Discussion
We characterized selenoproteins in Monopterus albus ’ genome by studying its homology to Danio rerio selenoproteins since it is the model organism that corresponds to the phylogenetically closest specie to Monopterus albus . Moreover, they both belong to the Actinopterygii class.
Danio rerio genome is well-sequenced and contains 53 different proteins in SelenoDB; 37 of them are characterized as selenoproteins while the other 16 belong to machinery proteins or proteins involved in metabolism.
Each protein was analyzed in Monopterus albus to describe how it evolved along lineages. We defined 35 proteins as selenoproteins and we also defined the machinery proteins and the ones involved in metabolism. Regarding the selenoproteins, only one of them was considered as an homolog for cys in Monopterus albus.
Selenoproteins in Monopterus albus
Sel15 Sel15, initially identified as a strongly Se-labeled selenoprotein in human T cells, shares homology and compartment characteristics with selenoprotein M. Sel15 is evolutionarily conserved in most animals and plants; however, only the vertebrate Sel15 homologs contain Sec. This protein has also been implicated in etiology of some types of cancer.[14]
Sel15 could be well-predicted in M. albus. The predicted protein contained a Sec residue and a grade A SECIS element described in the 3’-UTR. We must point out that we did not find a Methionine as the first aminoacid of our sequence. This is explained by the fact that the query protein of zebrafish that we got from SelenoDB didn’t have it either, probably because it was not well anotated. In the case of Sel15, we compared our t-coffee output with our Seblastian prediction and we realized we are missing an initial first exon, containing the starting Methionine and composed by aproximately 16 aminoacids. Analyzing the t-coffee result and comparing between Zebrafish and M. albus, an almost perfect homology was observed. This huge conservation seems to indicate the important role of this protein.
SelenoE Fish selenoprotein 15 (SELENOE) is an ER-resident selenoprotein of unknown function that is found only in fish. This selenoprotein has a specialized function that is distinct from those of Sel15 and SelM. [1]
SELENOE is absent in mammals, can be detected only in fish and is present in these organisms only in the selenoprotein form. In contrast with other members of the Sel 15 family, which contain a putative active site composed of Sec and cysteine, SELENOE has only Sec. When transiently expressed in mammalian cells, SELENOE incorporated Sec in an SECIS- and SBP2-dependent manner and was targeted to the endoplasmic reticulum by its N-terminal signal peptide. Phylogenetic analyses of Sel15 family members suggest that SELENOE evolved by gene duplication.[15]
SELENOE could be also identified in the M. albus’ genome with a really high homology with the zebrafish homonymous. Three different hits on the same scaffold were identified with tBLASTn, corresponding to three different exons. We can conclude, then, that there is only one copy of the gene, unlike some other species of fishes. A Sec residue was observed and one SECIs element was predicted in 3’-UTR. In addition, the Seblastian prediction of the protein confirmed the same sequence we had found. We noticed that this protein does start with a Methionine, so the reliability of the prediction is high.
GPx familyGlutathione peroxidases (GPxs) are a widespread protein superfamily found in many organisms throughout all kingdoms of life with peroxidase activity, whose main biological roles are to protect organisms from oxidative damage. Currently, 8 sub-members of GPxs have been identified in humans. GPxs may have evolved to such an extent in function without perturbing their fold and keeping the active site strictly conserved. Mariotti et al analysis of GPx1-8 families highlighted three evolutionarily related groups: GPx1/GPx2, GPx3/GPx5/GPx6 and GPx4/GPx7/GPx8. GPx4 appeared to be the most ancient GPx, whereas GPx5 and GPx6 were the most recently evolved GPx forms. [2][16][17]
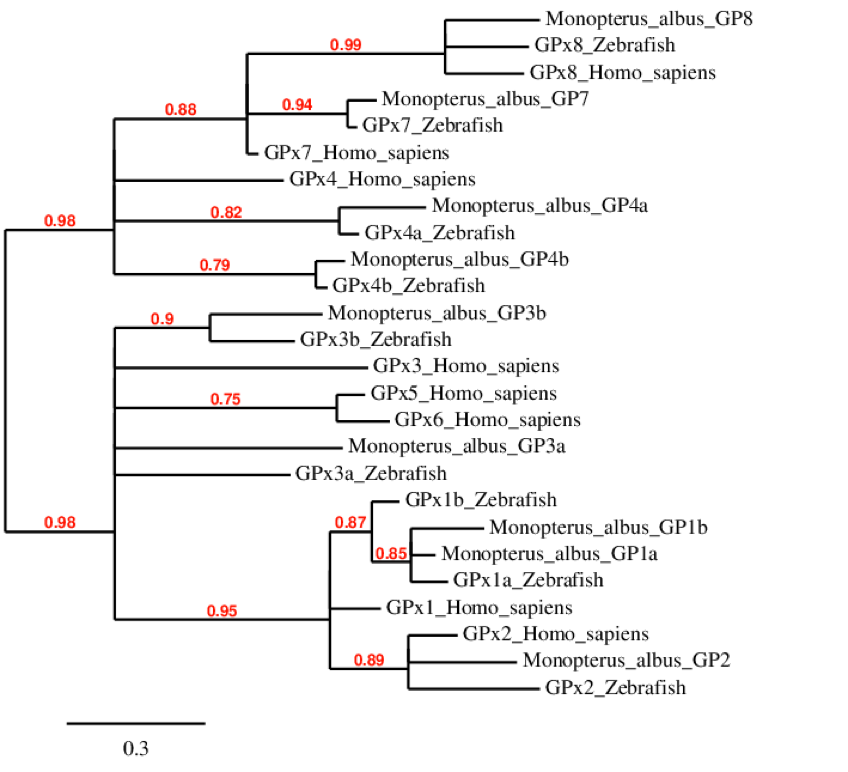
The phylogenetic tree of GPx family shows us that all GPx8 in the different species were originated from the same ancestor. We can see that GPx7 in human is less similar than this protein in Monopterus albus and zebrafish. For the GPx4, we can see a duplication that is only found in fishes. As expected, GPx5 and GPx6 are only found in Human, due to the duplication of GPx3. GPx1 is also duplicated only in fishes. Finally, all GPx2 in the different species come from the same ancestor.
GPx1a
We can confirm the presence of GPx1a in M. albus’ genome. As we can see, it clusters together with all the GP1 proteins, including those from Humans and Zebrafish. In this case, our reference protein from zebrafish starting codon was a Methionine, which makes the evidence more solid. Our prediction matches the Seblastian prediction, and we also found a SECIS structure in its 3’-UTR. To conclude alignment shows a great homology with zebrafish including the selenocysteine residue, and this coincides with the information found where GPx1 is a really conserved protein.
GPx1bThis protein could also be predicted in the genome of M. albus. Similarily to GPx1a, GPx1b clusters together with the GP1 group. It was probably formed in a duplication event. The t-coffee alignment with the zebrafish homonymous was almost perfect, and included the Selenocysteine residue. Our prediction, like that of zebrafish started with a Methionine as the first codon, and it matched perfectly the Seblastian prediction. Besides, a SECIS structure was predicted in the 3’-UTR of the gene.
GPx2GPx2 could be predicted in M. albus. It was found in KV884828.1 scaffold and was formed by 2 exons. The selenocysteine residue is conserved. Although we can observe that the alignment is good, and it covers most of the protein (including the first methionine and the last residue). We notice that our predicted protein contains a gap of around 25 aminoacidic positions filled by gaps and Xs. This is due to the poor anotation of M. albus’ genome, because we can see that that scaffold contains a lot of regions filled with N, meaning that the sequencing wasn’t correctly done. If we compare our prediction with that of Seblastian we can see that the protein predicted is exactly the same. A SECIS structure could also be predicted in the 3’UTR of the gene. We can assume this protein is found in M. albus genome because the alnment and the Seblastian prediction confirm it, and aditionally because we can see it clusters perfectly with the homonymous proteins of zebrafish and human.
GPx3aGPx3a could be predicted in the Monopterus albus ’ genome as well. The t-coffee alignment displayed a great similarity with zebrafish, and the selenocysteine residue was conserved. Although the protein we predicted is very similar to that of Seblastian, none of them started with a methionine residue. Nevertheless, the Seblastian prediction did start with a Lys residue and lysine has been reported to be able to funcion as a starting codon.[18]
Also, a SECIS structure was predicted in the 3’-UTR region of the gene. We can conclude that GPx3a does exist in the M. albus genome as it does in the zebrafish.
GPx3b protein was found in scaffold KV884757.1 and it is constituted by 5 exons located on the negative strand. The t-coffee alignment was good, the selenocysteine is conserved as it is also aligned with the zebrafish protein. No selenoprotein could be predicted for this sequence with Seblastian but one SECIS structure was found in the opposite strand, meaning that it is not part of the protein.
GPx3b could be predicted in M. albus. The result of the t-coffee showed a really good alignment between our prediction and the protein in zebrafish. The selenocysteine residue was conserved in the protein. Our predicted protein and the query we got from zebrafish both started eith a Methionine residue, so this adds reliability to our prediction. A SECIS structure could not be predicted using Seblastian. Although Seblastian could not predict the protein, the high conservation observed and the clustering in the phylogenetic tree prove that this protein does exist in M. albus.
GP4a was predicted in M. albus. The provided us with a highly conserved alignment, and the Selenocysteine residue was conserved as well. A SECIS structure was predicted in the 3’-UTR of the gene. Comparing our prediction with that of Seblastian we saw that the protein is mainly conserved and the predictions match, but some differences were observed.The Seblastian prediction starts 20 nucleotides before our prediction and does start with a methionine codon. We repeated the obtention process with an exhaustive exonerate but our results were exactly the same. Anyway, a SECIS structure was predicted in the 3’-UTR of the gene. We hypothesized that maybe our prediction is not considering the first exon as it is very small. In conclusion, we consider that GP4a does exist in M. albus because it aligns perfectly with the zebrafish selenoprotein and it the agrupation in the phylogenetic tree is really clear.
GPx4bGPx4b could be predicted within the genome of M. albus. The alignment of the t-coffee was almost a perfect match, showing a high homology with zebrafish. The selenocysteine residue was conserved, and a SECIS structure could be predicted in the 3’-UTR of the gene. GPx4b could not be predicted by Seblastian. Our query protein from zebrafish did not start by a methionine, because the selenoDB database contains proteins that do not start with a methionine. We consider that GPx4b does exist in M. albus, because the homology with zebrafish is extremely high, a SECIS has been predicted and the phylogenetic agrupation is really clear.
GPx5 and GPx6GPx5 and GPx6 are not found in the genome of zebrafish as these two proteins are the most recently evolved GPxs, which appeared to be the result of a tandem duplication of GPx3 at the root of placental mammals [ref article Drive]. Hence we did not expect M. albus to have them. However, in order to make sure that M. albus did not have these two selenoproteins, we compared GPx5 and GPx6 from Homo Sapiens genome to M. albus’ genome. We saw that there were some scaffolds with a low hit on the tBLASTn, although these ones were already found in other selenoproteins at the same position with better scores. Therefore, we concluded that these selenoproteins do not exist in M. albus’ genome, as expected.
GPx7 and GPx8Both GPx7 and GPx8 evolved from a selenoprotein, a GPx4 ancestor, but they lost their selenocysteine residue. This correlates perfectly with the agrupations in the phylogenetic tree. As expected, none of these proteins contained a Selenocysteine residue. The information from the t-coffee, which in both cases was a good alignment and the absence of predicted selenoproteins by the Seblastian confirmed our expectations. Considering thehigh homology between the sequences when compared to zebrafish and the clarity of the phylogenetic tree, we conclude that GPx7 and GPx8 are present in the M. albus’ genome.
DIO familyThis family is one of the most important families of selenoproteins. There are three DIO enzymes known in mammals which are involved in the regulation of the thyroid hormone activity by reductive deiodination, all of which contain Sec: DIO1, DIO2, DIO3. The deiodinases possess a thioredoxin-fold and show significant intrafamily homology. The protein DIO3 is duplicated in all bony fishes (DIO3b). [2][6]
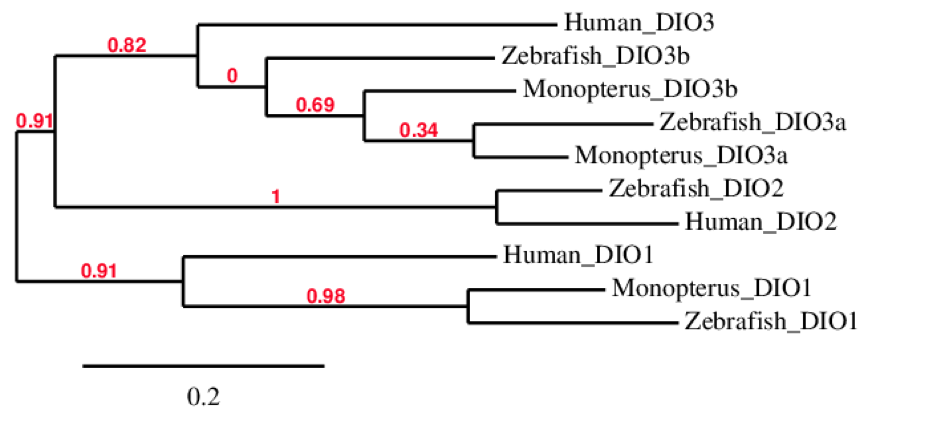
DIO1 is responsible for the deiodination of T4 into T3 and of T3 into T2 as it plays a role in providing a source of plasma T3 by deiodination of T4 in peripheral tissues such as liver and kidney.
DIO1 could be predicted in M. albus: it had one selenocysteine that was conserved but the alignment did not start with Methionine. When looking at Seblastian predicted protein we saw that this started with a Methionine just 2 aminoacids before from the t-coffee result, so we assumed that zebrafish protein from SelenoDB was not perfectly annotated as it did not start with Methionine either. The rest of our prediction matches the Seblastian prediction. Moreover, we also found two SECIS structures in 3’ UTR, being part of the protein. To conclude, the alignment shows great homology with zebrafish including selenocysteine residue and this coincides with the information found about this conserved protein.
DIO2DIO2 is also responsible for the deiodination of T4 into T3 and is essential for providing appropriate levels of T3 to the brain during the critical period of development.
This protein was not found in Monopterus albus ’ genome. It was found in some scaffolds but the e-value was high and the alignment that resulted from the t-coffee matched a very small fraction of the protein. Therefore, we concluded that this protein does not exist in M. albus.
DIO3 irreversibly inactivates the thyroid hormone by deiodination of the inner tyrosyl ring. Interestingly, all detected DIO3 genes (including DIO3b) are intronless. Moreover, this protein was duplicated only in all bony fishes, as said before. [2]
DIO3aWe could predict this protein in M. albus: t-coffee results were good although the protein did not start with Methionine. When looking at Seblastian predicted protein, we could see that the protein started with Methionine four amino acids before from the t-coffee alignment. The rest of our prediction matched the Seblastian prediction so we assumed that zebrafish protein from SelenoDB was not perfectly annotated as it did not start with Methionine either. There was a selenocysteine that matched with the zebrafish protein. Moreover, one SECIS structure was predicted in 3’ UTR. To conclude, the alignment shows homology with zebrafish and this coincides with the information found about this protein, resulted from a duplication in bony fishes.
DIO3bThis protein was predicted in M. albus: the alignment resulted from t-coffee after doing exhaustive exonerate filled the first part of the protein, starting with Methionine. When looking at Seblastian predicted protein, this also started with Methionine and matched the protein of the t-coffee. Moreover, the selenocysteine was also found and aligned and Seblastian predicted one SECIS structure found in 3’ UTR. To conclude, the alignment shows homology with zebrafish and this coincides with the information found of this protein, resulted from a duplication of DIO3 in bony fishes.
SEPHS familySelenophosphate synthetase (SEPHS) is essential for selenoprotein biosynthesis. This family consists in two SEPHS paralogues in higher eukaryotes called SEPHS and SEPHS 2. SEPHS, an enzyme, is not involved in Sec synthesis in mammals. SEPHS genes originated through a number of independent gene duplications from an ancestral metazoan selenoprotein SEPHS2 gene that most likely already carried the SEPHS function. Thus, in SEPHS genes, parallel duplications and subsequent convergent subfunctionalization have resulted in the segregation to different loci of functions initially carried by a single gene.[19]
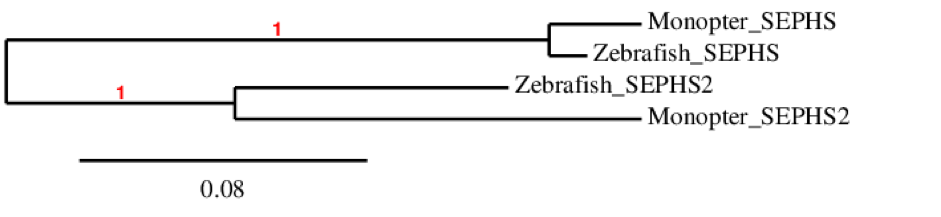
The phylogenetic tree of SEPHS family shows that this protein is only present in fishes as we could not find it in humans. SEPHS2 sequences are similar between zebrafish and Human. This means that they originated from the same ancestor protein. The same happens for SEPHS.
SEPHS This protein could be predicted in M. albus. A perfect homology was the result of the t-coffee comparison with its zebrafish homonymous. Our predicted protein did start with a Methionine residue, and neither selenocysteine nor SECIS were found, as expected because SEPHS is a protein involved in machinery. We then concluded that SEPHS does exist in M. albus’ genome.
SEPHS 2
The function of selenophosphate synthetase 2 (SEPHS 2) is to generate the Se donor compound (selenophosphate) necessary for Sec biosynthesis, and interestingly it is itself a selenoprotein. [2]
SEPHS 2 could be predicted within M. albus’ genome. The t-coffee result showed a great homology with the zebrafish protein. A selenocysteine residue was conserved. The prediction of seblastian coincided with ours, except from the first 6 aminoacids, which contained a Methionine. We performed an exhaustive exonerate, but no improvements could be observed. Also, a SECIS structure was predicted in the 3’-UTR of the gene. In conclusion, we consider that SEPHS 2 is conserved in M. albus.
SELENOHThis selenoprotein, widely distributed in all vertebrates, has a unique subcellular localization pattern and it is localised specifically to the nucleoli. It is sensitive to dietary Se intake. It specifically binds to sequences containing heat shock and stress response elements, and it possesses glutathione peroxidases activity implicated in the regulation of transcription.
This protein could be predicted in M. albus: with the first obtained alignment it seemed like there was an exon lacking (the first one) but this was not found in the tBLASTn. After repeating the obtention process using exhaustive exonerate, the new prediction we obtained made much more sense, as it covered the query protein since the 6th residue, starting with K (Lys). We compared it to our Seblastian prediction and we saw that the Seblastian predicted protein did not start with Methionine either because it started with G. We rely on our results as it has been found in some studies that some proteins can start with a Lysine codon. [18] To conclude, the alignment shows homology with zebrafish and this coincides with the information found of this protein, being conserved in M. albus.
SELENOISelenoprotein I is one of the least studied selenoproteins since it evolved recently and is only found in vertebrates. It catalyzes phosphatidylethanolamine biosynthesis from CDP-ethanolamine so it plays a central role in the formation and maintenance of vesicular membranes.
This protein could be predicted in M. albus: the t-coffee results showed a good alignment and selenocysteine residue was conserved and well aligned with the zebrafish one. However, the t-coffee alignment did not start with Methionine because the zebrafish protein extracted from SelenoDB was not well annotated as it did not start with Methionine either. When looking at Seblastian predicted protein we saw that the protein started with Methionine 3 amino acids before from the t-coffee result. Finally, one SECIS structure was found in 3’ UTR, as expected. To conclude, the alignment shows good homology with zebrafish and this coincides with the information found of this protein, it is conserved in M. albus.
SELENOJ1In contrast to known selenoproteins, SELENOJ1 appears to be restricted to actinopterygian fishes and sea urchin, with Cys homologues only found in cnidarians. This protein shows significant similarity to the jellyfish J1-crystallins and with them constitutes a distinct subfamily within the large family of ADP-ribosylation enzymes. Consistent with its potential role as a structural crystallin, SELENOJ1 has preferential and homogeneous expression in the eye lens in early stages of zebrafish development. [20]
This protein could be predicted in M. albus: the alignment with zebrafish protein was really good, it started with Methionine and the selenocysteine was conserved and well aligned. When predicted with Seblastian, one SECIS structure was found in 3’ UTR. To conclude, the alignment shows good homology with zebrafish and this coincides with the information found of this protein, SELENOJ1 is conserved in M. albus.
SELENOKThis protein is required for calcium flux in immune cells and plays a role in T-cell proliferation and in T-cell and neutrophil migration. It is involved in endoplasmic reticulum-associated degradation (ERAD) of soluble glycosylated proteins. SELENOK is also required for palmitoylation and cell surface expression of CD36 and involved in macrophage uptake of low-density lipoprotein and in foam cell formation.
This protein could be predicted in M. albus: the alignment with zebrafish protein was really good, it started with Methionine and the selenocysteine was conserved and well aligned. When predicted with Seblastian, two SECIS structures were found in 3’ UTR. To conclude, the alignment shows good homology with zebrafish and this coincides with the information found of this protein, SELENOK is conserved in M. albus.
SELENOLThis protein is only found in fishes, among vertebrates and its function remains still unknown. When looking at the results after analyzing this protein, it looked like some exons were lacking but they were not identified in any of the scaffolds. Anyway, the selenocysteine and the second half of the protein were highly conserved. Surprisingly, after performing an exhaustive exonerate, we got a new prediction that covered some of the blank space, starting in the 21st amino acid and aligning perfectly with our query, including the selenocysteine residues within the protein. However, it did not start with Methionine and this protein was well annotated in zebrafish. When looking at Seblastian prediction, no selenoprotein could be found but one SECIS structure was found in 3’ UTR. Therefore, we concluded that SELENOL does exist in M. albus.
SELENOMSELENOM is expressed in a variety of tissues, with increased levels in the brain, as it might have a neuroprotective action. It is localized to the perinuclear structures, and its N-terminal signal peptide is necessary for protein translocation.
This protein could be predicted in M. albus: the alignment with zebrafish protein was good, but it did not start with Methionine because the zebrafish protein extracted from selenoDB was not well annotated. The selenocysteine was conserved and well aligned. When predicted with Seblastian, we saw that the predicted protein started with Methionine. Moreover, one SECIS structure was found in 3’ UTR. To conclude, the alignment shows good homology with zebrafish and this coincides with the information found of this protein, so SELENOM is conserved in M. albus.
SELENONSelenoprotein N (SELENON) plays an important role in cell protection against oxidative stress and in the regulation of redox-related calcium homeostasis. It also acts as a modulator of ryanodine receptor (RyR) activity: protects RyR from oxidation due to increased oxidative stress, or directly controls the RyR redox state regulating the RyR-mediated calcium mobilization required for normal muscle development and differentiation.
This protein could be predicted in M. albus: the first alignment made with t-coffee was good and the selenocysteine was conserved and aligned. However, the predicted alignment only covered the positions 64 to 555 of the query protein. After repeating the obtention process using exhaustive exonerate, the new prediction we obtained made much more sense, as it covered the query protein since the 16th residue. However, the protein did not start with Methionine although the zebrafish protein was well annotated in this case. Seblastian could not predict any selenoprotein but it did predict one SECIS structure in 3’ UTR. Therefore, we concluded that this protein does exist in M. albus as the homology with zebrafish was good and the selenocysteine was well conserved and aligned.
SELENOO familySelenoprotein O is the largest mammalian selenoprotein with orthologs found in a wide range of organisms, including bacteria and yeast and has been hypothesized to have a kinase domain. [21]
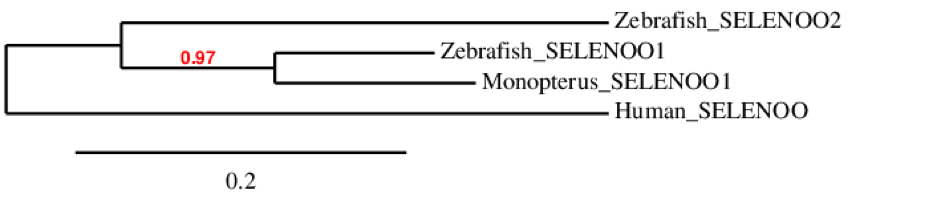
The phylogenetic tree of this family shows that SELENOO is duplicated in zebrafish. This probably means that this protein was duplicated in bony fishes. However, we could not predict SELENOO2 in M. albus, as explained below. SELENOO1 is in the same cluster for both zebrafish and M. albus, as we can see, so they were originated from the same ancestor protein. Finally, human has only one SELENOO protein and it differs sequentially from the ones of zebrafish and M. albus is low.
SELENOO1This protein could be predicted in M. albus: we tried to predict the protein using exhaustive exonerate because the first alignment had some parts of the protein with gaps but the second prediction was very similar to the one we got at the beginning with the first alignment. However, the alignment started with Methionine and the selenocysteine was well aligned and conserved with the one of zebrafish. When looking at Seblastian, this could predict a selenoprotein and one SECIS structure was also predicted in 3’ UTR. To conclude, the alignment shows good homology with zebrafish and this coincides with the information found of this protein, so we conclude that SELENOO1 is conserved in M. albus.
SELENOO2This protein could not be predicted in M. albus: it aligned the same region of the same scaffold as SELENOO1 with a much worse alignment. Therefore, we conclude that SELENOO2 is not found in M. albus.
SELENOP familySelenoprotein P (SELENOP) is an extracellular protein produced in many tissues but primarily by the liver. This protein transports selenium from the liver to extrahepatic tissues and protects against oxidative injury, this is the reason why it is the best indication for human selenium nutritional status. Moreover, it contains multiple Sec residues per protein molecule [22][23]
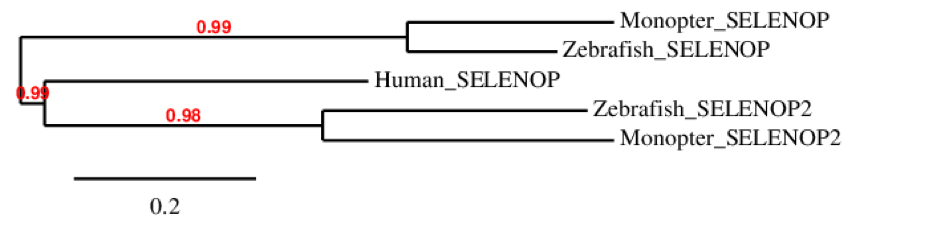
This protein could be predicted in M. albus: the alignment of this protein with the one of zebrafish was very bad although the “X” (selenocysteine) coincided. Huge gaps were observed, as we could see in t-coffee results. After repeating the process with an exhaustive exonerate, the alignment obtained was really good, covered most of the protein and coincided with the expected selenocysteine. However, it did not start with Methionine. When looking at Seblastian prediction, we could see that the predicted protein does start with a Methionine, as the sequence starts a few amino acids before from our sequence. Moreover, one SECIS structure was predicted in 3’ UTR. To conclude, the obtained alignment shows good homology with zebrafish and this coincides with the information found of this protein, so we conclude that SELENOP is conserved in M. albus.
SELENOP (2)This selenoprotein could be predicted in M. albus: SELENOP (2) contains 17 Selenocysteine residues in zebrafish and 10 in humans. The overall alignment obtained with t-coffee was good, but some of the selenocysteine residues did not match between them because there was a one position shift. We therefore hypothesized that this could be due to the value given by the algorism to the gap opening penalty, as it is very low. This would explain why it looks like the sequences are shifted to the right. When looking at Seblastian prediction, the selenocysteines are all well aligned and conserved with the zebrafish protein. Moreover, two SECIS were predicted in 3’ UTR.
To conclude, the alignment shows good homology with zebrafish and this coincides with the information found of this protein, so we conclude that SELENOP (2) is conserved in M. albus.
Methionine sulfoxide reductases (Msr) are antioxidant repair enzymes that have a role in the detoxification of reactive oxygen intermediates. Msr catalyze the reduction of oxidized methionine (Met-O) to methionine (Met) in free and protein-bound forms. Although two kinds of Msr namely MsrA and MsrB exist in both prokaryotes and eukaryotes, they share little identity between them either at primary sequence level or at structural level.[24]
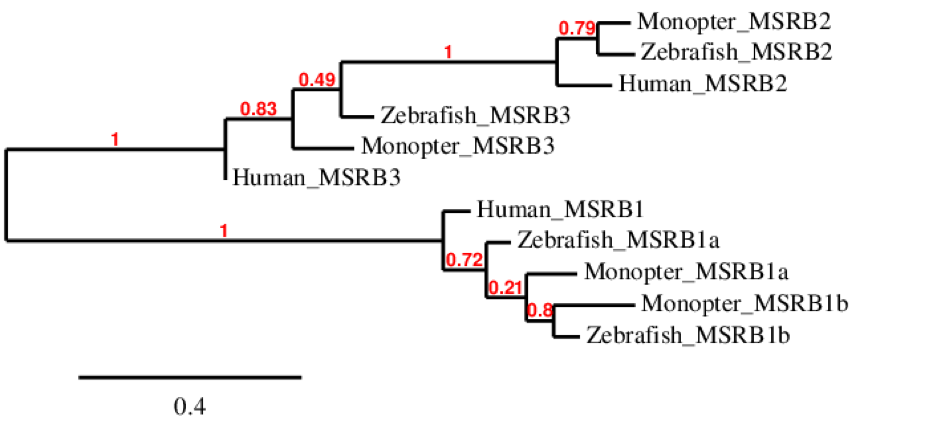
The phylogenetic tree of MSRB family is extended. As we can see, MSRB1 is only duplicated in fishes as human have only MSRB1. However, all these proteins belong to the same cluster. MSRB2 is similar between the tree species although the human one differs a little bit from the zebrafish and M. albus ones, as it is expected. The same happens for MSRB3.
MSRB1MSRB1 plays a role in innate immunity by reducing oxidized actin, leading to actin repolymerization in macrophages. Amongst the different isoforms of MSRB, MSRB1 is one of the ancestral vertebrate selenoproteins.
MSRB1aThis protein could be predicted in M. albus: the alignment made with t-coffee was perfect, as it started with Methionine and the selenocysteine residue was conserved and well aligned. When looking at Seblastian prediction, no selenoprotein was predicted but two SECIS structures of grade B were predicted in 3’ UTR. To conclude, the alignment shows good homology with zebrafish and this coincides with the information found of this protein, as it comes from a duplication that only happened in fishes, so we conclude that MSRB1a is conserved in M. albus.
MSRB1bThis protein could be predicted in M. albus: the alignment we obtained from the first t-coffee was perfect, from the first Methionine to the last amino acid, as Seblastian had given us. Moreover, one SECIS structure was predicted with Seblastian in 3’ UTR. To conclude, the alignment shows good homology with zebrafish and this coincides with the information found of this protein, as it comes from a duplication that only happened in fishes, so we conclude that MSRB1b is also conserved in M. albus .
MSRB2MSRB2 can repair oxidative damage to proteins due to reactive oxygen species, by reducing the methionine sulfoxide in proteins back to methionine. It is known to be a machinery protein.
This protein could be predicted in M. albus: the first alignment we obtained from the first t-coffee, performed with our automatic program, lacked the first 36 aminoacids compared with our query from zebrafish. After re-doing it with an exhaustive exonerate we obtained a great alignment of the whole sequence, from the first Methionine to the last amino acid. When looking at Seblastian prediction, no selenoprotein could be predicted as expected and no SECIS structure could be predicted either, as also expected because it is a machinery protein and has no selenocysteine residue. To conclude, the alignment shows good homology with zebrafish and this coincides with the information found of this protein, so we conclude that MSRB2 is conserved in M. albus.
MSRB3This protein could be predicted in M. albus: the first alignment we obtained from the first t-coffee was good, it started with Methionine and there was no selenocysteine residue, as expected because MSRB3 is also a machinery protein. When looking at Seblastian prediction, no selenoprotein could be predicted as expected and no SECIS structure could be predicted either, as also expected. To conclude, the alignment shows good homology with zebrafish and this coincides with the information found of this protein, so we conclude that MSRB3 is conserved in M. albus.
SELENOSSelenoprotein S is involved in the degradation process of misfolded endoplasmic reticulum (ER) luminal proteins. It participates in the transfer of misfolded proteins from the ER to cytosol, where they are destroyed by the proteasome in a ubiquitin-dependent manner. This protein is found in all vertebrates with a poorly characterized function. [25]
With the tBLASTn we only got one hit, with a relatively high e-value and a low percentage identity. The t-coffee result gave us an average good alignment, but we got an X where we did not expect one, aligned with a Valine. Our initial hypothesis was that we got a new selenocysteine residue in the protein, appearing de novo. That would imply that a punctual mutation originated the UGA codon, coding for Selenocysteine. We then performed an exhaustive exonerate and repeated the t-coffee. Our results were the same, but we found out that the X in our result was not caused by a UGA codon, but for a UAA codon. This information had to be contrasted with Genewise, where no selenocysteine residue was detected, and it was a bad alignment. We assumed that this protein does not exist in M. albus. First of all, it contains a Stop codon (UAA) inside the coding region, so the resulting protein would be incomplete and probably not functional. In addition to this, the prediction is incomplete and does not seem accurate. Finally, we must point out that Seblastian did not find any selenoprotein in our sequence. This makes us think that probably this protein was mutated in some point originating a Stop codon, so that the functionality of the protein would have been lost.
SELENOT familySELENOT contains a thioredoxin-like fold encompassing the Sec residue within a putative catalytic domain and has been shown to be involved in fibroblast cell adhesion and PC12 cell calcium homeostasis. Its gene is robustly and widely expressed during embryogenesis, but declines postnatally in most tissues, including the brain. This is the most conserved selenoprotein, with an impressive identity across all mammals even at the nucleotide sequence level. [2][26]

The phylogenetic tree of SELENOT family shows that SELENOT1 is duplicated in zebrafish. We could not find SELENOT1b in M. albus. We can also see that SELENOT1 from M. albus is more similar to SELENOT1b from zebrafish than to SELENOT1 also from zebrafish. We can see that both fishes have also SELENOT2 while human has only one SELENOT that corresponds to T1 in fishes.
SELENOT1SELENOT1 could be predicted within the M. albus’ genome. The alignment provided by the t-coffee was almost perfect, and our predicted protein did start with a Methionine. The Selenocysteine residue was conserved, as we expected. The protein predicted by Seblastian coincides with ours, missing just by 3 aminoacids, and a SECIS structure was predicted in 3’-UTR too. These results confirm that SELENOT1 is another protein of M. albus.
SELENOT1bThis protein is not found in M. albus. We found hits on the tBLASTn exactly in the same scaffolds and the same positions than SELENOT1, but with much higher e-values, lower percentage of identity and less coverage. Taking that into account we concluded that M. albus’ genome does not contain SELENOT1b.
SELENOT2SELENOT2 was also found in M. albus. The t-coffee alignment showed a high homology with its homonymous from zebrafish, where the selenocysteine residue was conserved. In our first attempt to examine it with our program, we found a gap of 39 aminoacids in the middle of the protein sequence in the t-coffee result. We performed a manual exhaustive exonerate afterwards and our new prediction covered all the gap and matched in an almost perfect alignment. The predicted protein from Seblastian matched ours perfectly, starting by methionine as well. Finally, a SECIS structure was predicted in the 3’-UTR. Taking all this into account, we consider that SELENOT2 was found in M. albus.
SELENOU familyIn all bony fishes, there was a duplication of selenoprotein gene SELENOU1. Selenoprotein U (SelU) was firstly found in fish and also reported in birds and unicellular eukaryotes, such as Chlamydomonas reinhardtii. In high mammalian species, such as humans and mice, all SelU proteins exist in Cys form. The Prx-like2 structure domain presented in these proteins implies that they belong to the thioredoxin-like superfamily. [3]
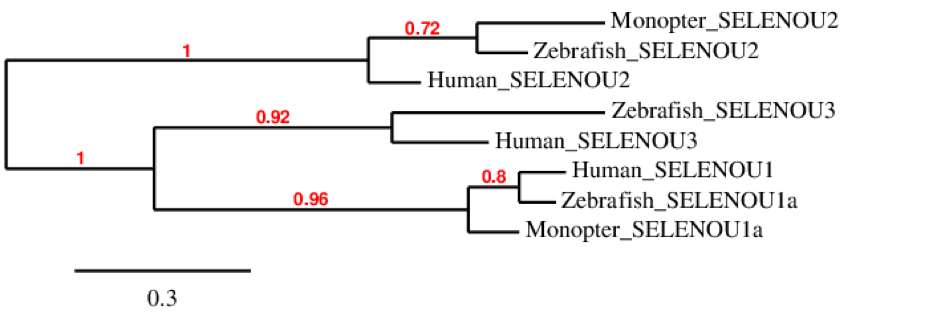
The phylogenetic tree of SELENOU family shows surprisingly that SELENOU1 is more similar between human and zebrafish than between M. albus and this two. For SELENOU2 we can see that is present in the three species but that it is more similar between M. albus and zebrafish. Finally, we could not find SELENOU3 in M. albus although it is present in zebrafish and human and they both come from the same ancestor protein. Moreover, we can see that SELENOU3 and SELENOU1 were originated from the same ancestral protein as they both belong to the same cluster.
SELENOU1aThis protein could be predicted in M. albus: the alignment resulted from t-coffee was good but it did not start with Methionine. This was because the zebrafish protein obtained from selenoDB was not well annotated either. When looking at Seblastian prediction, no selenoprotein could be predicted although one SECIS structure was predicted in 3’ UTR. To conclude, the alignment shows good homology with zebrafish and this coincides with the information found of this protein, so we conclude that SELENOU1a is conserved in M. albus.
SELENOU2This protein could be predicted in M. albus: t-coffee results showed a good alignment but it did not start with Methionine. The zebrafish protein obtained from selenoDB did not start with Methionine either, as it is not well annotated. No selenocysteine was found as expected, because SELENOU2 is a machinery protein. When looking at Seblastian prediction, no selenoprotein could be predicted. Moreover, no SECIS structure could be predicted for this protein as it was also expected. To conclude, the alignment shows good homology with zebrafish and this coincides with the information found of this protein, so we conclude that SELENOU2 is conserved in M. albus.
SELENOU3This protein had 0 hits in Monopterus albus ’ genome. Therefore, we concluded that SELENOU3 does not exist in M. albus.
SELENOW familyThis family of proteins has been reported in both the prokaryotic and eukaryotic kingdoms. Selenoprotein W (SELENOW) is expressed in various tissues, but it is especially high in the skeletal muscle of mammals. SELENOW is the smallest selenoprotein identified to date that contains the canonical amino acid selenocysteine (Sec). It is known to have an antioxidant effect on cells, as many other selenoproteins.[27]
SELENOW (1)This protein could be predicted in M. albus: when looking at t-coffee results we saw that it had a great alignment (starting with Methionine) and a selenocysteine was aligned and conserved. However, no selenoprotein could be predicted for this sequence with Seblastian but one SECIS structure was located in 3’ UTR. To conclude, the alignment shows good homology with zebrafish and this coincides with the information found of this protein, so we conclude that SELENOW (1) is conserved in M. albus.
SELENOW (2)This protein was not found in Monopterus albus as it was predicted in the same region of the scaffold as SELENOW (1) with a higher e-value. Therefore, we concluded that this protein is not present in M. albus.
SELENOW (3)This protein had 0 hits in Monopterus albus ’ genome. Therefore, we concluded that SELENOW (3) is not present in M. albus.
TXNRD familyThioredoxin Reductase (TXNRD) is an ubiquitous homodimeric flavoenzyme whose physiological role is the transfer of reducing equivalents from NADPH to thioredoxin. Two variants of TXNRD have evolved independently: the High molecular weight TXNRD found exclusively in higher eukarya and the Low molecular weight TXNRD, found in procarya and some eukarya including fungi and plants. Mammalian thioredoxin reductase 1 and thioredoxin-glutathione reductase evolved from an ancestral glutaredoxin-domain containing enzyme, still present in fish.[2] [28]
TXNRD2This protein could be predicted in M. albus: t-coffee results showed a perfect alignment with zebrafish protein and the selenocysteine was aligned and conserved between species. However, it did not start with Methionine, as the zebrafish protein obtained from selenoDB was not well annotated. Finally, when searched in Seblastian, the predicted protein did not start with Methionine either, instead, it started with Glycine. Furthermore, one SECIS structure was predicted in 3’ UTR. To conclude, the alignment shows good homology with zebrafish and this coincides with the information found of this protein, so we conclude that TXNRD2 is conserved in M. albus.
TXNRD3TXNRD3 could be predicted in M. albus as well. The result of the t-coffee alignment showed a high homology with the zebrafish homonymous. The selenocysteine residue was conserved. Our predicted protein started with a Methionine residue, making our prediction more reliable. A SECIS element was predicted in 3’-UTR. We conclude that TXNRD3 is conserved in M. albus, as the alignment showed a good homology with zebrafish and all this information coincides with the one that has been searched for this protein.
Machinery and metabolism
eEFsecThe elongation factor eEFsec could be predicted in M. albus. When the t-coffee was performed, a high identity was observed. No Selenocysteine residue was found in the protein, as expected for a machinery protein. Although the whole sequence was perfectly aligned, the first aminoacids of the zebrafish did not match, probably because the genome was not well anotated. As we expected, no SECIS structure nor selenoprotein was predicted by Seblastian. Considering all this information we consider that eEFsec can be found in M. albus’ genome.
MsrA familyThis is a family of enzymes, referred to as methionine sulfoxide reductases (Msr), and in recent years there has been considerable interest in MsrA. This enzyme has been shown to protect cells against oxidative damage as MsrA catalyzes the reduction of methionine-S-sulfoxide to methionine in different cellular compartments of mammalian cells. Two proteins are included in this family: [29]
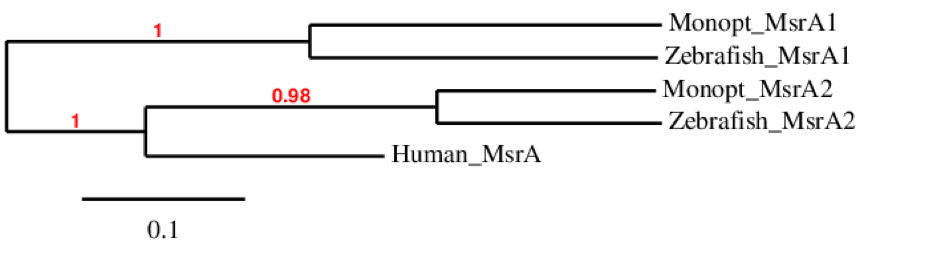
This phylogenetic tree of MsrA family shows that MsrA1 is only found in Monopterus and zebrafish, whereas MsrA2 is found in M. albus, zebrafish and human. This probably means that the duplication of MsrA only happened in fishes and that human MsrA comes from the same ancestor than MsrA2 of zebrafish and M. albus.
MsrA (1)This protein could be predicted in M. albus: the alignment resulted from t-coffee did not match the first few amino acids of the protein. When predicted with Seblastian no selenoprotein sequence could be found and no SECIS could be found either, since it is a protein involved in the selenium metabolism and Seblastian can only predict selenoproteins. Almost the whole protein matched with the zebrafish one. To conclude, the alignment shows good homology with zebrafish and this coincides with the information found of this protein, indicating a good conservation of it in M. albus.
MsrA (2)This protein could also be predicted in M. albus: the alignment resulted from t-coffee was perfect, the protein started with Methionine and there was no selenocysteine found as it is also involved in metabolism. Therefore, Seblastian could not predict any selenoprotein and any SECIS structure either. To conclude, the alignment shows good homology with zebrafish protein and this coincides with the information found about this protein, indicating that is well conserved in M. albus
PSTKPhosphoseryl-tRNA kinase (PSTK) belongs to the machinery proteins and is an intermediate product produced under oxidative stress conditions. PSTK is mainly involved in the synthesis of the antioxidant stress molecules, such as glutathione peroxidase (GSH-Px) and C-reaction protein (CRP). PSTK functions in the mitochondrial complex I assembly and participates in mitochondrial apoptosis and mitochondrial fatty acid beta-oxidation. [30]
PSTK could be predicted in M. albus. The alignment obtained using t-coffee was regular, as some parts of the query protein were absent in the alignment. No selenocysteine was observed in the sequence, as expected. We repeated the process using an exhaustive exonerate but no improvements were made. No selenoprotein could be predicted using Seblastian. One SECIS structure can be predicted but we assumed it was a false positive as it is a machinery protein. In conclusion, we believe this protein is conserved in M. albus’ genome, because we consider that the homology observed with the zebrafish homonymous is strong enough.
SBP2 familySBP2 (selenocysteine insertion sequence binding protein 2) is an essential factor in selenoprotein synthesis. Patients with SBP2 defects have a characteristic thyroid phenotype and additional manifestations such as growth delay, male infertility, impaired motor coordination and developmental delay. SBP2 may be the protein that combines and reacts with all of the SECIS elements in one organism. Thus, the core pattern of SECIS elements are more conserved in a single organism than in a single selenoprotein family. [3] [31]

The phylogenetic tree of SBP2 family shows that SBP2 was duplicated in zebrafish although Monopterus albus does not have SBP2b, the same happens for human. Moreover, we can see that zebrafish SBP2a, M. albus SBP2 and Human SBP2 were originated from the same ancestral protein.
SBP2 (1)This protein could be predicted in M. albus: the initial t-coffee alignment had 2 different gaps: one in the middle of the protein and the other one at the end. With the second prediction, made using exhaustive exonerate, the gaps were filled and the alignment was much more consistent and reasonable. There was no selenocysteine found, as expected, because this protein belongs to machinery. In addition, Seblastian could not predict any selenoprotein or SECIS structure, as also expected. To conclude, this protein showed good homology with zebrafish, coinciding with information found about it, and thus being conserved in M. albus.
SBP2 (2)This protein was not found in M. albus’ genome as the sequence predicted had a very bad alignment with zebrafish protein. To get a better alignment, we decided to analyze the protein again by doing an exhaustive exonerate. Here, we saw that all the alignments started in the position 340 and their homology was not very good. This way, the resulting protein would not be the same protein as our query. We then concluded that Monopterus albus does not have SBP2 (2) in its genome.
SecSSelenocysteine synthase (SecS) is an ubiquitously expressed enzyme that catalyzes the terminal reaction of selenocysteine synthesis. SecS converts the phosphoseryl group into the selenocysteinyl moiety in a mechanism that requires selenocysteine tRNA (tRNASec) and a cofactor. The product of SecS catalysis, Sec-tRNASec, is an obligate substrate for selenoprotein synthesis, thus suggesting that the catalytic activity of SecS is indispensable for the human selenoproteome integrity. [32]
This protein could be predicted in M. albus: t-coffee results showed a good alignment, starting with Methionine. There was no selenocysteine residue found in the sequence and Seblastian could not predict any selenoprotein as expected because SecS is a machinery protein. One SECIS structure can be predicted but we assumed it was a false positive as it is a machinery protein. To conclude, the alignment shows good homology with zebrafish protein and this coincides with the information found about this protein, indicating that is well conserved in M. albus.
SECp43 familySECp43, is a highly conserved protein with two ribonucleoprotein-binding domains and a polar/acidic carboxy terminus. The protein and corresponding mRNA are generally expressed in rat tissues and mammalian cell lines. It is involved in the early steps of selenocysteine biosynthesis and tRNA(Sec) charging to the later steps resulting in the cotranslational incorporation of selenocysteine into selenoproteins. Stabilizes the SECISBP2, EEFSEC and tRNA(Sec) complex. Therefore, enhances efficiency of selenoproteins synthesis. [32]
SECp43 (1)This protein could be predicted in M. albus: this protein had a good alignment with the one from zebrafish and it started with Methionine but no selenocysteine was found as we expected, because it is a machinery protein. Seblastian could not predict any selenoprotein as expected and no SECIS structure could be predicted in 3’ UTR, as also expected. To conclude, the alignment shows good homology with zebrafish and this coincides with the information found of this protein, so we conclude that SECp43 (1) is conserved in M. albus.
SECp43 (2)This protein could be predicted in M. albus: when looking at the results from t-coffee we saw that it had a good alignment as it started with Methionine but there was a region that had a lot of gaps. After doing an exhaustive exonerate, all the gaps were filled. No selenocysteine was found as it is a machinery protein. Seblastian could not predict any selenoprotein as expected and no SECIS structure could be predicted in 3’ UTR, as also expected. To conclude, the alignment shows good homology with zebrafish and this coincides with the information found of this protein, so we conclude that SECp43 (2) is conserved in M. albus.