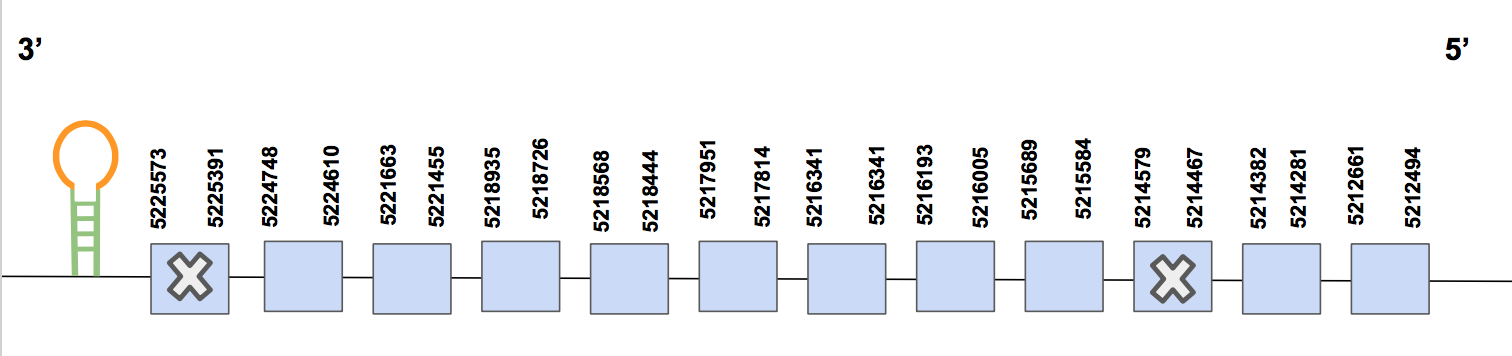
Regarding the Seblastian output, one SECIS structure of grade A was found in positions 5211329-5211399; it means that it is in 3’UTR and, also, in the reverse strand. Hence, the results obtained were the expected ones, we have annotated the complete sequence of SelN, which is present in U.parryii according to the results.
Selenoprotein O (SelO)Selenoprotein O’s main role is to regulate redox reactions, being located in the mitochondria.
SelORegarding this query, 9 hits were found for the same scaffold named as QVIC01000590.1. The T-coffee alignment has 988 score and the alignment is complete from the methionine to the end, there are 3 small gaps and several aa changes. The Sec found matches with the human’s Sec, located in the last exon (5’ → 3’ direction), as it is expected since SelO is a selenoprotein.
The predicted gene has 9 exons, located in the forward strand, and its coordinates are: 484262 - 502526, which means a total of 18264 nt [see figure below].
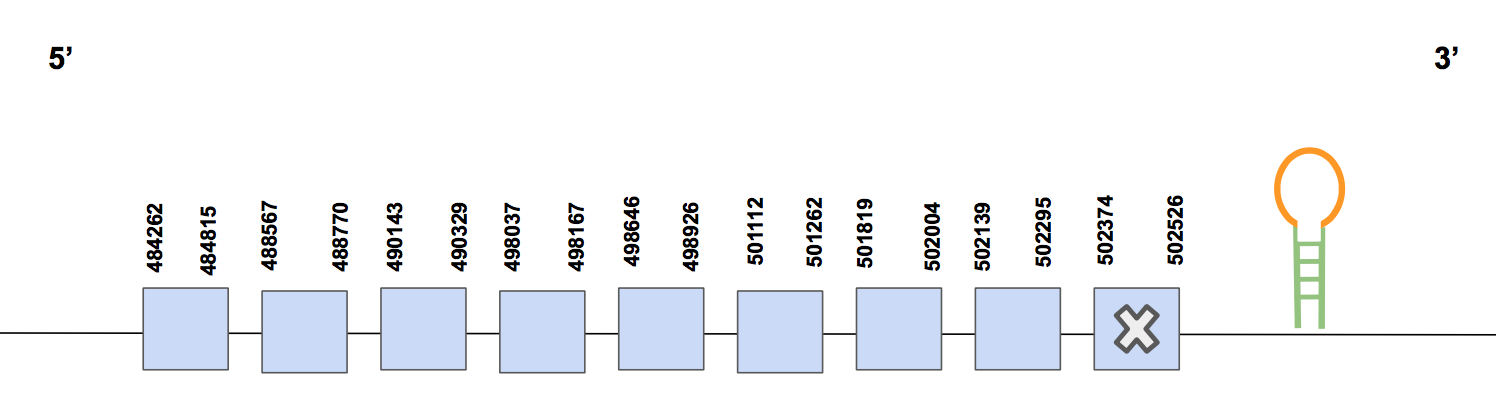
Seblastian provided a SECIS structure of grade A in the forward strand and 3’UTR (502589-502663 positions). To sum up, SelO is reported to be encoded by nine different exons and is present in our organism's genome.
Selenoprotein P (SelP)Selenoprotein P is associated with extracellular transport of the selenium and has also an essential role related to antioxidant effects.
SelPThere are 4 significant hits that refer only to one scaffold named QVIC01001039.1, whose identity percentages were not very high. The T-coffee alignment has a score of 982, including 2 gaps and some aa changes, but including the first methionine.
Regarding the selenocysteines’ analysis, there are 9 Secs that match with 9 human’s Sec spread in the different exons. Nevertheless, there are even more Sec predicted for the squirrel’s genome, specifically 4 Sec more that might be a result of an annotation error or may be it is because the squirrel has more Sec in this selenoprotein. Moreover, there is a Sec in the human’s query that matches with a Cysteine of the squirrel, which may be due to an aa substitution since both aa are quite similar.
The predicted protein has 9 exons, located in the forward strand between the positions 86117 and 92656 (length 6539 nt) [see figure below].
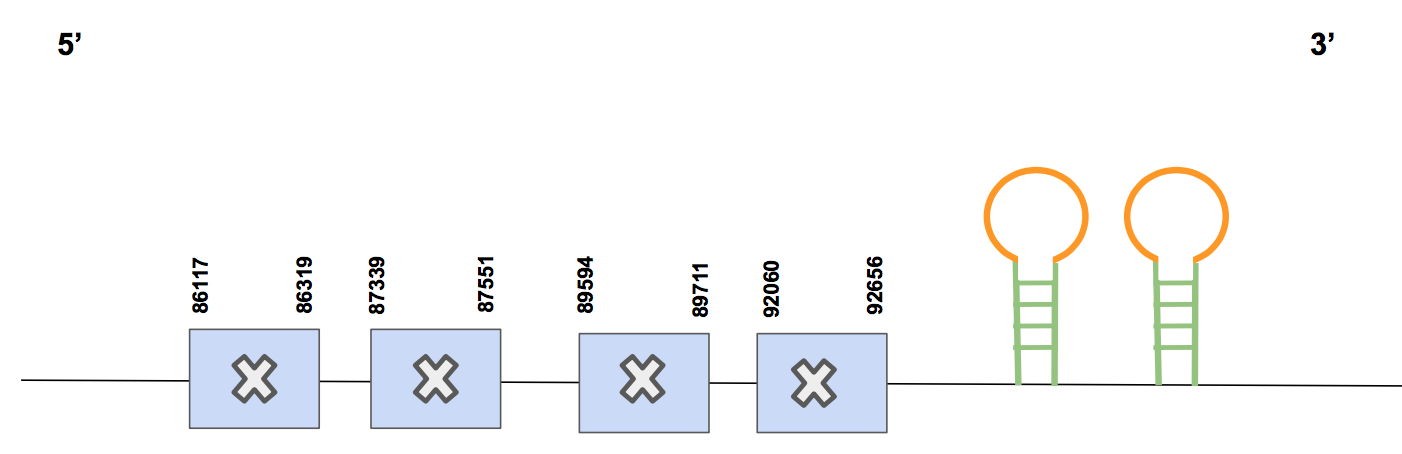
Finally, Seblastian predicted four valid SECIS structures, two of them located in the reverse strand and the others two in the forward strand. Only those located in the forward strand are valid and, also, they both are in 3’ UTR (coordinates 93384-93449 and 92916-92998). Considering that the Seblastian alignment also supports the other results, we can affirm that SelP is found in U. parryii genome, being a selenoprotein with two SECIS.
Selenoprotein R (MSRB)The enzymes belonging to this family are zinc-containing enzymes involved in the conversion of methionine sulfoxide to methionine.
SelR1The chosen scaffold was QVIC01000038.1, presenting 2 significant hits and an identity percentage of 75% out of 100. About the T-coffee results, we see that it aligns quite well, with matching selenocysteine and starting with methionine. Nevertheless, it has 3 gaps in the end of the alignment and a few aa changes.
We see that it is composed by 3 exons, and the Sec is found in the 1st exon, in the reverse strand. It’s location is between 2916500- 2918511, and composed by 2011 nt [see figure below].
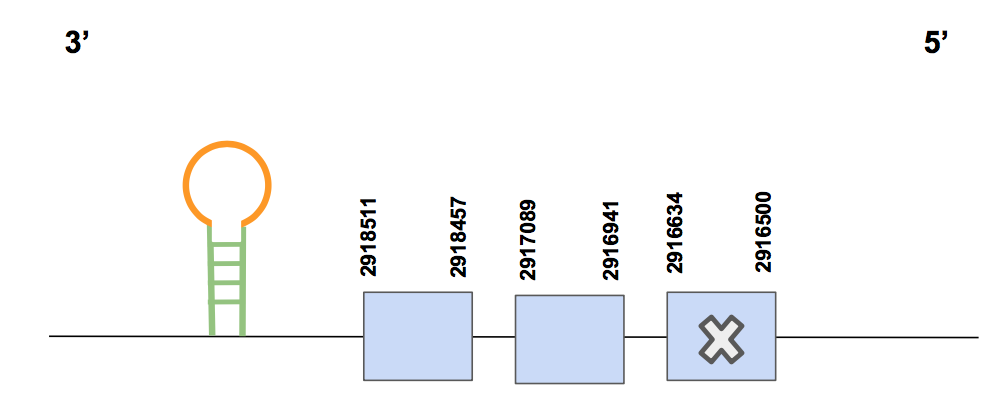
On the other hand, we had a SECIS prediction by Seblastian in 3’UTR in the positions 2914867-2914938, which fits with our expectations. So, we can conclude that SelR1 is a selenoprotein present in the Urocitellus parryii genome, and that we were able to recover almost the whole protein, missing 3 aa in the end.
SelR2The chosen scaffold was QVIC01000241.1, with 4 significant hits, and an identity percentage between a 77% to 84% out of 100. With relation to the T-coffee results, we can see that there is no Sec alignment, we can find two in U. parryii’s genome, but they do not correspond with any in the reference genome. Furthermore, we missed the starting part of the protein and were not able to retrieve it, since more scaffolds were not run and Seblastian did not give an output in this case.
Our predicted protein is constituted by 5 exons. The exact location is from 160987-181530, in the forward strand and composed by 20543 nt. The selenocysteines are located in exon 3 and in exon 5 [see figure below].
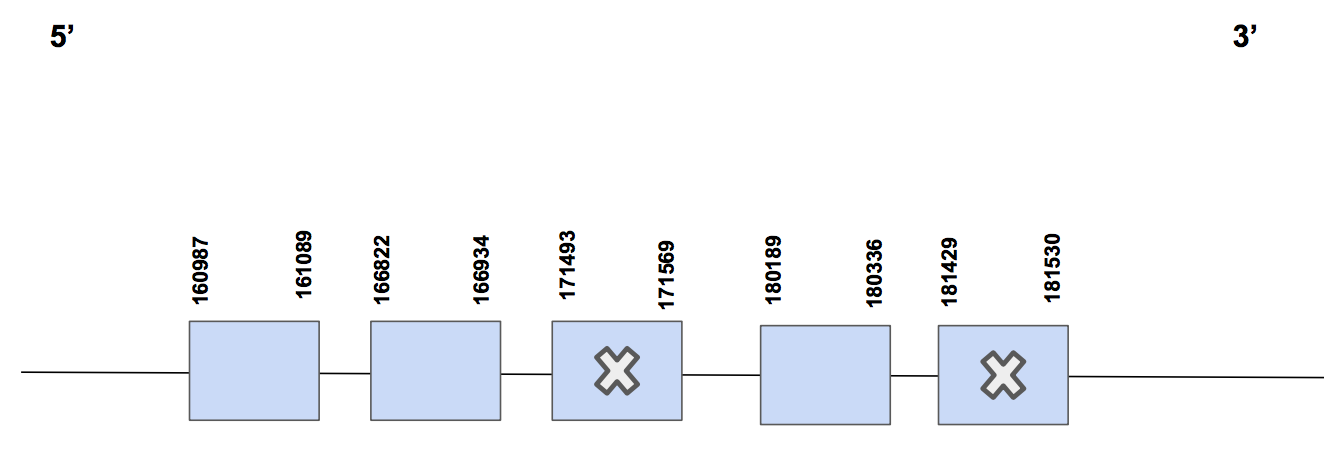
In this case, neither Seblastian nor SECISearch3 were able to predict any SECIS element. So we cannot conclude that SelR2 is a selenoprotein. First of all because no SECIS element has been found, so there is no way a Sec can be included in the sequence. Moreover, if the Sec cannot be included it means that the only way of reading a UAG codon is as a STOP codon. However, in this case we see that there are more amino acids present after this one, so we could affirm that an annotation error has happened and that SelR2 is not a selenoprotein.
SelR3In this case the scaffold QVIC01000046.1 was chosen to perform our protein prediction. It has four significant hits, with an identity percentage of 96%. The T-coffee alignment shows no Sec in human nor in Urocitellus parryii genomes; we found matching cysteines in different positions. The alignment is appropriate since it begins with methionine and has only two gaps plus a certain number of aa changes.
The predicted protein is constituted by six exons, more exactly from 162044-318665, in the forward strand and composed by 156621 nt [see figure below].
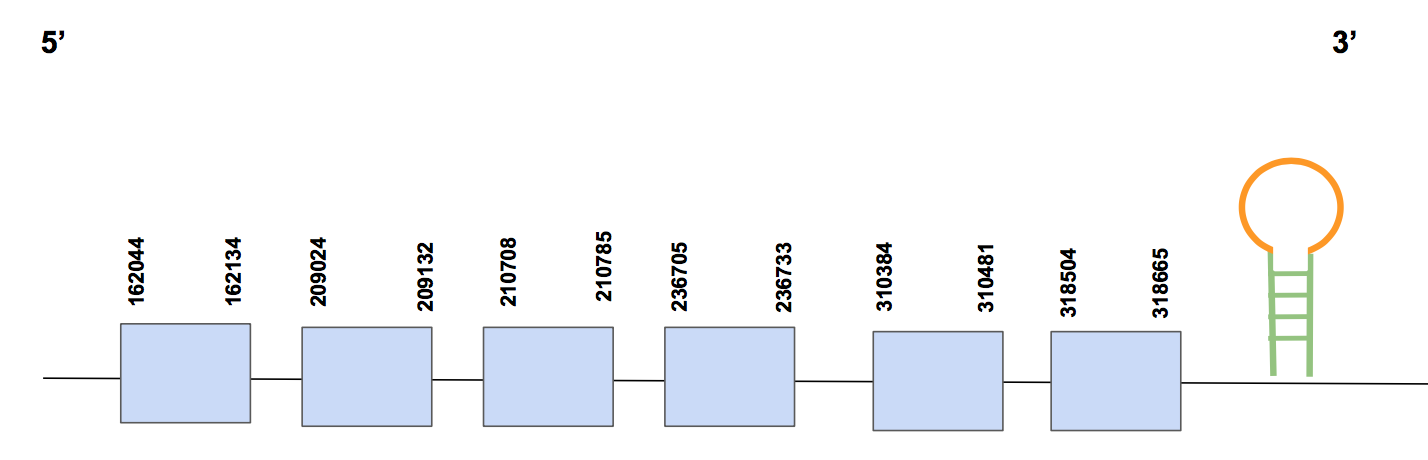
In this case, Seblastian did not predict any SECIS element. When running SECISearch3 we obtained a SECIS prediction in 3’UTR of the forward strand (323321-323397 positions). Considering all the evidence together, we can conclude that the protein SelR3 is a cysteine homolog, in both human and Urocitellus parryii, with still a remaining SECIS structure. We were able then to annotate properly and successfully the protein in U. parryii genome.
A phylogenetic tree was performed in order to double-check if our predicted proteins are accurate enough.
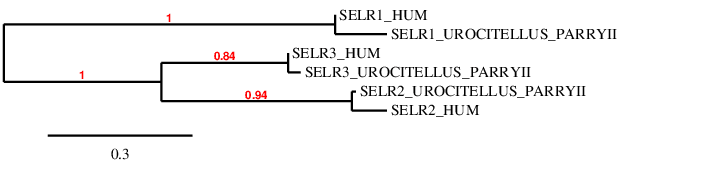
This protein is involved in the transfer of misfolded proteins from the endoplasmic reticulum to the cytosol.
SelSScaffold QVIC01000145.1, has four significant hits, with an identity percentage between 61% and 89%. The T-coffee alignment is good, it starts with methionine and has a few aa changes. However, at the Sec location in the human genome, we see a gap.
The predicted protein has six exons between positions 418327-442692, in the forward strand, and composed by 24365 nt [see figure below].
Seblastian did not give a prediction in this case. When running SECISearch3 we obtained four SECIS predictions, one of which was in 3’UTR of the forward strand and was of grade A and located in positions 443032-443119.
Considering all the evidence together, we can conclude that the genome of Urocitellus parryii has lost the Sec aa while keeping the SECIS. This selenoprotein is not present in the U. parryii genome, and we were able to successfully annotate it.
Selenoprotein T (SelT)The functional information about this family is limited, although we know that they are thioloxireductases.
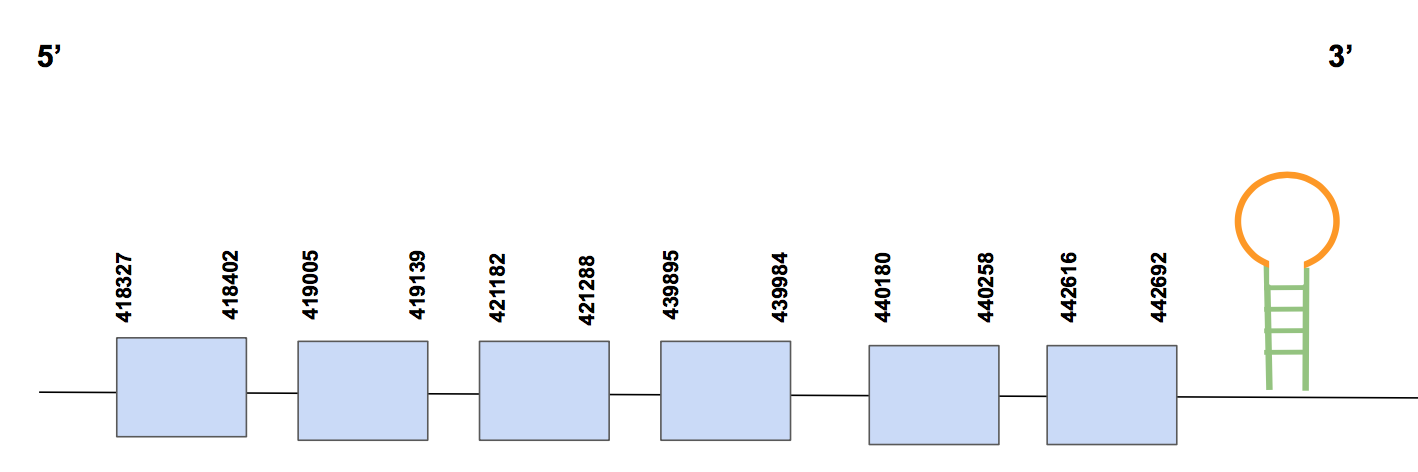
SelTScaffold QVIC01000380.1 has five hits with a percentage of identity of 84%. The T-coffee alignment starts with methionine, has 10 gaps at the end and a few aa changes. Interestingly, besides an aligned sec aa in human and Urocitellus parryii, there are 12 sec aa that do not match any human one.
The predicted protein has five exons, between positions 315906-334480. The Sec aa is in exon 2 (the Sec matching with the human genome), and in exon 3 (the repetitions of Sec) and composed by 334480 nt [see figure below].
When running SECISearch3 we found a SECIS in 3’UTR in the forward strand of grade B, between positions 341198-341278.
Considering all this evidence, we can conclude that SelT is a selenoprotein in Urocitellus parryii genome, although we observe some arrangement errors and the possibility of microsatellite duplications. We see that we were not able to recover the whole protein due the presence of 10 missing aa in the beginning.
Selenoprotein U (SelU)The function of this family is unclear, although it is believed that it is involved in the regulation of a large number of biological processes, through its redox function. It has several homologs in which the sec aa is replaced by cysteine. It consists in three proteins: SelU1, SelU2 and SelU3.
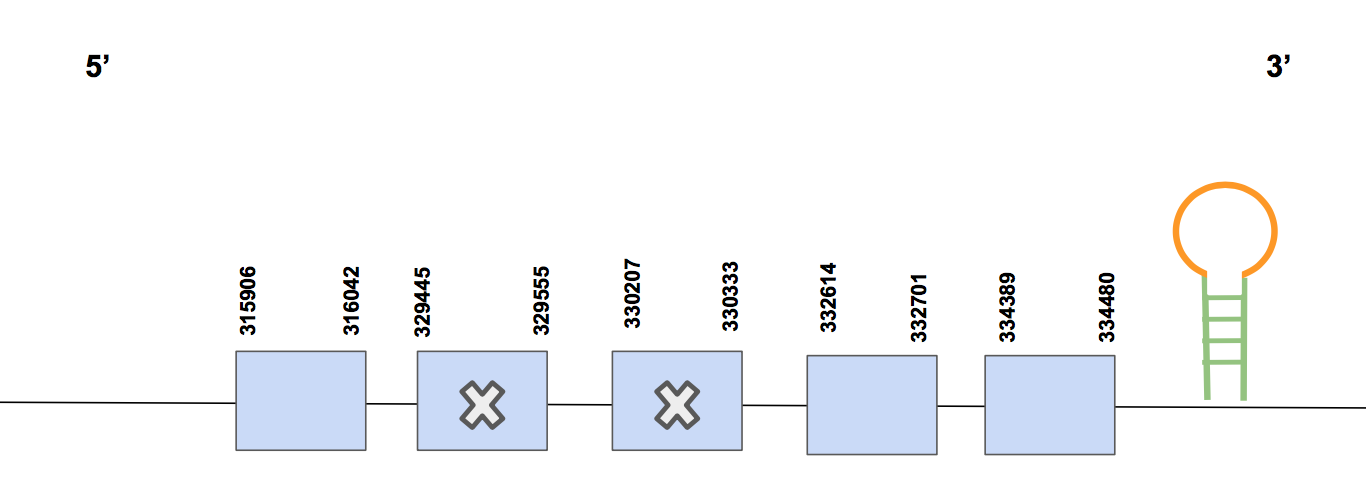
SelU1Scaffold QVIC01000340.1 has five significant hits with an identity percentage of 86.4%. The T-coffee alignment starts with methionine, it has no gaps and almost no aa changes, so it’s a quite good alignment. Besides, as expected, no Sec was found in the sequence.
The predicted protein has five exons, between positions 292138-301982 in the forward strand, and is composed by 9844 nt [see figure below].
In this case, the Seblastian did not give an output. When running SECISearch3 we found a SECIS in 3’UTR (positions 307806-307886), but it is in the reverse strand, so it is not valid. It has sense since SelU1 is a Cys-containing homolog.
Considering all this evidence, we can conclude that protein SelR3 is a cysteine-containing homolog which is present in Urocitellus parryii. We were able to annotate properly and successfully this protein in U. parryii genome.
SelU2Scaffold QVIC01000808.1 has three significative hits, with a identity percentage of 87%. The T-coffee alignment starts with methionine, has 1 gap of 4 aa at the beginning and a few aa changes. We find a sec in Urocitellus parryii, in exon 6, but not in human. One Sec was found in the predicted protein, but we attributed this fact to an error since this protein has no SECIS and, if not, it means that this codon will be codified as an STOP codon. On the other hand, there are three cystein matches, as it is expected.
The predicted protein has six exons between positions 61635-72278, located in the forward strand, and composed by 10643 nt [see figure below].
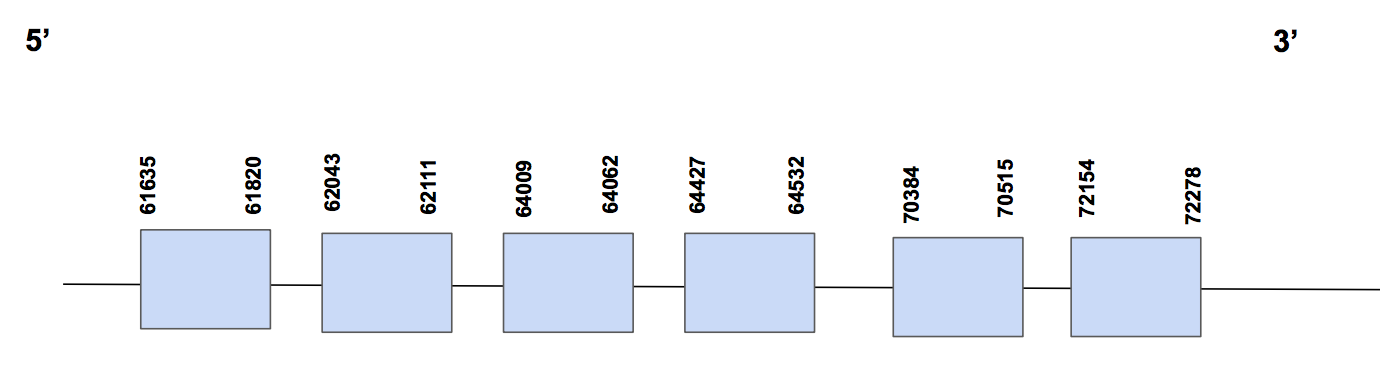
Seblastian was not able to give an output in this case. When running SECISearch3 we found a SECIS in 3’UTR (located in positions 117852-117935) of grade B, but it is in the reverse strand so it is not valid.
Considering all this evidence, we can conclude that SelU2 is a Cys-containing homolog present in U. parryii genome. We were able to recover the main part of the protein.
SelU3Scaffold QVIC01000810.1 has three significant hits, with a sequence identity of 64%. The T-coffee alignment starts with methionine, has 1 gap of 5 aa at the end and a few aa changes. There is no sec aa, neither in human nor in Urocitellus parryii. But, we could find matching cysteins in both genomes in different positions.
The predicted protein has six exons between positions 26228-28171, in the reverse strand, and composed by 1943 nt [see figure below].
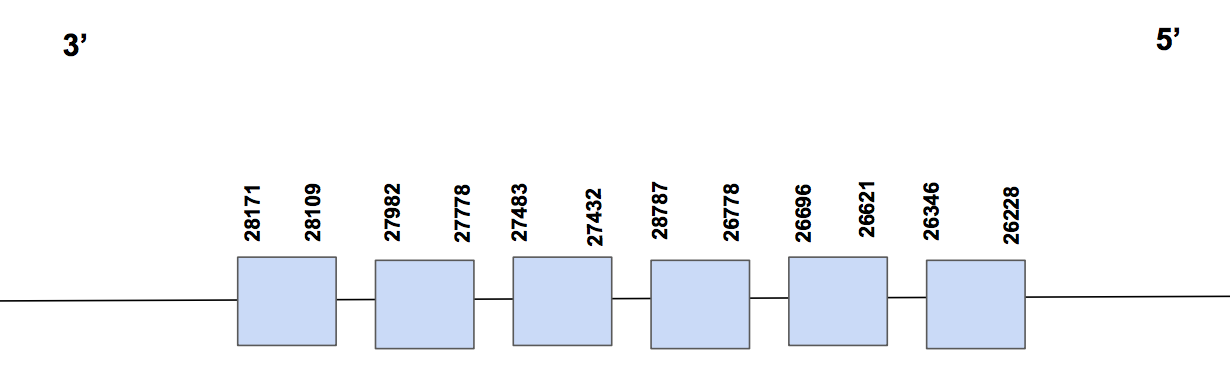
In this case, no SECIS was found, neither with SECISearch3, nor with Seblastian. Therefore, we can can conclude that it’s a cysteine-containing homolog, and that we’ve been able to recover the main part of the protein, considering that it presents five missing aa at the end.
A phylogenetic tree was performed in order to double-check if our predicted proteins are accurate enough.
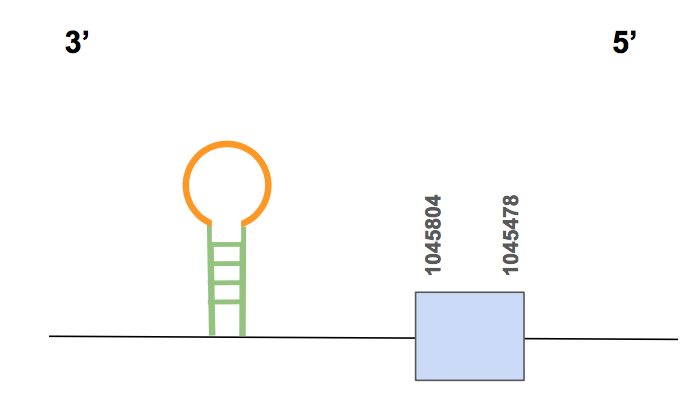
This protein is believed to be a duplication of selenoprotein SelW. Preserves a Cys-x-x-Sec motif, suggesting a redox function for this gene, as in SelW.
SelVIn this case the scaffold QVIC01000501.1 was the only one that presented significant hits, with an identity of 73% in general, which is not so high but is the only one. 9 genes were obtained for this scaffold, but none of them showed an alignment good enough since there are so many gaps, amino acid changes, etc. Nevertheless, SECISearch3 predicted one valid SECIS. Taking everything into account, we concluded that SelV is not present in U. parryii.
This family possesses a thioredoxin-like fold and a conserved CxxU (C is cysteine, U is Sec) motif. This facts suggests a redox function for this gene. It has been discovered that they play a role in muscle growth and differentiation in mouse. Moreover, it seems that they also play a key role in the protection of neurons from oxidative stress during neuronal development (also seen in mouse).
Selw1SPP00000030 shows just 1 significant hit and it is found in the scaffold QVIC01000457.1, whose identity is 92%. The T-coffee alignment score is equal to 999 and we can see that even if there are just small changes of aa, there is a big gap at the beginning that means that a part of the protein has not been found and may be at another part of the genome. But the tBLASTn results did not find any other hits in the genome, so there is no other way to find out where is the lacking part of the protein. In these cases, we check the Seblastian results to see if query has been found in any other specie so that we can find out what should be the beginning of the protein. Running this program, no SECIS where predicted so we could not find out the missing part of the protein.
Furthermore, the Sec found in the the query does not match with any position of the predicted protein as it matches with the part of the protein that is missing. Owing to this fact, we can not say that it is a Selenoprotein or not. Actually, we expected it to be as it is a Selenoprotein in humans.
The protein has 3 different exons, is found in the forward strand and has 480 nt in total (positions: 1339581- 1340061) [see figure below].
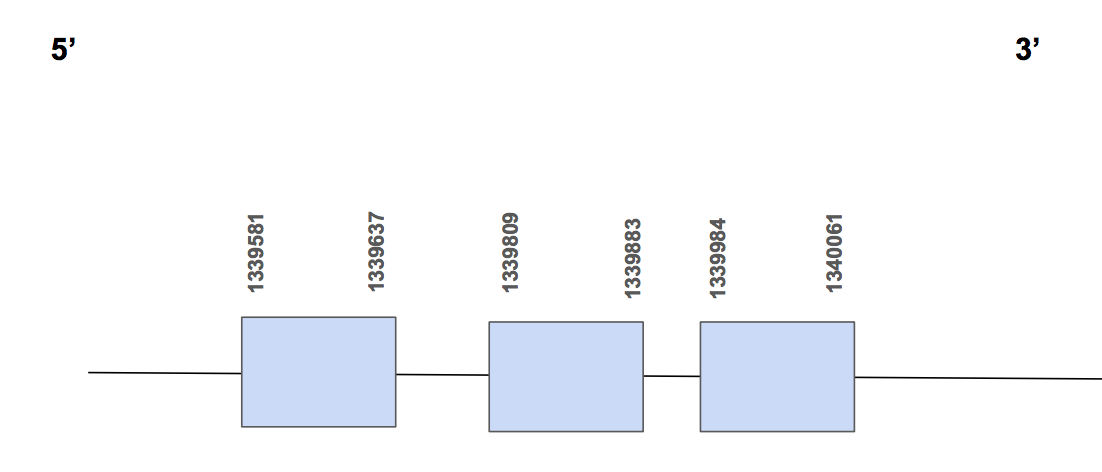
Seblastian was not able to predict a SECIS element in this case. SECISearch3 predicted one SECIS of grade B located in the forward strand but in 5’UTR (1295924 - 1296001 positions). Therefore, we can say that SelW1 may not be a Selenoprotein in U. parryii genome as the SECIS should be found in the 3’UTR. Above all, we conclude that this selenoprotein may not be found in U. Parryii.
Selw2SPP00000031 shows 4 hits and all of them are found in the scaffold QVIC01000047.1, whose identity is between 86-96%. The T-coffee alignment score is equal to 100 and we can see just small changes of aa. We also observe that there is a very good homology with the cys-homolog human query and that the predicted protein starts with a methionine (M).
Furthermore, in humans this protein is not a selenoprotein, it is a cysteine, as we said in the introduction part. We can see that there are 3 cysteins found and that all of them match with a cysteine in the U. Parryii’s genome. So even if we do not know which of them was a selenocysteine in the past, as all of them match, we can conclude that in U. Parryii this protein is not a selenocysteine, it has been converted to a cysteine.
The protein has 4 different exons, is found in the reverse strand and has 978 nt in total (positions: 5058218 - 5057240) [see figure below].
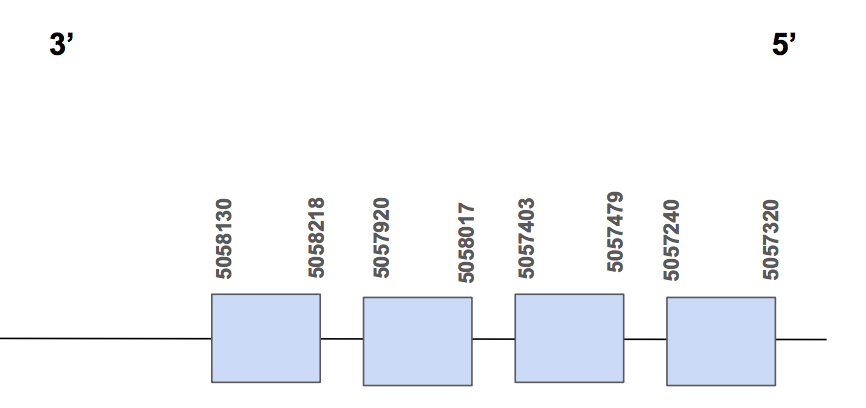
As Seblastian was not able to predict any SECIS element, we ran SECISearch3 predicted one SECIS of grade B located in 5’UTR (5090808 - 5090877 positions) and in the forward strand, so it is not valid, but anyway no SECIS is needed for a Cys-containing homolog. Therefore, we can say that cys-homolog SelW2 is found in U. Parryii genome. Above all, the SelW2 of U. Parryii was completely and successfully annotated.
Selenophosphate synthetase (SPS)Its function is to generate the selenophosphate necessary to incorporate selenium into selenocysteine.
SPS1At the beginning, we were not sure about the scaffold that we should use. Because of that, we checked QVIC01000450.1 and QVIC01000220.1. Once we got the results, we saw that the T-coffee obtained with the saffold QVIC01000450.1 matched perfectly with our query. So in order to do our discussion, we are going to focus on this one.
Regarding the significant results of the tblastn (e-value smaller than 0.001), the scaffold QVIC01000450.1 showed 8 hits with a high percentage of identity, between 68 and 100%. The T-coffee results show a 1000 score and no gaps neither amino acid changes were found in the alignment. We finally chose this as the scaffold where our predicted protein would be located in. We can see that it starts with methionine (M).
Furthermore, in humans this protein is not a selenoprotein, it is an other amino acid containing homologous. So we can not find any selenocysteine but we can say that the same type of protein is found in U. Parryii as the homology is perfect.
The protein has 7 different exons, are found in the forward strand and has 28901 nt in total (positions: 1363449 - 1392350) [see figure below].
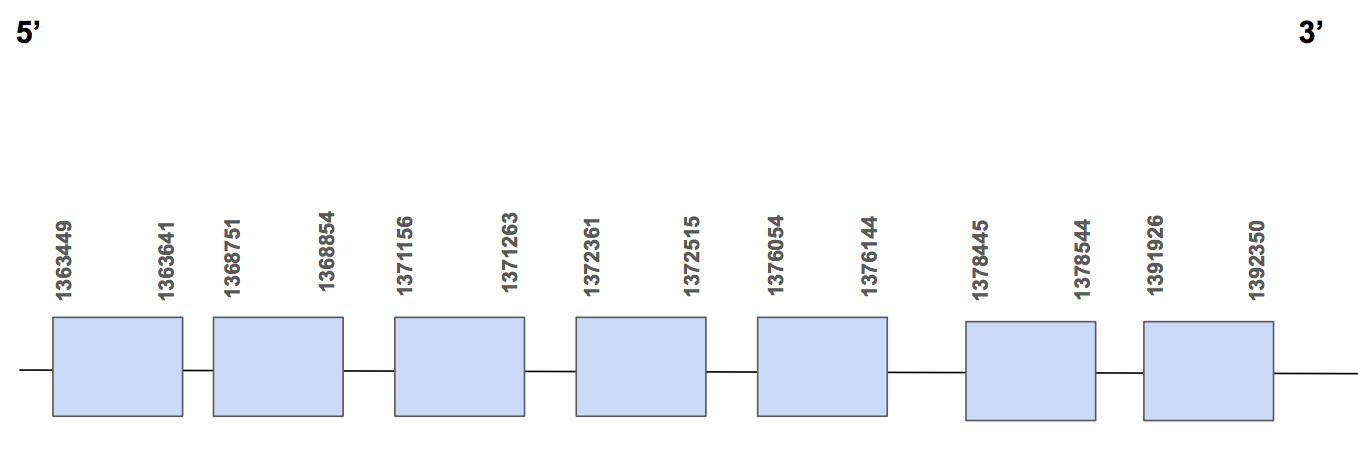
Neither Seblastian nor SECISearch3 could found SECIS elements. Therefore, we can say that an amino acid homolog of SPS1 is found in U. Parryii’s genome. Above all, the SPS of U. parryii was completely and successfully annotated.
SPS2SPP00000033 shows 13 hits and they are distributed in different scaffolds. The higher percentages of identity and the lower e-values where found in QVIC01000230.1, whose identity is 93% and we can just observe one hit. The T-coffee alignment score is equal to 998 and we can see a small amount of amino acid changes. We also observe that there is a very good homology except from the beginning, where we can see that there is a small gap of 4 amino acids so that the predicted protein does not start with a methionine (M). As this scaffold was not the only one that was found with hits in tBLASTn, maybe we could find this 4 aa in another part of the genome. With the results that we obtained running Seblastian we can see that the first 4 amino acids do not match perfectly with the human genome (the 4rt changes) but this is normal as they are comparing the query with other animals. Anyway, we are gonna use this 4 amino acids added to our prediction to do the phylogenetic tree. See the document attached.
Apart from this, we can see that the selenocysteine found in the query matches with a selenocysteine found in the U. Parryii’s genome. Furthermore, in humans this protein is a selenoprotein so we actually found what we expected.
The protein has 1 exon, it is found in the forward strand and has 1340 nt in total (positions: 3087827 -3089167) [see figure below].
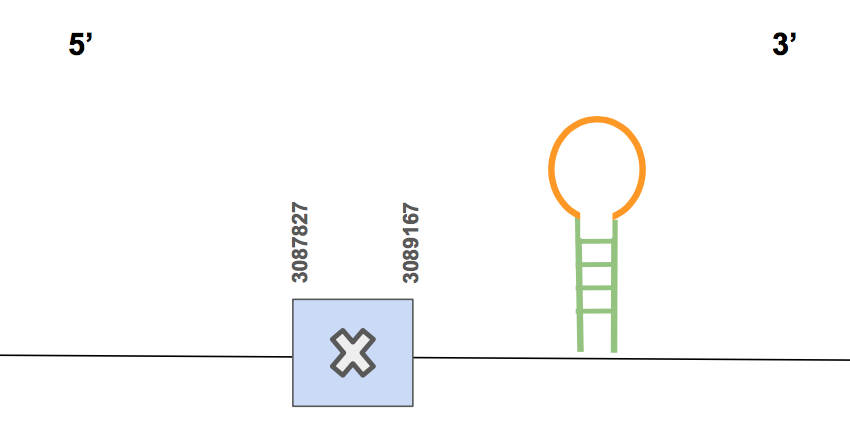
Seblastian predicted one SECIS of grade A located in 3’UTR in the forward strand (3089722 - 3089798 positions); thus, this SECIS is valid. Therefore, we can say that the selenocysteine SPS2 is found in U. Parryii genome. Above all, the SPS2 of U. Parryii was completely and successfully annotated as a selenoprotein of the squirrel.
A phylogenetic tree was performed in order to double-check if our predicted proteins are accurate enough.
These proteins reduce thioredoxins among other substrates and they also play an important role in the metabolism of selenium and the protection against oxidative stress.
TR1SPP00000034 shows 28 hits and they are distributed in different scaffolds. At the beginning, we were not sure about the scaffold that we should use. Because of that, we checked QVIC01000007.1 (with 10 hits and 59-92% of identity) and QVIC01000475.1 (with 8 hits and 53-81% of identity). Once we got the results, we saw that the t-coffee obtained QVIC01000007.1 had a much better homology. The T-coffee alignment’ score is equal to 1000 and we can see just a few amino acid changes and no gaps. Moreover, there is a very good homology and the predicted protein starts with a methionine (M).
Furthermore, we can see that the selenocysteine found in the query matches with a selenocysteine found in the U. Parryii’s genome. In humans, this protein is a selenoprotein so we actually found what we expected.
The protein has 13 exons, it is found in the reverse strand and has 38083 nt in total (positions: 3537207 - 3499124). The selenocysteine is found in the exon 13 [see figure below].
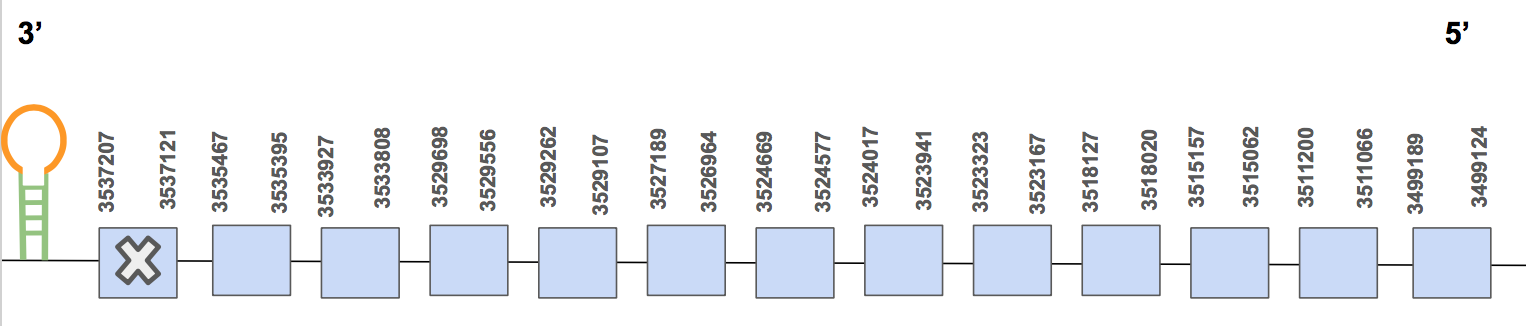
Seblastian predicted a valid SECIS structure of grade A located in the reverse strand and 3’UTR (3498896 - 3498818 positions). Furthermore, the Seblastian alignment supports our hypothesis. Hence, we can say that the selenoprotein TR1 is found in the U. Parryii’s genome. Above all, the TR1 of U. Parryii was completely and successfully annotated.
TR2SPP00000035 shows 32 hits and they are distributed in different scaffolds. At the beginning, we were not sure about the scaffold that we should use. Because of that, we checked QVIC01000007.1 (with 6 hits and 37-72% of identity), QVIC01000475.1 (with 6 hits and 42-76% of identity) and QVIC01000253.1 (with 12 hits and 53-100% of identity). Once we got the results, we saw that the t-coffee that showed better results was using QVIC01000253.1. The T-coffee alignment’ score is equal to 999 and we can see a few amino acid changes and a gap in the first amino acid, so that the predicted protein does not start with a methionine. With the results that we obtained running Seblastian, we can see that the first amino acid found in the target is a methionine so that we can recover it and add it to our prediction to do the phylogenetic tree (attached doccument).
Furthermore, we can see that the selenocysteine found in the query matches with a selenocysteine found in last exon of U. Parryii’s genome. In humans, this protein is a selenoprotein so we actually found what we expected.
The protein has 17 exon, it is found in the forward strand and has 55578 nt in total (positions: 645475 - 701053). The selenocysteine is found in the exon 17 [see figure below].
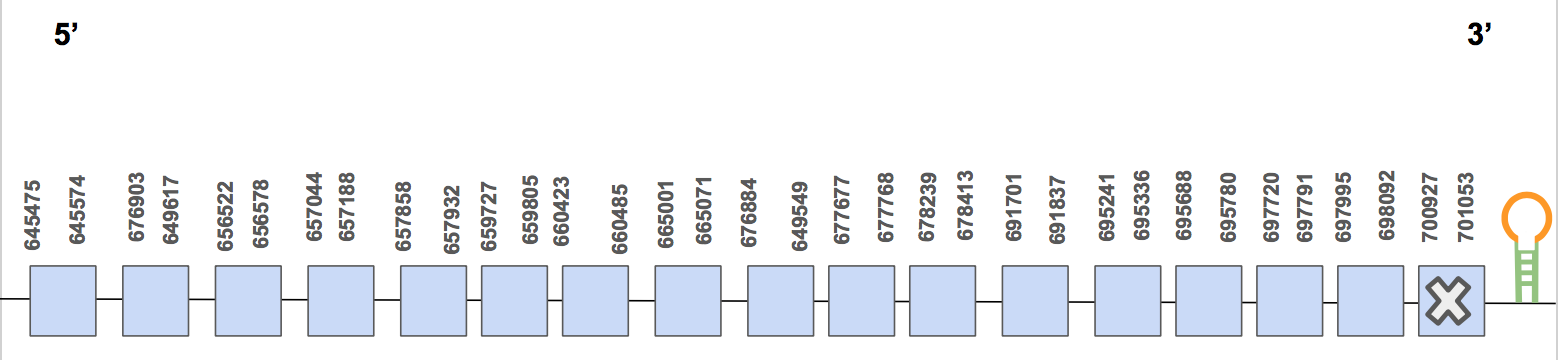
Seblastian predicted one valid SECIS of grade A located in 3’UTR of the forward strand (702421 - 702494 positions). Therefore, we can say that the selenocysteine TR2 is found in the U. Parryii’s genome. Above all, the TR2 of U. Parryii was completely and successfully annotated.
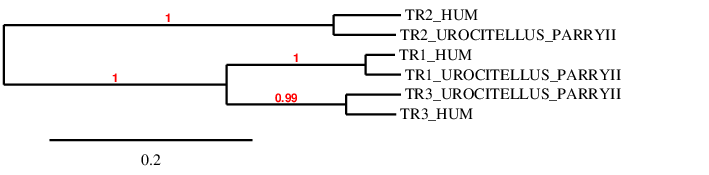
TR3SPP00000036 shows 33 hits and they are distributed in different scaffolds. At the beginning, we were not sure about the scaffold that we should analyze. Because of that, we checked QVIC01000007.1 (with 9 hits and 45-88% of identity), QVIC01000475.1 (with 10 hits and 62-91% of identity) and QVIC01000137.1 (with 6 hits and 33-85% of identity). Once we got the results, we saw that the t-coffee that showed better results was using QVIC01000475.1. The T-coffee alignment’ score is equal to 999 and we can see a few amino acid changes but a huge gap at the beginning of the alignment. This could be owing to the fact that this part of the protein could be found in another scaffold that we did not run with the program (we run 3 out of 6), as all the scaffolds that we analyzed have a similar gap at the beginning of the prediction. We did not use more scaffolds because their percentage of identity was lower and because their homology with other proteins from the same family was bigger than with this one. In this case the results that we obtained running Seblastian did not help us predicting the part of the protein that was missing.
However, we can see that the selenocysteine found in the query matches with a selenocysteine found in the last exon of U. Parryii’s genome. In humans, this protein is a selenoprotein so we actually found what we expected.
The protein has 15 exons, it is found in the forward strand and has 30731 nt in total (positions: 1165232 - 1195963). The selenocysteine is found in the exon 8 [see figure below].
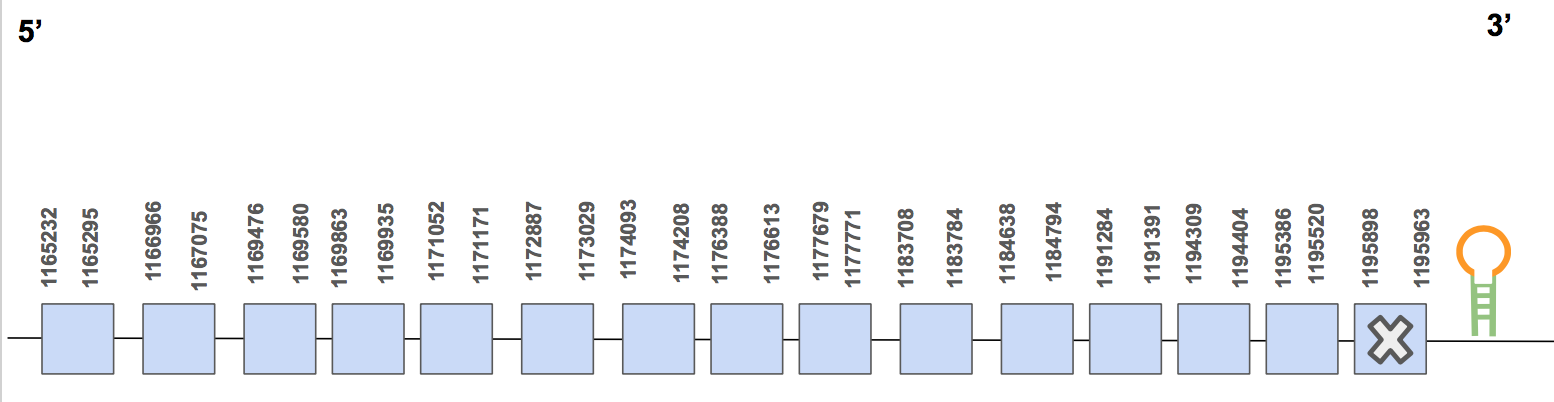
Seblastian predicted one SECIS of grade B located in 3’UTR (1196163 - 1196217 positions). Therefore, we can say that the selenocysteine TR3 is found in the U. Parryii’s genome. Above all, the TR3 of U. Parryii has been partially annotated.
A phylogenetic tree was performed in order to double-check if our predicted proteins are accurate enough.
Selenoprotein Machinery
SECIS binding protein 2 (SBP2)In humans, his protein plays a role in the selenium molecular machinery. This protein binds to the SECIS element which has been specifically stimulated by a Sec-specific elongation factor.
SBP2SPP00000037 shows 18 hits and they are distributed in different scaffolds, but most of them are found in QVIC01000360.1 (with 14 hits and 56-97% of identity) so we only run this one. The T-coffee alignment’ score is equal to 990 and we can see some amino acid changes and a gap that includes the first 12 amino acids. This could be owing to the fact that this part of the protein could be found in another scaffold that we did not run with the program.
Moreover, we can see that nor the query neither the prediction had a selenocysteine. This matches with our previous idea because in humans it plays a role in the selenoprotein synthesis process but it is not a selenocysteine, the same that it seems to be in U. parryii.
The protein has 16 exons, it is found in the reverse strand and has 35203 nt in total (positions: 1069642 - 1034439) [see figure below].
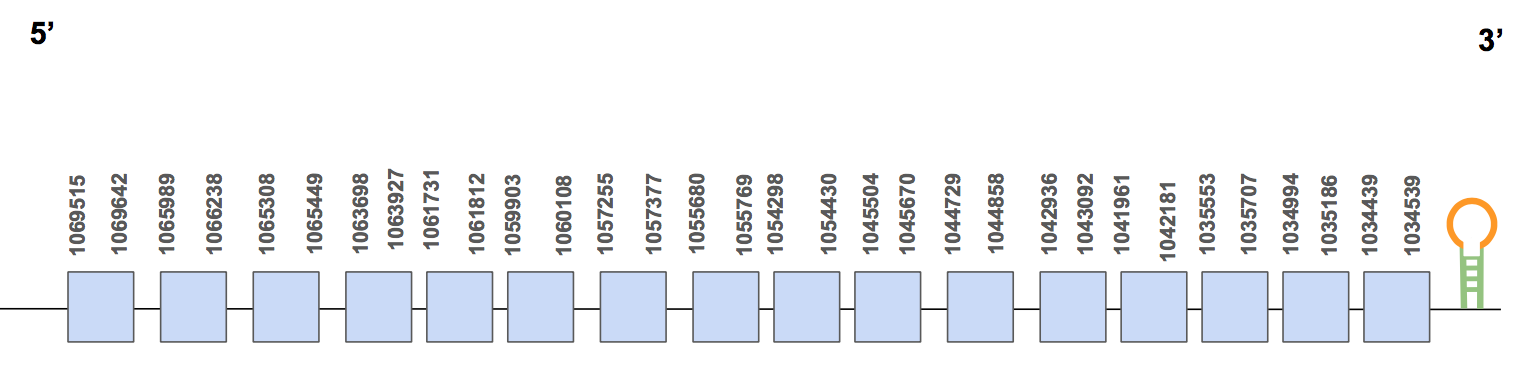
SECISearch3 predicted one SECIS of grade B located in 5’UTR (1066790 - 1066863 positions) and in the forward strand, since it is not valid, as it is expected because the main function of the protein is to bind to SECIS. Therefore, we can say that the protein SBP2 is found in the U. Parryii’s genome and that it does not have a selenocysteine.
Eukaryotic elongation factor (eEFsec)In eukaryotes, his protein plays a role in the selenium molecular machinery. This protein takes on the role of elongation factor specific to selenocysteines.
eEFSecIn this case, 6 significant hits were shown for the scaffold QVIC01000475.1, with an identity percentage of 75-95%. This scaffold contains two different genes that were predicted, but it is important to mention that they are contiguous and it can be clearly appreciated in both T-coffee alignments. We are going to focus in gene number 1 because it has a higher score, much more homology and a longer alignment. Nevertheless, a considerable part of the N-terminal extrem is lacking. No Sec was found in the sequence, which makes sense since it belongs to the family of selenoprotein synthesis machinery.
The predicted protein has 7 exons, located in the reverse strand, positions from 398640 to 513874 (115234 nt length) [see figure below].
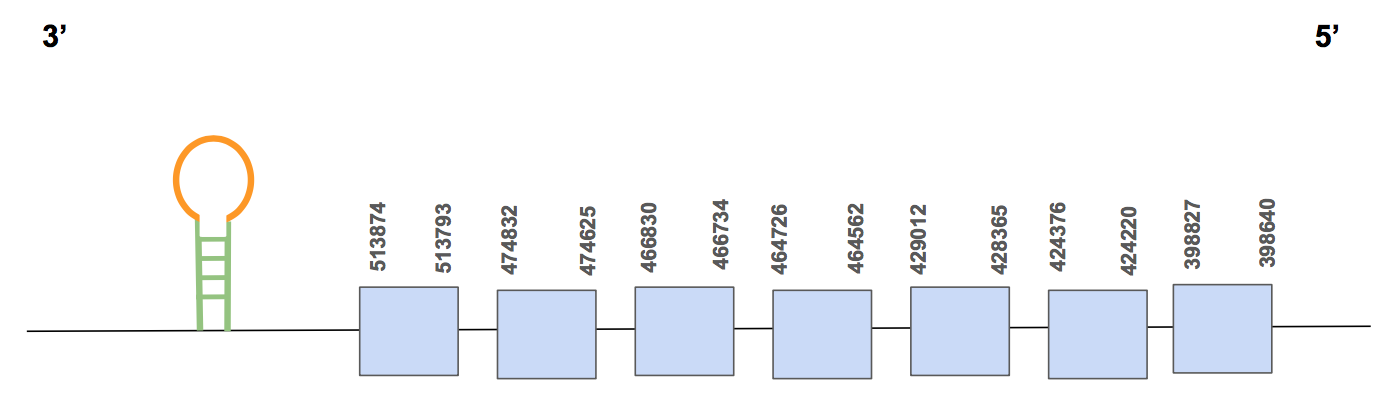
Here, 5 predictions were found through SECISearch3, with different grades. Two of them were in the forward strand, so they are not valid. Of the other two, only one is valid since it is located in 3’UTR (positions 381569-381495). Although it seems that there is a valid SECIS, there is not Sec, so it makes sense because eEFsec is a protein of selenoproteins machinery.
Finally, for presenting the best prediction, we decided to join both genes of this scaffold, so a more accurate protein can be shown [see eEFsec.finalprediction.txt]. Therefore, we conclude that eEFsec was found in Urocitellus parryii, even though a short part of the N-terminal part is still lacking.