 With
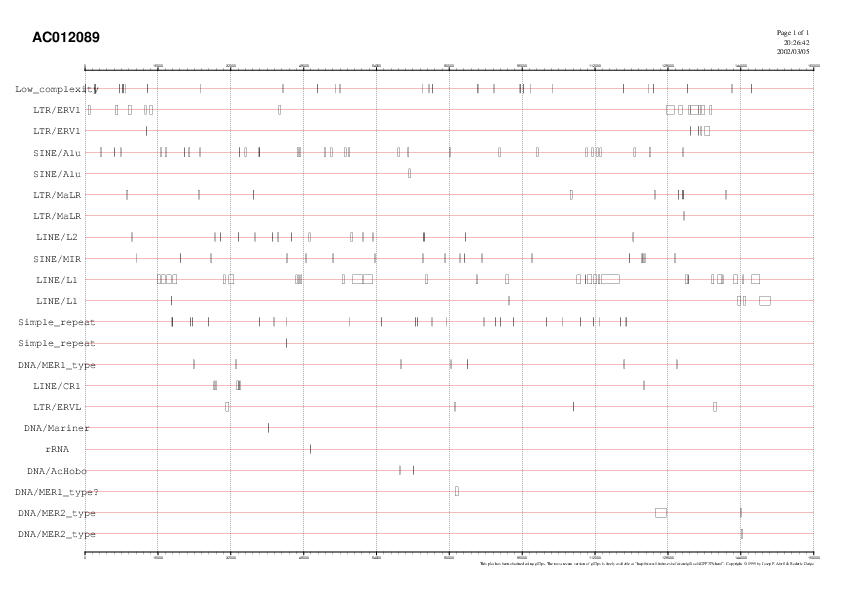
RepeatMasker a graphic of repeat elements was drawn (fig. 1).
With
RepeatMasker a graphic of repeat elements was drawn (fig. 1).
Fig. 1. It is important to highlight
the presence of three LINE1 clusters along the sequence (around nucleotide
number 18,000, number 62,000, and between nucleotides 110,000-120,000).
Alus are spred uniformly over all the sequence, and LTRs are mostly concentrated
at the end of the DNA fragment (28,000-40,000 nucleotides).
After having predicted the genes,
a variety of exons is obtained. For a start, GeneMark and Grail do not
predict exact genes, but exons with no connection. Moreover, the predictions
made by GeneMark have nothing to do with the ones solved by the other three
programs (see fig. 2). GeneId and Genscan offer possibilities of exons/genes
that support each other. Three of them appear to match, though they differ
in the number of genes.
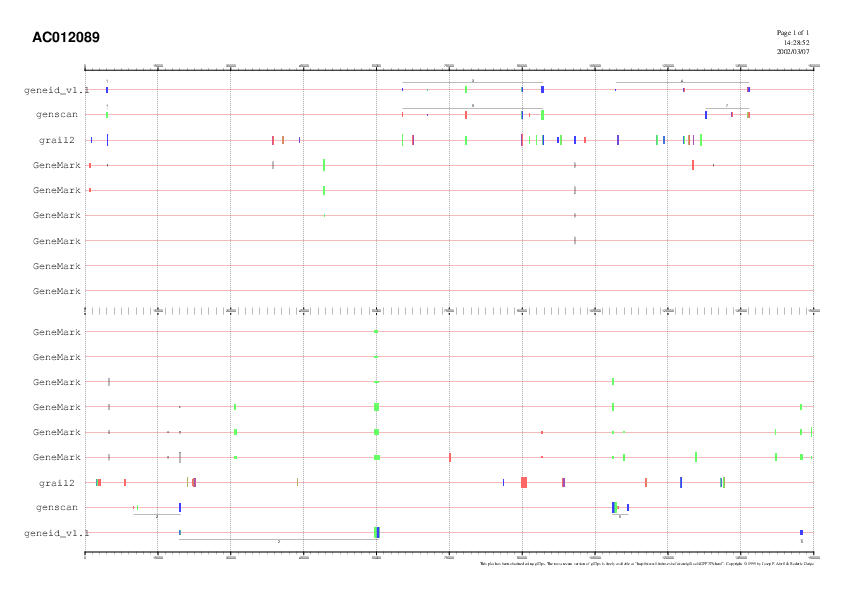 Fig.
2. GeneId, Genscan, GeneMark and Grail gene prediction. The first exon
that both of them predict is located at the very begining of the sequence,
and it is quite isolated from the others; the two programs agree that it
as a terminal exon. The predicting programes differ in the second gene:
the terminal exon for Genscan becomes the first exon for GeneId. However,
it is consensus that this gene is coded reversely. According to Genscan
results, there are two genes that do not appear in the graphic but in the
table: the third and the fourth, both reversed and nearly touching each
other (table Genscan). The third gene
predicted by GeneId is equivalent to the fifth predicted by Genscan. The
only difference is the existence of an internal exon according to Genscan.
This last program predicts another gene, reversed, with a quite strong
terminal exon. The next gene matches again in both programmes, but, although
it ends at the same level, Genscan's prediction considers it half long
than GeneId's. There is another exon according to GeneId at the end of
the sequence, placed reversely.
Fig.
2. GeneId, Genscan, GeneMark and Grail gene prediction. The first exon
that both of them predict is located at the very begining of the sequence,
and it is quite isolated from the others; the two programs agree that it
as a terminal exon. The predicting programes differ in the second gene:
the terminal exon for Genscan becomes the first exon for GeneId. However,
it is consensus that this gene is coded reversely. According to Genscan
results, there are two genes that do not appear in the graphic but in the
table: the third and the fourth, both reversed and nearly touching each
other (table Genscan). The third gene
predicted by GeneId is equivalent to the fifth predicted by Genscan. The
only difference is the existence of an internal exon according to Genscan.
This last program predicts another gene, reversed, with a quite strong
terminal exon. The next gene matches again in both programmes, but, although
it ends at the same level, Genscan's prediction considers it half long
than GeneId's. There is another exon according to GeneId at the end of
the sequence, placed reversely.
Only a few ESTs were found when
running a Megablast against Human ESTs (see fig. 3). All of them validate
the same gene: the third according to GeneId and the fifth for Genscan.
As there were no ESTs to confirm the other genes, their sequences were
blasted directly against GenBank's database. The first gene -the
terminal exon at the beginning of the sequence- was excluded since it was
only a fragment.
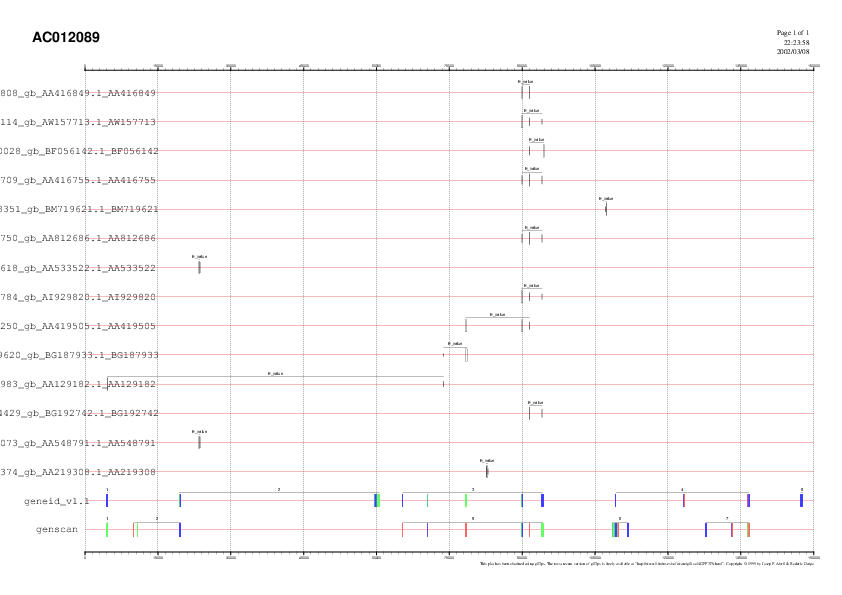
Fig. 3. Image showing the ESTs
found and the genepredictions for Genscan and GeneId.
After translating to protein the second gene predicted by Genscan and doing some searches withBlastp, it was found that all the resulting proteins were homologous with fragments of reverse transcriptase from LINE1. No matches were found using a protein domain database. Similar results were obtained with the second gene predicted by Geneid.
The Blast run after the isolation of the region containing the supposed third gene returned the hypothetical protein FLJ22419 (ID FLJ22419 in GenBank) and a protein similar to the hypothetical protein (ID LOC131464 in GenBank). The database InterPro was used to find patterns in the proteins predicted by GeneId, Genescan and FLJ22419. The results are shown in figure number 3: the three proteins have one, two and three Zn finger, respectively. The sequence of the protein was used again to find the exact sites for the exons that codify for it (Blast 2 sequence). The mRNA sequence of the FLJ22419 protein was obtained from the entry in GenBank and then it was used to run another Blast. Another BAC sequence was obtained. The new sequence was found to match with the begining of the mRNA of the protein FLJ22419. The next step was to find out if the sequence of the two BACs matched each other in order to inquire if there was overlapping. The results showed alignment between the two strands in both senses, forward and reverse.
The mRNA sequence of the hypothetical
protein FLJ22419 was blasted to the sequences of the two BACs independently.
The results desestimated the prestablished hypothesis (results).
As a consequence it was considered that the previous alignments were due
to the abundance of repetitive regions in both sequences.
A Blast using FLJ22419 mRNA to the human genome sequence revealed the existence of eight exons and seven introns (fig. 4). The five last exons were found to belong to the BAC RP11-758R3, placed between the nucleotide 25,320,000 and the nuclotide 25,467,000 in the third human chromosome. On the other hand, the first two exons were placed in the other BAC, RP11-598P2, between the nucleotides 25,746,000 and 25,596,000, reverse position, in chromosome number three. The third exon was somewhere between the nucleotides 25,467,000 and 25,596,000 in the same chromosome, region located between the two BACs. The exact location of the BACs in the region 3p24.1 in the third chromosome was obtained by running a Blast against the human genome.
Fig. 4 Map of FLJ22419
protein
The fourth gene, predicted
by GeneId, is a forward one about 27,000 bp which contains three exons.
The Blastp
results showed homology between the protein and three common envelope proteins
from HERV-H retroviruses: HERV-H/env62, HERV-H/env60, and HERV-H/env59.
However, when it was analyzed by Interpro no matches were found. As the
seventhgene
predicted by Genscan is located inside the fourth gene region predicted
by Geneid, the results are exactly the same in both cases.
Finally, the sixth gene, predicted by Genscan and located in reverse strand, contains three exons, a promoter region and a polyadenilation signal. It is about 17,000 bp long and Interpro found an homology with an homeobox domain. In addition, using Blastp we found an homologous haemopoietic progenitor homeobox protein, called VENTX2. This protein is expressed in bone marrow and is located in chromosome number 10. Comparisons between the VENTX2 gene and its mRNA with the BAC and the protein, using Blastx and Clustalw, were made in order to guess the origin of this match.
Tblastx was used to determine if all the predicted genes were functional in other organisms. The only similarities obtained were with DNA clones in different chromosomes from Homo Sapiens.