Introduction
Selenium
Selenium (Se) is an essential micronutrient with important functions in human health and relevance to several pathophysiological conditions. This micronutrient supports various important cellular and organismal functions. Its deficiency has been recognized as a contributing factor to pathophysiological conditions such as heart disease, neuromuscular disorders, cancer, male infertility and inflammation. The biological effects of Se are mediated by selenium-containing proteins (selenoproteins) present in all three domains of life. [1]
Selenoproteins
A selenoprotein is a protein which has one or more selenocysteine residues. The selenocysteine (Sec) is an amino acid known as the 21st “naturally ocurring” amino acid in the genetic code and has a very similar structure to the cysteine. Instead of a Sulphur atom the selenocystein has a Selenium atom:
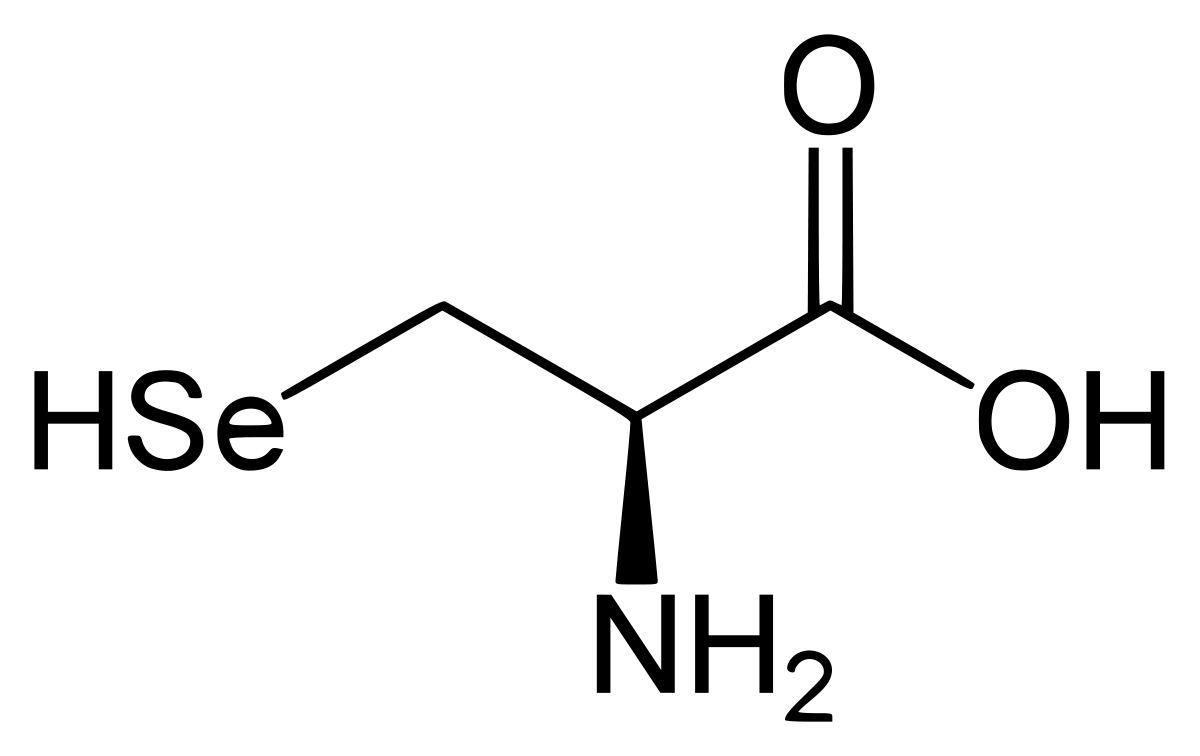
All selenoproteins contain at least one selenocysteine and most serve oxidoreductase functions. Sec is inserted into polypeptide chains in response to the UGA codon, which is normally read as a stop codon and used for termination of protein synthesis. To decode UGA as Sec, organisms evolved the Sec insertion machinery that allows incorporation of this amino acid at specific UGA codons in a process requiring a Sec insertion sequence element (SECIS). After comparison of different organisms and due to evolution, it has been seen that cysteines have replaced selenocysteines in some selenoproteins. Despite the fact that basic mechanisms of Sec synthesis have been studied in great detail, the identity and function of many selenoproteins remain still unknown.[1] [2]
There are around 45 selenoproteins described in vertebrates but only 25 are found in humans:
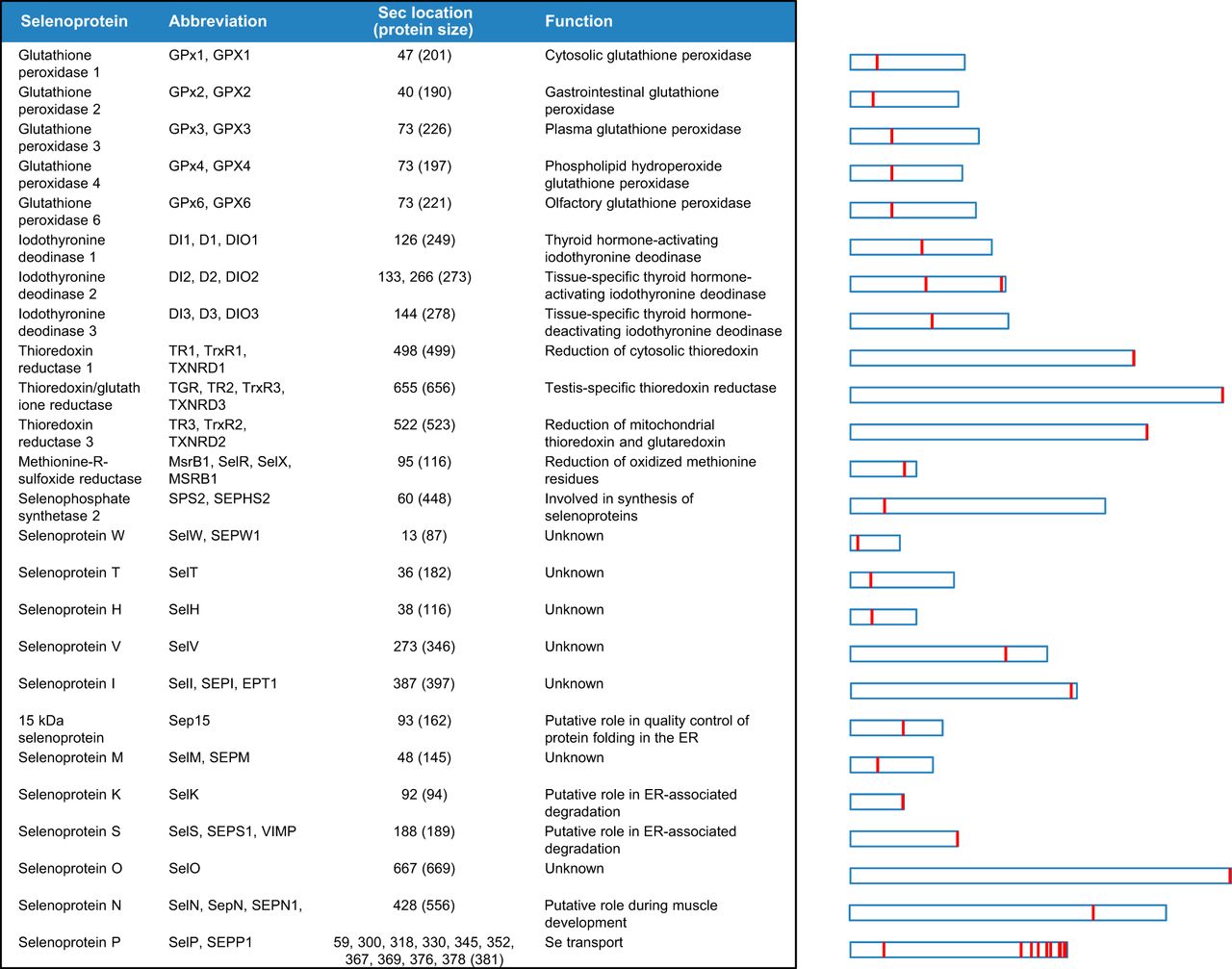
Evolution of Selenoproteins in the metazoan
Selenoproteins are present throughout all 3 kingdoms of life. Studies of primitive invertebrate selenoproteomes are rarely reported outside of insects and nematodes, however, selenoproteins in vertebrates have been fully investigated. Even the most primitive animals possess a selenoproteome range and variety similar to humans. During evolutionary history, very few selenoproteins have emerged and another few have been lost (totally lost or changed to Cystein forms). Moreover, the loss of selenoproteins in insects and nematodes ocurred independently in isolated evolutionary branches.[3]
Selenoprotein families
Selenoproteins are usually classified in different families. Although most of them have been deeply studied some of them have to be still characterized. Different species have some changes on their selenoproteins when we compare them to the human selenoproteome. Here we present a summary of the human selenoproteome:
15-kDa Selenoprotein and Selenoprotein M
Both proteins are involved in redox reactions associated with the formation of disulfide bonds and contain an NH2-terminal signal peptide, consistent with their ER localisation.
15-kDa Selenoprotein (Sel15): Sel 15 was initially identified as a strongly Se-labeled selenoprotein in human T cells, shares homology and compartment characteristics with selenoprotein M. It may contribute to the quality control of protein folding in the endoplasmic reticulum. [4]
Selenoprotein M (SelenoM): SelenoM is expressed in a variety of tissues, with increased levels in the brain. It is localized to the perinuclear structures, and its N-terminal signal peptide is necessary for protein translocation. [4] [5]
Selenoproteins W, V, T and H
These proteins belong to the Rdx family of selenoproteins and all of them possess a thioredoxin-like fold. Based on its motifs, it is proposed that they are thiol-based oxidoreductases, but their exact function remains unknown.
Selenoprotein W (SelenoW): plays a role as a glutathione (GSH)-dependent antioxidant. May be involved in a redox-related process. May play a role in the myopathies of selenium deficiency. [4]
Selenoprotein T (SelenoT): it is a selenoprotein with thioredoxin reductase-like oxidoreductase activity. Protects dopaminergic neurons against oxidative stress and cell death and it is involved in ADCYAP1/PACAP-induced calcium mobilization and neuroendocrine secretion. SelenoT also plays a role in fibroblast anchorage and redox regulation. [4]
Selenoprotein H (SelenoH): this selenoprotein has a unique subcellular localization pattern and it is localised specifically to the nucleoli. Similar to SelenoW, it is sensitive to dietary Se intake. It specifically binds to sequences containing heat shock and stress response elements, and it possesses glutathione peroxidases activity implicated in the regulation of transcription. [4]
Selenoprotein I
Selenoprotein I (SelenoI) is one of the least studied selenoproteins since it evolved recently and is only found in vertebrates. It catalyzes phosphatidylethanolamine biosynthesis from CDP-ethanolamine so it plays a central role in the formation and maintenance of vesicular membranes. It is known to be involved in the formation of phosphatidylethanolamine via “Kennedy” pathway. [4]
Selenoproteins K and S
These two selenoproteins have no significant sequence similarity. However, they can be assigned as related selenoproteins based on their topology, including a single transmembrane domain in the NH2-terminal sequence, the presence of a glycine-rich segment and a characteristic location of Sec residues in the COOH-terminal end of the protein.
Selenoprotein S (SelenoS): is involved in the degradation process of misfolded endoplasmic reticulum (ER) luminal proteins. It participates in the transfer of misfolded proteins from the ER to cytosol, where they are destroyed by the proteasome in a ubiquitin-dependent manner. This selenoprotein probably acts by serving as a linker between DERL1 (which mediates the retrotranslocation of misfolded proteins into the cytosol) and the ATPase complex VCP (which mediates the translocation and ubiquitination. [4]
Selenoprotein K (SelenoK): this protein is required for calcium flux in immune cells and plays a role in T-cell proliferation and in T-cell and neutrophil migration. It is involved in endoplasmic reticulum-associated degradation (ERAD) of soluble glycosylated proteins. SelenoK is also required for palmitoylation and cell surface expression of CD36 and involved in macrophage uptake of low-density lipoprotein and in foam cell formation. Therefore, it plays a role in protecting cells from ER stress-induced apoptosis and it also protects cells from oxidative stress when is overexpressed in cardiomyocytes. [4]
Selenoprotein O
Selenoprotein O (SelenoO) is one of the least characterised human selenoproteins. It contains a single Sec residue located in the antepenultimate position at the COOH-terminal end of the protein. However, the majority of SelenoO homologous contains a Cys residue instead of Sec. It has a mitochondrial targeting peptide and a putative protein kinase domain therefore it may be a redox-active mitochondrial selenoprotein which interacts with a redox target protein. [4]
Selenoprotein N
Selenoprotein N (SelenoN) was one of the first selenoproteins identified through bioinformatics approaches. SelenoN plays an important role in cell protection against oxidative stress and in the regulation of redox-related calcium homeostasis. Acts as a modulator of ryanodine receptor (RyR) activity: protects RyR from oxidation due to increased oxidative stress, or directly controls the RyR redox state regulating the RyR-mediated calcium mobilization required for normal muscle development and differentiation. [4]
Selenoprotein P
Selenoprotein P (SelenoP) is an abundantly expressed and secreted selenoprotein that accounts for almost 50% of the total Se in plasma. It might be responsible for some of the extracellular antioxidant defense properties of selenium or might be involved in the transport of selenium. Therefore, SelenoP may supply selenium to tissues such as brain and testis. [4]
Gluthatione Peroxidases family
Gluthatione Peroxidase (GPx) contains selenium and was discovered in 1957 by Gordon C. Mills. There have been eight different members of the GPx family that have been identified in humans: GPx1, GPx2, GPx3, GPx4, GPx5, Gpx6, GPx7, and finally, GPx8. All eight have the same free radical scavenging functions but are tissue specific.
For example, GPx1 (the most abundant of all eight) is found throughout all the tissues in the human body, whereas GPx2 is mainly found in the intestines. Interestingly, GPx5 is specifically expressed in the tissues of the male reproductive tract as it seems to play an important role in protecting the delicate membranes of the spermatozoa (sperm) from the oxidative damage of lipid peroxidation. So it actually promotes healthy male sperm.
GPx1: it is the most abundant selenoprotein in mammals. It catalyses glutathione (GSH)-dependent reduction of hydrogen peroxide to water. It is expressed in all cell types, protects the hemoglobin in erythrocytes from oxidative breakdown. [4]
GPx2: it could play a major role in protecting mammals from the toxicity of ingested organic hydroperoxides. Tert-butyl hydroperoxide, cumene hydroperoxide and linoleic acid hydroperoxide but not phosphatidycholine hydroperoxide, can act as acceptors. [4]
GPx3: protects cells and enzymes from oxidative damage, by catalyzing the reduction of hydrogen peroxide, lipid peroxides and organic hydroperoxide, by glutathione. [4]
GPx4: it protects cells against membrane lipid peroxidation and cell death. It is required for normal sperm development and male fertility so it is an essential protein for embryonic development. Moreover, GPx4 protects from radiation and oxidative damage so it is known to be essential for maturation and survival of photoreceptor cells. It also plays a role in a primary T cell response to viral and parasitic infection by protecting T cells from ferroptosis. [4]
GPx5: as well as GPx3, it protects cells from oxidative reactions. They catalyze reduction of hydrogen peroxid, lipid peroxides and organic hydroperoxide by glutathione. It may be a glutathione-peroxidase like protective system against peroxide damage in sperm membrane lipids. [4]
GPx6: it is only found in the olfactory epithelium during embryonic development. Like all other gluthatione peroxidase it has response to oxidative stress. [4]
GPx7: it protects esophageal epithelia from hydrogen peroxide-induced oxidative stress. It protects against oxidative damage in DNA, double-strand breaks and eliminates acidic bile acid-induced reactive oxigen species (ROS). [4]
GPx8: it reduces H202 content and oxidative stress in the ER. [4]
Iodothyronine Deodinases family
Iodothyronine Deodinases (DIO) family: this family is the second most important family of selenoproteins. It consists of three paralogous proteins in mammals (DIO1, DIO2 and DIO3) which are involved in the regulation of the thyroid hormone activity by reductive deiodination. [6]
DIO1: is responsible for the deiodination of T4 into T3 and of T3 into T2 as it plays a role in providing a source of plasma T3 by deiodination of T4 in peripheral tissues such as liver and kidney.
[4] [6]
DIO2: is also responsible for the deiodination of T4 into T3 and is essential for providing appropriate levels of T3 to the brain during the critical period of development.
[4] [6]
DIO3: deiodinates T4 into RT3 and T3 into T2. RT3 and T2 are inactive metabolites. This selenoprotein may play a role in preventing premature exposure of developing fetal tissues to adult levels of thyroid hormones. It can regulate fetal thyroid hormone concentrations throughout gestation, this is the reason why it has an essential role for the regulation of thyroid hormone inactivation during embryological development.
[4][6]
Thioredoxin reductase family
Thioredoxin reductase (TXNRD) family are oxidoreductases that together with thioredoxin (Trx) catalyse the reduction of thioredoxin dependent of NADPH, play a regulatory role in metabolic activity and are associated with the prevention of some cancers. In mammals, there are three TXNRD isoenzymes (TXNRD1, TXNRD2 and TXNRD3). These proteins contain a Sec residue in the COOH-terminal penultimate position.
TXNRD1: this protein has different isoforms. The first isoform may possess glutaredoxin activity as well as thioredoxin reductase activity apart from inducing actin and tubulin polymerization, leading to formation of cell membrane protrusions. Isoform 4 enhances the transcriptional activity of estrogen receptors alpha and beta while isoform 5 enhances the transcriptional activity of beta receptor only. This last isoform mediates cell death induced by a combination of interferon-beta and retinoic acid.
[4]
TXNRD2: it is localised in the mitochondria, as it maintains thioredoxin in a reduced state. It is implicated in the defenses against oxidative stress and may play a role i redox-regulated cell signalling.
[4]
TXNRD3: this thioredoxin reductase displays also glutaredoxin and glutathione reductase activities. Moreover, it catalyzes disulfide bond isomerization and promotes disulfide bond formation between GPx4 and various sperm proteins. For that, it may play a role in sperm maturation by promoting fomation of sperm structural components.
[4]
Selenoprotein R or Methionine-R-Sulfoxide Reductase 1
Methionine-R-Sulfoxide Reductase 1 (MSRB1) is a zinc-containing enzyme that was initially identified as selenoprotein R and selenoprotein X by searching for putative SECIS element structure using bioinformatics tools. Later, this protein was found to function as a Methionine-R-sulfoxide reductase, specifically reduces methionine (R)-sulfoxide back to methionine. Methionine oxidation is also a post-translational modification that takes place on specific residue. It acts as a regulator of actine assembly by reducing methionine-sulfoxide mediated by MICALs on actin, thereby promoting filament repolymerization. MSRB1 plays a role in innate immunity by reducing oxidized actin, leading to actin repolymerization in macrophages.
[4]
Selenoprotein synthesis
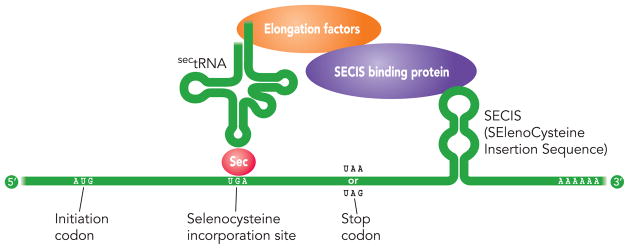
Unlikely most aminoacids, selenocystein is directly synthesized on the tRNA.
The tRNASec recognizes the codon UGA, which is usually used as a stop codon. Seryl-tRNA synthetase conjugates Ser to tRNASec and Phosphoseryl-tRNA kinase modifies Serine to phosphoserine. Concurrently, Selenophosphate synthetase 2 (SPS2) adds a phosphate to Selenium and is added to phosphoserine by Selenocysteine synthetase to produce Selenocysteine.
In order to insert Sec in the protein translation, an element SECIS in the 3’UTR of the mRNA is needed. SECIS elements are the binding sites for SECIS-Binding protein 2 (SBP2), a protein with five isomers. The assembly of both factors (SECIS and SBP2) and recruitment of the Sec tRNA-specific elongation factor, EFsec, bound to tRNASec on selenoprotein mRNA in the nucleus, allows decodification of UGA as Sec and exportation outside the nucleus. Even so, additional cofactors such as SECp43, nucleolin or NSEP1 might contribute to selenoprotein synthesis and other may be still not discovered.
[7]
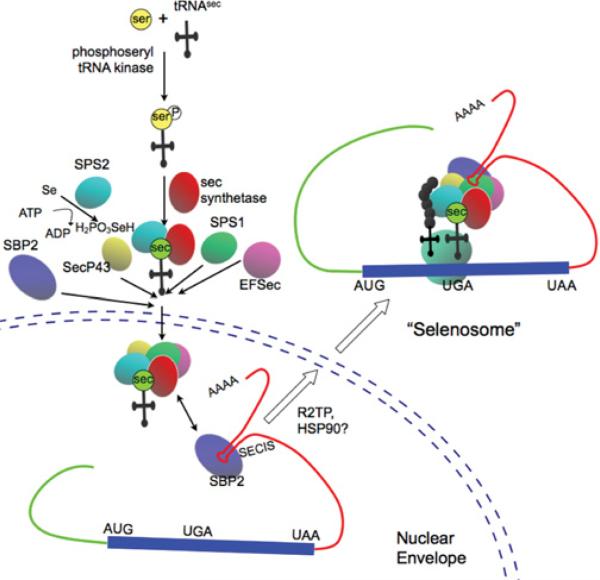
We have already remarked that the incorporation of Sec occurs in a unique and peculiar way. As shown in the upper figure, the recognition of Sec codon and the insertion of Sec requires an amount of trans-acting elements including tRNASec, which is an specific elongation factor, and SECIS-binding proteins which are proteins that bind to the Sec insertion sequence. The targeted deletion of tRNASec gene results in an embryonic lethal phenotype in mice. Since tRNASec gene governs the production of all selenoproteins, this suggests that at least some selenoproteins are crucial for early embryonic development.[8]
Monopterus albus
Classification

Phylum: Chordata
Class: Actinopterygii
Order: Synbranchiformes
Family: Synbranchidae
Subfamily: Neopterygii
Genus: Monopterus
Species: M. albus
Morphology
Asian swamp eel, also known as rice swamp eel (Monoperus albus), is an air-breathing specie of fish. It has an anguilliform body without scales or fins. Most individuals have a slippery olive-drab brown skin with light-orange bellies although some individuals have color variation of orange, black and gold spots which allows them to camouflage in the aquatic environment. They have a blunt nose, a large mouth with tiny teeth and one V-shapped gill opening below the head. [9]. Generally they grow between 25 and 40 cm,[10] although they can reach 1 meter length.[11] They are morphologically similar to two American native fishes: American eel and lampreys. Monoperus albus is distinguised by the absence of pectoral fins or the morphology of the gill opening and teeth. Swamp eels are also similar to two native salamanders but they differ in the presence or absence of front and hind limbs.[11]
Native and current distribution
M. albus lives in swamps, large rivers, muddy ponds and rice fields where it digs in damp earth during dry seasons in order to survive without water. They are nocturnal predators that eat fishes, worm crustaceans and other small aquatic animals. [10] They are able to survive in cold climates, in a wide range of oxygen levels and months without a food source if necessary.[9] Originally found in the waters of East and Southeast Asia. However is very widespread around Asia and Australia, being found in India, China, Thailand or Vietnam among other countries.[11] It also has been identified as an invasive specie in North America since around 1900 they were brought to Hawaii by Asian immigrants as a food fish and release into the wild with purpose. Later on, species of M. albus were found near Georgia and Florida.[9] Althought the impact is uncertain, they are likely to affect the population size of their prey as well as the availability of native fish, frogs and aquatic invertebrates considering that there is no known of natural M. albus predators in United States, have complex adaptative skills and insatiable eating habits.[10][11]
M. albus is considered a food fish delicacy in Asia or United States as well as is sold in some pet stores. [10]
Lifecycle
M. albus is hermaphroditic. All young are females and when they begin to mature, some of them transform into males.[11][12]. Moreover, males can change sex to recover female populations when female densities are too low. Females produce up to 1000 eggs every spawning and can lay eggs throughout the year. Eggs are laid in bubble nests which float above superficial waters. Usually, the male escort the eggs and the young.
[10][12]
Disease
Asian swamp eels are host for Gnathostoma spinigerum so eating raw or undercooked swamp eel can cause gnathostomiasis, a common disease in Thailand and Vietnam.[13]
More information about Monopterus albus can be found in Viquipèdia (information written in Catalan).