Selenium
Selenium is an essential micronutrient of major metabolic significance since it is required by selenium-dependent proteins in the form of selenocysteine [1,2,3,4]. The entry point of selenium in animals is via plants, by absorption of the element in its inorganic form from the soil [5]. Under physiological conditions, the selenium is almost fully ionised providing an efficient biological catalysis.
This chemical element plays an important role in biological processes, including cellular response to oxidative stress, redox signalling, cellular differentiation, protein folding and immune response[2]. Furthermore, it has also been proved that evinces antioxidant activity, anti-inflammatory, antimutagenic, anticarcinogenic, chemopreventive, antiviral, antibacterial, antifungal and antiparasitic effects[6].
Nevertheless, there is a small window between the deficiency of selenium and toxicity. This proves that the effects of this micronutrient depends to a great extent on its levels of it in the organism [7]. Specifically, selenium deficiency in chicken was manifested as hepatic malnutrition, deformation and necrosis. On the other hand, an excess of selenium could result in reduced growth, and anemia, reduced egg production and an impaired immune function[8].
Selenoproteins
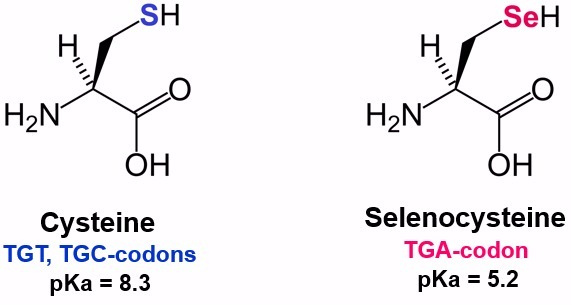
Selenoproteins are proteins with enzymatic activity that incorporate selenium in the form of selenocysteines, which are proteinogenic aminoacids homologues of cysteines [9]. They can be found in the eubacterial, archaea and eukaryotic domains of life[4]. The selenoprotein family is formed by, at least, 30 proteins between the domains. Selenoproteomes among species are usually small. This size can be explained by the limited selenium supply in nature and its energy-expensive biosynthesis [5].
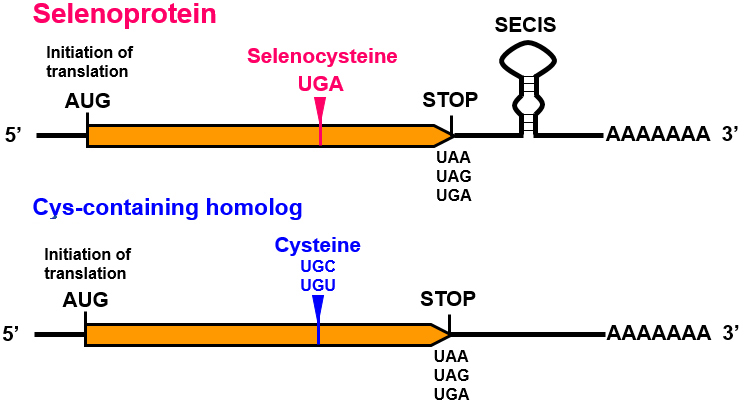
A unique mechanism is in charge of the incorporation of selenocysteine into selenoproteins. Nevertheless, some differences have been found in the mechanisms of selenoprotein synthesis between the different domains [6]. The common features to all the organisms are the decodification of the codon UGA in mRNA, involved in the termination of translation, the specific tRNA, the selenocysteine insertion sequence (SECIS) element and protein factors[5].
Different studies have shown the important role of selenoproteins in chickens. These proteins seem to be involved in several biological processes such as growth performance, central nervous system function, male fertility and muscle development. Selenoproteins also present oxidoreductase function[8].
Selenoproteins biosynthesis
As previously mentioned, selenoproteins contain a selenocysteine (Sec), which is an amino acid coded by an UGA codon[10]. The mechanism by which the ribosome incorporates selenocysteines at different UGA is still under study[5]. UGA codon is usually in charge of stopping the translation[5]. Nevertheless, there are a variety of cis- and trans- elements that control this process in order not to stop the translation and proceed with the addition of the selenocysteine[11].
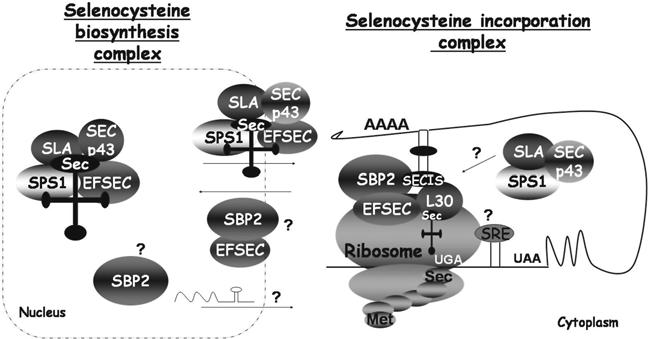
Regarding cis- elements, the UGA codon and a stem-loop structure known as SECIS element are the main ones. When referring to trans-acting elements, we find machinery proteins such as SECIS-binding protein 2 (SBP2), eukaryotic elongation factor selenocysteine-tRNA (eEFsec), selenocysteine synthase (SecS), selenophosphate synthase (SPS2 or SEPHS), or phosphoseryl-tRNA (PSTK)[10].
The biosynthesis starts with the charge of the serine on the specific tRNAsec. This serine is phosphorylated by the PSTK. At the same time, the SPS2 prepares the selenium to be incorporated. Then, the SecS adds the phosphorylated selenium to the phosphoserine and produces the selenocysteine[10].
Subsequently, the protein SBP2 binds to the SECIS element and, at the same time, it recruits the eEFsec. This elongation factor allows incorporation of the specific Sec tRNA into the ribosome[11]. All this gives places to the decoding of the UGA codon as a selenocysteine[10]. The sec-tRNA interacts with the UGA codon and then Sec is incorporated to the selenoprotein[11].
Selenoproteins families
15 kDa selenoprotein
Sel15 was one of the first identified selenoproteins and was originally named the 15-kDa selenoprotein based on its molecular mass. Recent studies showed that it is a member of the thioredoxin,like fold family of proteins and it localized in the ER [5]. It has been revealed that it contains a thioredoxin-like domain and a NH2-terminal signal peptide, which is suggestive of a redox activity[13]. Its function is based on the quality control of protein folding[7].
Glutathione peroxidase (GPx)
Glutathione peroxidases (GPx) are the larges selenoproteins family in vertebrates [8]. It is thought to be associated with the antioxidant activity of selenium. Glutathione peroxidases are in charge of catalyzing the reduction of hydrogen peroxide and organic hydroperoxides. The common feature among selenium-dependent GPxs is the conserved catalytic triad containing Sec, Gln, and Trp, which acts by successive oxidation and reduction of Sec during the catalytic cycle[13].
This family is integrated by 8 GPx homologs (GPx1-GPx8). GPx1-4 and GPx6 are selenoproteins in humans and in macaque, whereas GPx5, GPx7 and GPx8 are cysteine-containing homologues. Unlike mRNAs for other characterized glutathione peroxidases, the mRNA of these homologues does not contain a selenocysteine (UGA) codon. Thus, the encoded protein is selenium-independent [8].
- Glutathione peroxidase 1 (GPx1) plays a role in the Glutathione (GSH)-dependent detoxification of hydrogen peroxide[13].
- Glutathione peroxidase 2 (GPx2) was identified as an epithelium specific gastrointestinal glutathione peroxidase with a similar structure and function to GPx1[13].
- Glutathione peroxidase 3 (GPx3) is a glycosylated protein secreted to extracellular compartments. It is an efficient antioxidant in the plasma[13].
- Glutathione peroxidase 4 (GPx4) is able to reduce phospholipid and cholesterol hydroperoxides by using electrons from protein thiols. It is present in cytosolic, mitochondrial and nuclear isoforms with differential tissue distribution[13].
- Glutathione peroxidase 6 (GPx6) function is the detoxification of hydrogen peroxide. It is only seen in the olfactory epithelium and during embryo development.
- Glutathione peroxidase 7 and 8 (GPx7 and GPx8) have also been discovered but more research is needed in order to fully understand its function[6,7].
Iodothyronine deiodinase (DIO)
The iodothyronine deiodinases (DIOs) play an essential role in the regulation of thyroid hormone activity by the production and degradation of receptor-active 3,3,5-triiodothyronine (T3). Specifically, those proteins are responsible for the deiodination of T4 (3,5,3',5'-tetraiodothyronine) into its active form T3 (3,5,3'-triiodothyronine) and of T3 into T2 (3,3'-diiodothyronine) [8].
There are 3 different DIO subfamilies that contain selenocysteine residues: DIO1, DIO2 and DIO3. All of them have the typical selenocysteine in their catalytic site, as well as two conserved histidines, which are important for enzyme activity, but only DIO2 contains 2 Secs [8]. DIO1 and DIO2 are in charge of the activation of the thyroid hormones T4, T3 and rT3. DIO1 is expressed in the liver, kidney, thyroid and in the pituitary gland and DIO2 can be found in the thyroid, the CNS, the pituitary gland and skeletal muscle. Whereas, DIO3's function is to inactivate the thyroid hormones previously mentioned. It is more expressed in the uterus, placenta, embryonic liver and neonatal brain.[6,7].
Methionine sulfoxide reductase
Methionine-R-sulfoxide reductase (Msr), also known as SelR, is a member of the Msr family of proteins, which are in charge of catalyzing the reduction of oxidized methionine residues. SelR is localized in the nucleus and the cytoplasm of cells, and it presents a specific enzymatic function [6,7]. It was found to bind with zinc through four cysteines residues [8].
We distinguish MsrA and MsrB. MsrB works as a stereospecific methionine-R-sulfoxide reductase whereas MsrA catalyzes the reduction of the other isomer[15].
Selenoprotein H (SELENOH)
SelH is a very conserved selenoprotein among most vertebrate's which presents a subcellular localization pattern in the nucleolus [16]. It is predominantly present during the embryonic development. It has the capacity of binding to DNA sequences with heatshocks and stress-related elements. It is also involved in the transcription's regulation of gens related to the glutathione synthesis [6,7].
Selenoprotein I (SELENOI)
It is one of the most unknown proteins since it was one of the latest finding in the selenoprotein family. It is present only in vertebrates and it is thought to play an important role in the maintenance of the ER membrane shape and composition through its CDP-alcohol phosphatidyltranspherase domain, which is highly conserved [8,17]. Nonetheless, studies shown that the Sec residue is not involved in this function. That is why SelI's function is still unknown [6,7]. It has been suggested that the Sec residue may alter the high specificity of the CDP-alcohol phosphatidyl-transpherase domain [18]. Also, an evolutionary study noted that selI has no homologs in which the selenocystein is substituted by a cystein[8].
Selenoprotein K (SELENOK)
SelK is one of the most widespread selenoproteins among eukariotic seelnoproteins [20]. It is part of the same family as SelS since both of them present a transmembrane domain, a glycine-rich segment and positively charged amino acids. It is expressed predominantly in the heart and skeletal muscle even though it can be find in other tissues such as the pancreas, liver or placenta. It is usually located in the ER and the plasma membrane and it is thought to have an antioxidant function in the heart, to play a role in calcium flux during the activation of immune cells, as well as in the maintenance of ER homeostasis [20]. As SelS it is thought to degrade ER-associated misfolded proteins [8,10].
Selenoprotein M (SELENOM)
It is a thioredoxin-like fold mitochondria-resident protein [8]. It is a distant homolog of Sel15 since they share 31% sequence identity and demonstrate a similar distribution. As Sel15, it contains a thioredoxin-like domain and a NH2-terminal signal peptide, but in addition it possesses a COOH-terminal extension with an ER retention signal[13]. Its function is based on the rearregement of disulfide bonds in the ER-localized proteins [14].
Selenoprotein N (SELENON)
SelN is ubiquitously expressed in tissues. It generates a 65-kDa glycoprotein localized in the ER membrane, with its N-terminus domain facing the cytosol, and the rest of the protein inside the lumen of the ER. It is found in all vertebrates, although it may not be limited to vertebrates [17, 21]. It is said to play a crucial role in the regulation of intracellular calcium mobilization.
It is also required for muscle development and differentiation, and it seems to be related to the regeneration of skeletal muscle tissue[6,7].
In addition, its Sec residue points to a possible antioxidant function, possibly protecting mature fibers from oxidative stress [22]. Several studies have shown that SelN absence causes an increased basal oxidative activity in myotubes in vitro [23,24], as well as an increased sensitivity to H2O2 and the inhibition of the up-regulation of antioxidant enzymes [24].
Selenoprotein O (SELENOO)
SelO is a selenoprotein redox-active mitochondrial selenoprotein. It is found in all vertebrates and it is the largets of the 25 selenoproteins described in the mammalian genome. It is thought to have an antioxidant role, which is supported by the presence of a CXXU domain (Cys and Sec residues separated by two amino acids), which is common in oxidoreductases [5,8]. Nevertheless, more research needs to be done since its specific function is still unknown [6,7].
Selenoprotein P (SELENOP)
SelP is a secretory transport extracellular selenoprotein which constitutes the 50% of the selenoprotein in plasma. Its function has been conserved during evolution, but its structure may have variations. SelP usually present 13 selenocysteine residues and 2 SECIS elements [8]. Nevertheless, these numbers can vary depending on the specie. The main role of SelP seems to be the transport of selenium to peripheral tissues and antioxidant function[6,7].
Selenoprotein S (SELENOS)
SelS contains a transmembrane domain, a segment rich in glycine and positive amino acids. It also contains an unknown domain and a unique selenocysteine in the last five nucleotides of the C-terminal region, which is thought to stabilize the protein. Its expression is induced in response to ER stress, via an ER stress-response element within the SelS promoter. The pathway used in this response is the nuclear factor-B, thus it causes the degradation of misfolded proteins [6,7]. Studies described the importance of the separation between selenocystein and its cystein partner in the stability of the seleno sulfide bond. It has been shown that in Gallus gallus, these two are separated for 13 amino acids [8].
Selenoprotein T (SELENOT)
SelT is mainly located in the ER and the Golgi apparatus and it is expressed both during the embryonic development and adult tissues. Gallus gallus contains two thioredoxin-like folds and the selenocystein was located in the first thioredoxin-like fold[8]. It is expressed attached to an ubiquitin during the embryonic development and adult tissues. Its function is related to the regulation of the Ca2+ homeostasis and cellular adhesion[6,7].
Selenoprotein U (SELENOU)
The SelU family is widely distributed in the eukaryotic lineage as a Sec- or Cys- containing proteins. It contains a thioredoxin-like fold. is believed to be an important member of the avian selenoprotein family but its function is still unknown [19].
Thioredoxin reductase (TXNRD)
It is the only enzyme capable of reducing thioredoxin. It catalyzes thioredoxin reduction using NADPH and it mediates the final step in the electron transference pathway for the reduction of nucleoside diphosphate.
There are three types of thioredoxin reductases in animals. TrxR1 is located in the cytosol and nucleus. It is in charge of the NADPH-dependent reduction of Trx1. TrxR2 seems to contribute to cytosolic glutathione disulfide reduction[25]. TrxR3 is found in the mitochondria and it is involved in the reduction of mitochondrial thioredoxin (Trx2) and glutaredoxin 2 (Grx2)[6,26].
Selenoproteins machinery
Selenophosphate synthetase 1 (SEPHS1)
SPS1 is an enzyme involved in the catalysis of the selenophosphate's synthesis from selenide and ATP. Together with SPS2 and SECp43 binds the selenophosphate group to the Cys residue[10]. It is well conserved in all prokaryotic and eukaryotic genomes encoding selenoproteins.
Selenophosphate synthetase 2 (SEPHS2)
SPS2, also referred to as SEPHS, is a selenoprotein involved in the selenocysteine byosinthesis required for its own synthesis. It catalyzes the conversion of selenide to selenophosphate [6,7].
Phosphoseryl-tRNA (PSTK)
PSTK is in charge of the phosphorilation of the precursor Serine located on the specific tRNAsec[10].
Sec synthase (SecS)
The SecS adds a phosphorylated selenium to the phosphoserine and produces the selenocysteine[10].
SECIS binding protein 2 (SBP2)
SBP2 function is to bind itself to the SECIS element and, at the same time, to recruit the eEFsec. It is associated with ribosomes[11].
Eukaryiotic elongation factor (eEFsec)
The eEFsec allows the incorporation of the specific Sec tRNA into the ribosome, thus the selenocysteine is incorporated into the selenoprotein[11].
Anas zonorhyncha
- Morphology and classification
The eastern spot-billed duck or Chinese spot-billed duck, is a dabbling duck. It was formerly treated as a subspecies of the Indian spot-billed duck (A. poecilorhyncha). The name is derived from the yellow spot on the bill.
- Taxonomy


| Kingdom | Animalia |
| Phylum | Chordata |
| Class | Aves |
| Order | Anseriformes |
| Family | Anatidae |
| Genus | Anas |
| Species | A. zonorhyncha |
- Distribution
Eastern Spot-billed Ducks are species breeds almost throughout Japan and is generally a year-round resident south of Honshu (the largest main island).
- Habitat
Eastern Spot-billed Ducks prefer lowlands and usually occur in lakes, marshes, wetlands, rice fields, tidal flats and rivers. They nest separately from late April to July in meadows and reed beds on lakes, ponds and rivers, but they may also breed in a colony in island-like places such as sandbars in a river. In the wintering period, they are found in most bodies of water, such as lakes, marshes, ponds and rivers. They also form a flock with Mallards in littoral regions.
- Diet
The diet of Eastern Spot-billed Ducks consists primarily of the seeds and leaves of various plants including rice. They glean, filter food out of the water and feed upside down in the water depending on the feeding habitat[27].
Click here for more information