Discussion
Anas zonorhyncha's selenoproteins were characterized by the comparison of Gallus gallus (chicken) and Homo Sapiens (Human) selenoproteins. Our team chose Gallus gallus since its genome is phylogenetically close to Anas zonorhyncha - both of them belong to the birds class - and because its selenoproteome was annotated in SelenoDB 2.0. In regard to Homo Sapiens, we considered this specie due to the fact that its genome was completely sequenced. In addition, the human’s genome can be found in Seleno DB 1.0 which is much more reliable than SelenoDB 2.0.
The comparison of Gallus gallus and Homo Sapiens selenoproteins with Anas zonorhyncha's genome was realized with the queries obtained from SelenoDB. Nevertheless, the vast majority of the sequences obtained did not start with the amino acid Methionine (Met). As most of the proteins need this Met residue at the beginning of the sequence to start the translation, the fact that the transcript does not contain this amino acid evidences a bad annotation that could affect the whole sequence. Therefore, in those cases we first ran the program using the Homo sapiens sequences found in SelenoDB 1.0 as our initial proteins. In the cases where the alignment with the human protein was of poor quality, or the initial human protein did not start with a Met residue, we used Ensembl Gallus gallus database.
Then, we introduced the fastasubseq into Seblastian, which is a tool that predicts SECIS and known selenoproteins, allowing us to confirm the presence of the studied selenoproteins in Anas zonorhyncha.
Selenoproteins
15kDa selenoprotein
15 kDa selenoprotein (Sel15) is a zinc-containing protein whose function is to control the protein folding. It belongs to the thioredoxin-like fold family and its location is the ER [5,13].
The query from Gallus gallus did not start with a methionine, so we decided to compare this part of the genome in Ensembl. This genome browser showed the beginning of the sequence starting with methionine, but it had a missing part at the end of the sequence. As the codon that codifies for Sec is TGA, which is also a stop codon, we considered that Ensembl stopped the protein sequence there. Therefore, we decided to join the first part of the sequence from Ensembl and the sequence that follows the Sec residue in SelenoDB, and use the result as our query sequence.
The program T-Coffee showed that the alignment of the query and the predicted protein was correct. Nevertheless, the predicted one did not start with a methionine. That could be justified as the protein is not functional because it can not be translated but the major part of the sequence is conserved. This incorrect alignment of the beginning of the sequence could also be explained with Sel15 having a different N-terminus domain compared to the query, due to changes emerged during evolution, which would not enable the program to align them.
Then, we analyzed the sequence obtained in Seblastian. This program showed that the predicted genome presents one selenoprotein with a Sec residue and a SECIS element located in 3'UTR.
With these results, we can not confirm the presence of Sel15 in Anas zonorhyncha because the obtained sequence does not start with a methionine residue. However, Seblastian exhibits the presence of important structures characterized in these proteins, leading to the hypothesis that Anas zonorhyncha does have this protein in its genome.
The final T-Coffee alignment can be found here.
Glutathione peroxidase family
Glutathione peroxidases are the largest selenoprotein family in vertebrates. GPxs do not limit their function to antioxidant defense of the cell, but rather participate in complex signaling cascades. This family is composed by 8 GPx homologs (GPx1-GPx8) [8,13].
In our case, we have studied only three GPx proteins from Gallus gallus that we have obtained from SelenoDB 2.0: GPx3 which is a selenoprotein, and GPx7 and GPx8 which are selenium-independent cysteine-containing homologs.
We have generated a phylogenetic tree with Phylogeny.fr [32,33] for the GPx family, in order to check whether each predicted protein is closer to its query than to the rest of the predictions and queries. The result is shown below:

As the tree shows, each predicted protein is closer to its initial query than to any other proteins of the family. Interestingly, GPx7 and GPx8 seem to be closer to each other than to GPx3.
- Glutathione peroxidase 3
The first query that we have used from Gallus gallus from SelenoDB 2.0 did not start with a methionine. To minimize the error we decided to compare this genome with the one obtained from Ensembl. After running the program, T-Coffee’s results exhibit a good alignment with Anas zonorhyncha's genome but the predicted sequence did not start with a Methionine because there is a gap of two amino acids.
The explanation of this could be that there was a good conservation of the sequence of the protein but it has remained as a non-functional protein. Another explanation could be that the transcript has not been annotated correctly. It could also be a modification or an expansion occurred during Anas zonorhyncha's evolution which causes an unaccurate alignment with the Gallus gallus genome, even if the protein exists.
In spite of the analysis with Seblastian that showed no prediction of selenoproteins, it could predict one SECIS element of grade A, as we have expected because GPx3 is a selenoproteine. Thus, we can not ensure that GPx3 is found in Anas zonorhyncha's genome.
The final T-Coffee alignment can be found here.
- Glutathione peroxidase 7
As Gpx7 is a Cys-containing homologous, it does not contain any Sec in its sequence, as we could see in the results obtained in the T-Coffee program. Moreover, these T-Coffee results showed that there was a good alignment with the query but it did not start with a Methionine, so we chose the chicken sequence from Ensembl as a query.
We can observe in the T-Coffee alignment with the Ensembl’s query that there is a good alignment and that the first amino acid is a Methionine. As expected, Seblastian could not predict any SECIS or selenoprotein. Thus, we can confirm that GPx7 exists in Anas zonorhyncha's genome.
The final T-Coffee alignment can be found here.
- Glutathione peroxidase 8
Regarding GPx8, the T-Coffee results show that there was a good alignment between our sequence and the Gallus gallus query but both sequences start with a Lysine. As probably the sequence obtained from SelenoDB is not well annotated we have decided to choose the sequence from Ensembl. The obtained alignment was good and the predicted protein started with a Methionine residue.
As Gpx8 is a Cys-containing homologous, Seblastian has not predicted any selenoprotein. For this same reason, no SECIS elements have been predicted neither. To conclude, we can say that Gpx8 is present in the proteogenome of Anas zonorhyncha.
Iodothyronine deiodinase family
Iodothyronine deiodinases play an essential role in the regulation of thyroid hormone activity. There are 3 different DIO subfamilies that contain selenocysteine residues: DIO1, DIO2 and DIO3. All three chicken deiodinases have the typical selenocysteine in their catalytic site, as well as the two conserved histidines, which are important for enzyme activity. However, only DIO2 contains 2 Secs in the DIO family [8].
We created a phylogenetic tree with Phylogeny.fr [32,33] in order to check if every predicted protein in the DIO was closer to its query than to the rest of the analysed proteins of this family. In the case of DIO3, we have included the queries and predictions from the analysis of both Gallus gallus and Homo sapiens, as we have compared the duck’s genome with both of these species. The resulting tree is shown below:
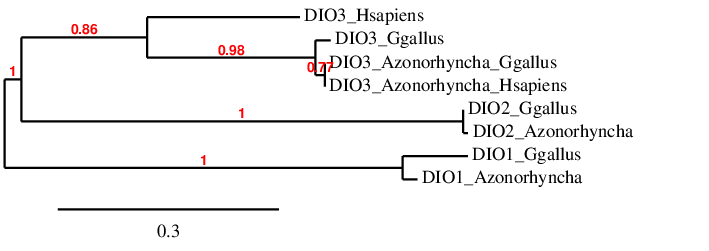
As the tree shows, both DIO1 and DIO2 are closer to its corresponding query than to any other sequences. Regarding DIO3, the tree shows that the two proteins predicted based on the genome of Anas zonorhyncha are closer to each other than to any other protein. This makes sense, as they are both proteins obtained from the same genome. These two predictions are closer to the Gallus gallus query protein than to the Homo sapiens genome, which also makes sense because chicken and duck are phylogenetically closer than human and duck. Interestingly, DIO2 and DIO3 appear to be closer to each other than to DIO1.
- Iodothyronine deiodinase 1
The results obtained in the T-Coffee program show that there is almost a perfect alignment with the Gallus gallus' sequence with the exception of a few amino acids.
Our sequence contains a Sec residue in the second exon and starts with a Methionine. Furthermore, also the protein that has been predicted with Seblastian correlates exactly with our result: it has the same number of exons, the Sec is in the second exon, and it also starts with a Methionine.
As we expected, Seblastian has also predicted one SECIS of grade A located in the 3’UTR region. All these results demonstrate that Anas zonorhyncha contains DIO1 in its genome.
- Iodothyronine deiodinase 2
Regarding DIO2, we have seen that the selenoprotein we have obtained in Anas zonorhyncha has a perfect alignment with Gallus gallus' sequence, with the exception of only five amino acids.
The T-Coffee results show that the protein starts with a Methionine and contains a Sec residue in the second exon.
Taking into account that only DIO2 contains 2 Secs in DIO family, we expected to find this second Sec residue, but neither the Gallus gallus nor Anas zonorhyncha sequences have it.
Theoretically, chicken DIO2 contains 2 Secs (Sec-132 and Sec-265). Recent studies[8] have reported that the second UGA codon would insert a Sec only if the first UGA codon is mutated. Then, we can conclude that the first TGA codon is not mutated and is followed by a pyrimidine (so, it is more likely to be translated as a Sec) and probably the second TGA codon is followed by a purine that makes it termination codon.
With this information, we can think that the first Sec plays a central role in the deiodination process whereas the second Sec can not be connected with any known biological process in birds. Finally, Seblastian was not able to predict any selenoprotein but could predict a SECIS of grade A. All in all, these results confirm that DIO2 protein is present in the Anas zonorhyncha's genome.
- Iodothyronine deiodinase 3
Concerning DIO3, the alignment with the Gallus gallus' sequence was good. Nevertheless, neither the predicted sequence nor the query of Gallus gallus start with a methionine residue.
Therefore, we decided to compare Anas zonorhyncha with Homo sapiens in order to predict a better biologically protein with a Methionine as the first amino acid. The results of the T-Coffee show that both sequences, the one of Anas zonorhyncha and the other of Homo sapiens, started with Methionine and there was a quite good alignment.
Seblastian could predict a selenoprotein and one SECIS element of grade A, as expected. Taking this into account and considering how phylogenetically far these two species are, we can firmly state that the protein exists in Anas zonorhyncha.
Methionine sulfoxide reductase family
This selenoprotein is in charge of catalyzing the reduction of oxidized methionine residues. In Anas zonorhyncha we found MSRB1, which contains one selenocysteine. This isoform can be found in both the cytoplasm and the nucleus. We also found an homologous, named MSRB3, which contains a cysteine instead of a selenocysteine [6,7].
We have generated a phylogenetic tree with Phylogeny.fr for the MSRB family, in order to check whether each predicted protein is closer to its query than to the rest of the predictions and queries. The result is shown below:
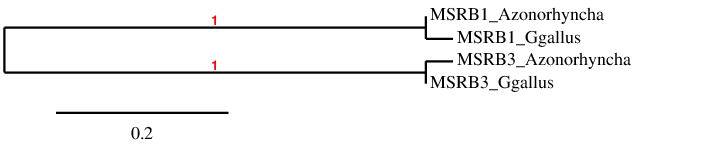
As the tree shows, each predicted protein is closer to its initial query than to any other proteins of the family.
- Methionine R-sulfoxide reductase 1
Concerning MSRB1, the alignment that we have obtained with the T-Coffee program was good. Nevertheless, the predicted sequence did not start with a methionine residue whereas the Gallus gallus' sequence did. This could mean that there is a good conservation of the sequence but a non-functional protein, since the vast majority of proteins need a methionine residue at the beginning of the sequence to start the translation.
We can observe that there is a gap of 17 amino acids at the beginning of the sequence, which is responsible for not having the methionine residue in the first position. This gap could be explained with the protein having lost a part of its sequence, or maybe it is because of a low sensitivity of the exon search with the Exonerate.
Surprisingly, the Seblastian's prediction shows up exactly the same sequence as the one that we have obtained, meaning that the prediction does not have a methionine at the beginning of the sequence either, and has a proline instead, which is what our alignment shows. In fact, in this prediction there is an extra exon, although its location is at the end of the protein.
Finally, Seblastian has also predict a SECIS element of grade A, as MSRB1 needs a SECIS for being a functional selenoprotein.
Probably, MSRB1 exists in Anas zonorhyncha's genome because in recent studies[28] it was observed that some proteins could start with different amino acids (Pro, Ala, Gly, Ser, Thr, Val), which makes our predicted MSRB1 a plausible selenoprotein.
- Methionine R-sulfoxide reductase 3
Regarding MRSB3 from Anas zonorhyncha, it is a protein that does not contain a selenocystein since it is a cysteine-containing homologous.
The query from the Gallus gallus from SelenoDB did not start with a methionine. To minimize the error we decided to compare this genome with the one from Ensembl. Results showed that there was an additional part at the beginning of the sequence missing in the genome from SelenoDB, so we decided to use Ensembl in order to predict our sequence. After running the program, results exhibit a good alignment with Anas zonorhyncha's genome, although the distance between the exons of the predicted protein is very large. Nevertheless, the predicted sequence does not start with a methionine. This could be explained as a good conservation of the sequence but a non-functional protein, or because the transcript is not correctly annotated either. Another explanation of this lack of methionine at the origin of the sequence is that a modification or an expansion occurred during evolution in Anas zonorhyncha so it could not be aligned to the Gallus gallus genome, even if the protein exists.
The analysis with Seblastian showed no prediction of selenoproteins. No SECIS elements were identified either. This agrees with the expected results since, as previously said, it is a cysteine-containing homologous.
Due to all the difficulties when analyzing this protein, we can not assure the presence of MSRB3 in the Eastern spot-billed duck.
Selenoprotein H
Selenoprotein H is a selenoprotein that is conserved among most vertebrates' genomes. Although its function remains unclear, some studies have shown that it is localized in the nucleolus, and that it exhibits gluthatione peroxidase activity [16]. Moreover, it has been suggested that it may play a role in organ development [26].
Selenoprotein H was described in the Gallus gallus genome, and we decided to compare its sequence with the genome from Anas zonorhyncha. However, the tblastn did not result in any valid hits. Therefore, we conclude that this protein is not found in the Anas Zonorhyncha genome.
Selenoprotein I
Selenoprotein I (SelI) is a selenoprotein present only in vertebrates [17]. Little is known about the function of SelI. In Gallus gallus, SelI is an endoplasmic reticulum (ER) transmembrane protein, having its selenocysteine residue in its C-terminal (cytosolic) domain, and it is thought to play an important role in the maintenance of the ER membrane shape and composition [8, 17]. However, the Sec residue is not involved in this function [8], even though the Sec residue is considered essential for a correct selenoprotein function. Therefore, SelI's function as a selenoprotein remains unknown.
Our search has resulted in an acceptable alignment when comparing the SelenoDB chicken SelI sequence with the Anas zonorhyncha genome. However, the N-terminus domain has not been correctly aligned. The predicted Sel I protein does not show methionine (M) as its first amino acid, and has many gaps in this domain. Nonetheless, the rest of the sequence shows a very high homology with the chicken Sel I sequence. Moreover, it contains a Sec residue in its C-terminal domain. Therefore, although it cannot be confirmed that Sel i is found as a functional selenoprotein in the genome of Anas zonorhyncha due to the low-quality alignment of the N-terminus domain, it is highly probable that it is present, as the quality of the alignment for the rest of the sequence is very high. Possibly, the N-terminus domain of Anas zonorhyncha has undergone crucial changes during the speciation process, thus making it impossible for our program to align it with the corresponding domain of Gallus gallus. This incongruence may also be due to our automatization process, as our program may not have been able to align the N-terminus domain due to a low-sensibility when finding exons, for instance.
Seblastian was not able to predict neither the selenoprotein nor a SECIS element. It is possible that the selenoprotein was not predicted due to the changes in the N-terminus domain, as mentioned above. Regarding the SECIS element, it is important to mention that no SECIS element for SelI has been confirmed in Gallus gallus either [8]. Thus, the fact that no SECIS has been predicted for SelI does not imply that the protein is not found in the duck’s genome; on the contrary, it may be possible that many bird species have lost this SECIS element. All in all, SelI does not seem to be an ordinary selenoprotein, and further studies regarding its functions, synthesis and structure should be performed.
Selenoprotein K
Selenoprotein K (SelK) belongs to the same family as selenoprotein S (SelS). Although its function is not fully understood, some studies have suggested that it plays an important role in calcium flux control. It may also be involved in ER stress-related cellular apoptosis, as well as in ER-mediated degradation of many misfolded proteins [20].
We conclude that SelK is present in the genome of Anas zonorhyncha. The selected alignment for SelK shows a whole protein. It starts with methionine in its N-terminus domain and has a Sec residue close to its C-terminus domain. Additionally, Seblastian was able to predict our protein, even though its prediction did not include the first exon that was obtained with our program. Moreover, it predicted a valid, high-quality SECIS element.
Selenoprotein N
Selenoprotein N (SelN) is an endoplasmic reticulum resident protein, with its N-terminus domain facing the cytosol, and the rest of the protein inside the lumen of the ER. It is found in all vertebrates, although it may not be limited to vertebrates [21], as it is overexpressed in human fetal embryos but its expression strongly decreases during the differentiation of muscle cells into myotubes. To date, it is the only selenoprotein whose missing or malfunctioning causes inherited disorders in humans, all of them related to muscular dysfunction [22]. Several studies have shown that SelN absence causes an increased basal oxidative activity in myotubes in vitro [23].
We can confirm that SelN is present in Anas zonorhyncha. Our search has resulted in a full SelN protein that aligns almost perfectly with the sequence obtained from the Gallus gallus SelenoDB page, with the exception of a few amino acid changes that correspond to the natural changes during the speciation process. Seblastian was able to predict the selenoprotein as well as a high-quality SECIS element, which further strengthens our hypothesis that SelN is present in the eastern spot-billed duck’s genome. This high conservation of the protein between chicken and duck is not due to coincidence. As mentioned earlier, SelN is thought to play an essential role in muscle contraction and protection against oxidative stress in muscle fibers, and its absence leads to neuromuscular disorders.
Selenoprotein O
Selenoprotein O (SelO) is found in all vertebrates and it is the largest of the 25 selenoproteins described in the mammalian genome [5]. It may play an important role in preventing oxidative stress to damage the mitochondria by targeting a specific redox target protein [5]. Its exact function, however, remains unknown.
According to our results, SelO has been found in the genome of Anas zonorhyncha. However, the protein sequence obtained from the Gallus gallus selenoproteome in SelenoDB did not yield a satisfactory alignment. This may be due to the fact that most selenoproteins in SelenoDB are not correctly annotated, as it is a database built on an automatic program. In order to check whether another annotation could yield a better alignment, we ran the program using the Ensembl SelO sequence as our initial protein, as it started with Met, although it was a shorter protein. However, the Ensembl sequence did not include a Sec residue. In the SelenoDB SelO sequence, the Sec residue is found in the third-to-last position, and the last residue found in the sequence of Ensembl is the one prior to the Sec residue, possibly because the UGA codon that codifies for the Sec residue in SelO had been taken as a stop codon. Therefore, we added the 3 last residues (USS) to the Ensembl sequence before running the program.
Our results show a satisfactory alignment between our predicted SelO and the Ensembl sequence. Both the initial and the predicted protein start with a Met residue, and contain a Sec residue in the last exon. Seblastian was also able to predict our protein, though it included some major changes. It added two extra exons at the beginning of the sequence, and also added a few residues at the beginning of the third exon. Seblastian predicted this protein based on a SelO found in the genome of another duck species, Anas platyrhyncho, which is phylogenetically closer to our species than Gallus gallus. This predicted protein also starts with a Met residue. Additionally, Seblastian predicted a high-quality SECIS element. Therefore, we can confirm that SelO is found in the genome of Anas zonorhyncha.
Selenoprotein P
Seleno P is an extracellular secretory-transport selenoprotein that comprises 50% of the selenoprotein in plasma. Its function has been fairly conserved during the evolution, but its structure can have variations: even if it is common for them to have 13 selenocysteine residues and 2 SECIS elements, these numbers can vary between different species. In Gallus gallus, two SelenoP have been described: SelenoPa, which contains 13 selenocysteines and 12 of them are located in the C-terminal domain, and SelenoPb, which lacks the C-terminal domains of the original protein and as a result contains just one selenocysteine residue [6,7].
The chicken’s SelenoP extracted form selenoDB 2.0 had just one selenocysteine, which indicates that is the SelenoPb isoform. The alignment obtained was very accurate.
In order to know whether the equivalent to SelenoPa’s Gallus gallus exists in Anas zonorhyncha, we got a second query from SelenoDB 1.0: it was a human SelenoP with 13 selenocysteines. In addition, we considered it a good query as it started with a methionine. The alignment obtained was quite good considering how far these two species are in the phylogenetic scale and the predicted protein conserved the 12 selenocysteines.
Considering that the two queries used were quite different, both proteins predicted were very similar and the sequence positions almost matched, which indicates that they are probably codified by the same gene. However, while the exonerate program detected five exons in SelPa, just four were detected in SelPb. We have hypothesized that the last exon is the C-terminal domain and that SelPb does not present it due to post traductional modifications.
For both sequences, Seblastian predicted the same two SECIS and the same selenoprotein. We expected to obtain the same results for both sequences, as the fasta sequence introduced in Seblastian was exactly the same. It has 13 selenocysteines and five exons, so it matches with the SelenoPa protein. The reason why Seblastian could not predict the SelPb protein might be the great similarity between both proteins.
Actually, this great similarity could also have been the reason why our designed program found two different proteins when just one really exists. However, as Gallus gallus is the closest specie in which SelP has been properly described and both proteins have been found in its genome, it is reasonable to believe that both proteins, SelPa and SelPb, have been conserved in Anas zonorhyncha’s genome.
Selenoprotein S
SelS is a membrane-bound selenoprotein found in Gallus gallus which plays an important role in maintaining membrane integrity and has redox properties. It is from the same protein family as SelenoK, and it contains an unknown domain and usually a unique selenocysteine in the last five nucleotides of the C-terminal region, which is an isoelectric point whose function is stabilizing the protein. It has been described the importance of the separation between selenocysteine and its cysteine partner in the stability of the selenide sulfide bond. In Gallus gallus these residues are separated by 13 amino acids [8].
The alignment obtained when comparing our problem genome with the chicken's one is quite poor at the beginning of the sequence, although the rest of it has an almost perfect alignment and the overall values are enough to assume that the protein is present in Anas zonorhyncha. In addition, the correct prediction of SECIS and selenoprotein by Seblastian strengthen this statement.
We have hypothesized that the first part of the gene encodes for the unknown domain and so it might be a very variable and poor conserved domain. Nevertheless, this is not an explanation for the lack of the first methionine, and that is why even Seblastian predicted a SECIS element and the selenoprotein, we cannot confirm that SelenoS is present in Anas zonorhyncha's genome.
Selenoprotein T
Seleno T is a selenoprotein which is mainly localized in the ER and Golgi Complex. It is expressed both during embryonic development and adult tissues. Seleno T belongs to the Rdx family together with SelO, SelH, SelW and SelT, which are characterized for possessing a thioredoxin-like fold codified by a conserved C-XX-U motif. However, SelT has been found to have not one but two thioredoxin-like folds in Gallus gallus, containing the unique selenocysteine residue in the motif that encodes for the first one of them [6,8].
The alignment obtained is accurate and Seblastian predicted a valid SECIS in the sequence. Nevertheless, the tool was not able to predict a selenoprotein. It is clearly due to a Seblastian intrinsic issue and, as there are more evidences that strengthen the hypothesis of the presence of SelenoT in Anas zonorhyncha rather than evidences that strengthen its lack, we confirm the existence of this protein in our problem genome.
Selenoprotein U
SelenoU1 is a selenoprotein containing the common thioredoxin-like fold. It has been described to be located in the mitochondria[19].
The query we have used to find this protein in our genome is the chicken SelenoU1 transcript extracted from SelenoDB 2.0. The annotation seemed a valid one as it started by a methionine. After running the program, the alignment obtained was correct, so we proceed to analyse the predicted protein. It had three amino acids before the methionine start of the sequence, but when using the Seblastian tool, a known selenoprotein was predicted, which was exactly the same without those three amino acids.
We have decided to consider their presence a program or database mistake and we have erased them, doing the proper exon position modification afterwards. Seblastian has also predicted a valid SECIS element, so we conclude this protein is present in Anas zonorhyncha, and its hypothesized initial position is 50006.
Thiodoredoxin reductase family
Thioredoxin reductases 1-3 are selenoproteins located in both the mitochondria and the cytoplasm. Those proteins contain FAD/NAD(P) binding domains and its function is to catalyze the formation of thioredoxin from thioredoxin disulfide [6,26]. We have extracted the three TXNRD1-3 protein transcripts of chicken from SelenoDB 2.0.
We have generated a phylogenetic tree with Phylogeny.fr for the TXNRD family, in order to check whether each predicted protein is closer to its query than to the rest of the predictions and queries. The result is shown below:

As the tree shows, each predicted protein is closer to its initial query than to any other proteins of the family. Interestingly, TXNRD and TXNRD3 seem to be closer to each other than to GPx3.
- Thiodoredoxin reductase
We realized that the query obtained from SelenoDB was not well annotated, as it did not started by a methionine. We have searched for it in Ensembl, and we have found the same sequence but without the first 14 amino acids, which makes the sequence start with methionine.
This methionine residue and the rest of the beginning of the sequence appear in the query we used in the first place, although they are not in the same position. Therefore, it does not affect the alignment of our predicted protein, but it does affect the length of it. We have modified the start position of the first exon after erasing these 14 amino acids, so the new start is located 42 nucleotides away, in the position 56424.
All in all, the final alignment was correct, and a SECIS element and a selenoprotein was predicted by Seblastian. We can conclude this protein is present in Anas zonorhyncha.
- Thiodoredoxin reductase 2
Not only the obtained alignment was good, but the query, the protein predicted by the program and the protein predicted by Seblastian started with methione, which strengthens our statement that TXNRD2 exists in Anas zonorhyncha. Seblastian also predicted a SECIS element in this sequence.
- Thiodoredoxin reductase 3
The chicken query obtained from SelenoDB started by methionine and the alignment was almost perfect, but this first methionine was missing in the predicted protein. The SECIS element was correctly predicted by Seblastian, as well as the selenoprotein, which matched with our predicted one and included the first methionine that our predicted protein was missing. Therefore, we have assumed that is a program mistake not to include it in our predicted protein, and we have added it manually, modificating the start position of the first exon.
Selenoproteins machinery
Eukariotic elongation factor
eEFsec is a machinery protein related to the synthesis of selenoproteins that has a glutathione peroxidase function in the detoxification of hydrogen peroxide.
We have decided that the best alignment was the one with the sequence of the chicken from Ensembl, because the one with the SelenoDB sequence show two large gaps: one at the beginning and the other at the end of the sequence. However the chosen alignment also has a big gap at the beginning of the sequence which makes the protein not start with Methionine.
This gap could be due to either a good conservation of the sequence but a resulting non-functional protein or a change in the beginning of the sequence during evolution. Furthermore, it is also possible that this is caused by a low sensitivity of the exon search with the Exonerate, due to the big dimension of the gap.
Finally, as we have expected, neither the protein or any SECIS element was predicted by Seblastian since it is a machinery protein. Despite not having obtained the expected alignment, we confirm that the predicted protein is present in Anas zonorhyncha's genome, since it is an essential machinery protein for the synthesis of selenoproteins.
The final T-Coffee alignment can be found here.
Phosphoseryl-tRna kinase
Phosphoseryl-tRNA kinase (PSTK) is a highly conserved machinery protein, which implies an important function in biological processes. Concretely, its main role is to phosphorylate the precursor Serine located on the specific tRNAsec [10].
The alignment of the genome with the query was correct. However, we saw two gaps that could not be aligned with the query from Gallus gallus. In conclusion, we can assert that the predicted sequence is, in general, conserved but it has some regions that were lost during evolution. Another reason why we obtain 2 gaps in the alignment could be explained as a low sensitivity of the exon search with Exonerate.
PSTK from Anas zonorhyncha was not predicted by Seblastian. No SECIS element were found either. This result agrees with the expected findings, since it is a machinery protein, and these do not usually present any selenoproteins nor SECIS elements. Thus, we conclude that this machinery protein is found in the Eastern spot-billed duck.
Selenophospate synthetase
Selenophosphate synthase (SEPHS) is required for the synthesis of selenoproteins, so it is considered part of the selenium cell machinery. Its important role is the reason why it has been conserved in all prokaryotic and eukaryotic genomes encoding selenoproteins.
SEPHS is found in many species, but in some of them the selenocysteine has been replaced by a cysteine. Despite losing the selenocysteine residue, cysteine-containing homologs are expected to have the same function [10].
In SelenoDB 2.0, chicken SEPHS was a cysteine-containing homolog/selenium machinery gene with no selenocysteine residue. The annotation starts with a methionine and is practically identical as the one in Ensembl, so we used this sequence as the query for comparing our problem genome. After running our designed program, we have obtained a very good alignment that predicted a complete protein. We can conclude that SEPHS is present in the genome of Anas zonorhyncha.
As expected, no selenoprotein has been predicted by Seblastian, although the tool did predict a SECIS though. It might be a conserved region from the ancestor protein, which cointained a selenocysteine residue.
SECIS binding protein 2
The SECIS-binding protein 2 is a machinery protein which function is to bind itself to the SECIS element and, at the same time, to recruit the eEFsec [11].
The query of Gallus gallus did start with a methionine residue so we could run the program without making any modifications.
The alignment with the query was correct. Nevertheless, the predicted sequence did not start with a methionine residue. This could be interpreted as a good conservation of the sequence but a non-functional protein, since the vast majority of proteins need a methionine residue at the beginning of the sequence to start the translation. Also, this impossibility to align the beginning of the query with the predicted one, could happen because the initial fragment is too different in comparison with the reference query. It should also be mentioned that it exists a gap of alignment in the middle of the sequence, which could be explained as the loss of a part of a protein in Anas zonorhyncha.
Then, we proceeded to analyze the results with Seblastian. SBP2 from Anas zonorhyncha did not show any selenoprotein in this program. Nonetheless, one SECIS element could be predicted. Its position exhibits that it is located in the middle of the protein, so we discard the prediction, as SECIS elements are found in 3'UTR. Thus, we assume that Seblastian predicted this characteristical feature of selenoproteins from a SECIS-like structure. These findings are in accordance with the expected results, since most machinery proteins do not present neither selenoproteins nor SECIS.
All in all, although the alignment for this protein is not optimal, we conclude that the predicted SBP2 is present in the Anas zonorhyncha's genome, as it is a machinery protein essential for the synthesis of selenoproteins, some of which have been confirmed in this duck's genome.
Sec synthase
SecS is a machinery protein that plays a role in the addition of the phosphorylated selenium to the phosphoserine and produces the selenocysteine [10]. The query from Gallus gallus seems to be well annotated since the sequence starts with a methionine residue.
The alignment of Anas zonorhyncha's genome with the query was correct so we conclude that SecS protein is found in the Eastern spot-billed duck.
Seblastian program was not able to predict a selenoprotein. Nonetheless, it predicted a SECIS element, which was located in 5'UTR. Thus, we assume that the program made an error when predicting this characteristical feature from selenoproteins, as SecS is a machinery protein that does not need a SECIS element in its sequence.
SECp43
SECp43 is a machinery protein that, together with SEPHS1 and SEPHS2, binds the selenophosphate group to the Cys residue during the synthesis of selenoproteins [10].
The query of Gallus gallus did not start with a methionine. Nevertheless, the methionine was located four residues away from the beginning of the sequence. After comparing the sequence from Ensembl with the one from SelenoDB, we confirmed that the sequence from Ensembl was identical, although it was missing these first four amino acids. Therefore, we chose the query from Ensembl as the optimal sequence. The alignment of the predicted protein with the query was optimal, but the exon started twelve nucleotides earlier (position 44919 instead of 44931).
Regarding Seblastian, results showed no prediction of selenoproteins. However, 3 different SECIS elements were found. Two of these SECIS elements were discarded since they were located in the opposite strand, but one of the SECIS elements was selected as suitable. Since SECp43 is a machinery protein, no SECIS nor selenoproteins were expected. This prediction of the SECIS elements could be due to the sequence having a SECIS-like sequence. Thus, we confirm the presence of SECp43 in Anas zonorhyncha.
The final T-Coffee alignment can be found here.