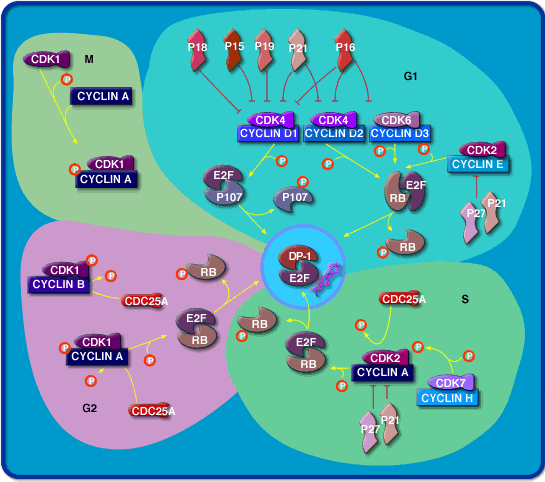
Through the cell cycle: cyclin dependent kinases
To get further information about this web page, contact us:
mariajose.escriva01@campus.upf.edu
susana.gomis01@campus.upf.edu
1. Introduction to the Cdks:The aim of our study
2.Analysis of the protein sequence
3.Phylogenetic distribution of the protein subject of our study2.1 Identification of the family members.
2.2 Alignment of the homologue sequences and phylogenetic evolution of the members.
2.3 Characterization of the consensus patterns reported in the protein family.
2.4 Proteins interacting with Cdk2
2.5 Common Blocks in the human Cdk's family
4.Analysis of the genes encoding the protein we study
4.1 Genomic location of the Cdk2 gene
4.2 Exonic compositions of the Cdk2 gene
4.3 Looking for undiscovered members of the family ...
4.4 Transcription of the Cdk2 gene: its promoter
4.5 Study of the Cdk2 splicing variants
4.6 SNPs analysis
5.1 Structure presentation
5.2 Protein domains and phosphorylation sites
In
the yeasts S. pombe and S. cerevisiae, all cell cycle events are controlled
by a single essential Cdk, which are named as cdc2 and csc28 respectively.
Cell cycle events in multicellular eukaryotes are controlled by two Cdks,
known as Cdk1 and Cdk2, which operate primarily in M phase and S phase,
respectively. Animal cells also contain two Cdks (Cdk4 and Cdk6) that are
not essential components of the core cell cycle control system but are
important regulators of entry into and exit from the cell cycle in G1.
Cdk
function
has been remarkably well conserved during evolution. It is possible,
for example, for yeast cells to divide normally when their cdc2 / cdc28
gene is replaced with the human Cdk1 gene. This and other evidence clearly
illustrates that Cdk function, and thus the function of the cell cycle
control system, has remained fundamentally unchanged over hundreds of millions
of years of eukaryotic evolution.
The
aim of our study is to understand the relationships among the different
Cdk members, both among the human but also different species along the
evolutionary trend. Taking as a starting point the human Cdk2, we will
first study the human homologues, and later on, the homology existent in
further well characterized species. the most part of the study will be
perform upon the protein sequences, due to the higher functional constrictions
that occur on them. Furthermore, we will also study the location
of the human homologues within the whole human genome, the splicing variants
and a few topics about the 3d structure of the Cdk2 as a model of the whole
family
Click in this link
to visit the NCBI entrance
to the original protein sequence that we used in order to start our
work or here
to access to the FASTA file.
2.1
Identification of the family members.
In these initial
studies, we just considered the human proteins which clearly belong to
the family. We set the appearance of false positive results at the blast
search around the following E-values:
1.
The first human protein without Cdk function is KPT1_HUMAN Q00536
a SER/THR–PROTEIN KINASE from the PCTAIRE family, which shows to
have an E-value= 2e-86 with a score = 317 in our blast search.
2.
The last human protein which belongs to the Cdk family is CDK8_HUMAN
E-value=
2e-47 score = 178.
3.
In addition, we also located the first non human protein without Cdk
function, which is called CC2H_DICDI P34117 CDC2-LIKE SER/THR–PROTEIN
KINASE CRP. Its E-value = e-111 and presents an score = 298. (further information
about the phylogenetic distribution of the Cdk family protein in the second
part of our work).
2.2 Alignment
of the homologue sequences and phylogenetic evolution of the human members.
The
most representative sequences selected from the protein blast search, allowed
us to see that the family of human Cdk is formed by 10 homologous proteins
which have different roles during the cell cycle.
The
next step was, to perform the alignment of these ten sequences named above.
The multiple alignment of the human protein sequences was done with clustalW
- note that we used the GONNET series matrices to generate the multiple
sequence alignment. These matrices were derived using almost the same procedure
as the PAM matrices (Dayhoff) but are much more up to date and are based
on a far larger data set. They appear to be more sensitive than the Dayhoff
series and therefore, we selected them as the most adequate for our porpoise.
The
results
of this alignment show the most conserved residues present in the proteins.
Based on the results
of the clustalW alignment, we built a neighbour phylogeny
tree with the 10 homologue sequences found. This tree was done with
PHYLIP tools. We used the program Neighbour in order to build the tree,
and then, further work was done to evaluate the strength of the branches
in it. The results of the bootstrap analysis do not support the ones obtained
with Neighbour. Nevertheless, we suggest bootstrap as the most trustworthy
analysis (click here to see the resulting results
and tree).
The
consensus patterns for these family sequences were searched in the
InterPro
page. Surprisingly, there are no specific consensus for the members of
the Cdk family. However, two consensus sequences were obtained:
[LIVMFYC]-x-[HY]-x-D-[LIVMFY]-K-x(2)-N-[LIVMFYCT](3).
To see which proteins
were interacting with the Cdk2, we consulted the Database
of interacting proteins. (Click here to
consult the results and the experiments done in order to confirm such an
interaction).
2.5 Common Blocks
in the human Cdk's family
The results
of this search reported us a list of common sequences in our 10 members
Cdk family which can be used to identify them as an homologous group.
3. PHYLOGENETIC
DISTRIBUTION OF THE PROTEIN SUBJECT OF OUR STUDY In
order to find out the presence of our protein family among the biological
kingdoms, we performed a blast search against
the reference organisms whose genome have been already sequenced (or
with organisms from which we could access to the draft sequence of their
genome). As we did before with the human sequences, the alignment
was performed taking in account those showing the greater E-values. The tree clearly
shows the existence of a higher homology in each Cdk type, beyond the taxonomic
differences. Thus, the Cdks present in many of our organisms of study are
more similar among them, than two given Cdks in a determinate organism.
Therefore, we claim the existence of a high conserved orthologous homology
among the Cdk family. 4. ANALYSIS OF
THE GENES ENCODING THE PROTEIN THAT WE STUDY 4.1 Genomic location
of the Cdk2 gene
4.2 Exonic compositions
of the Cdk2 gene
We next determined
the exons composition of the gene. With this objective we performed a search
in the Human Genome Project Working Draft.
The results of this search showed us the predictions of the introns and
exons present in our sequence. Acembly,
Ensembl,
Fgenesg++
and Genscan, were used
to develop these predictions. An alignment of the
different programs predictions is attached. All of them display the
same 7 exons distribution which is, in fact, supported by the already known
genes reported in RefSeq
from NCBI. Only the Ensembl prediction presents one more exon. Which can
in this conditions be excluded.
Moreover we run the
Geneid.
This results
(see results plot) were compared with
the previously linked alignment in order to obtain further conclusions.
As it can be seen, Geneid identifies only 6 from the 7 real exons from
Cdk2_human gene.
A slightly comparison
of the exon distribution among the different homologues present in human
and yeast show that exon I is longer in CDK2 than in the characterized
CDC2 genes and is conserved in X. laevis Cdk2. Other differences between
the CDK2 and the CDC 2 gene structure include two additional introns located
at amino acids 105 and 196 of the human CDK2 gene that are not present
in the Sacchromyces pombe CDC2 gene.
4.3 Looking for
undiscovered members of the family ...
We finally tried
to answer the question: would it be possible that further members of this
family have not been yet characterized?
Once located the
Cdk2 gene in the human genome, we looked for homologous genes to the Cdk2
in the hole genome draft. The results showed us many significant alignments.
If the identification number (example with the first sequence obtained
AC025162.31.80081.104328) of this DNA sequences is clicked, you will be
directly delivered to the respective alignments page where the score of
the different fragments can be evaluated. Once there, by clicking again
in the same ID number, you will be shown the complete information known
about the homologous fragments obtained in the search as exactly location
of the DNA region, it's gene content, the identified RNAs and the proteins
translated.
In order to view
quickly all these results, consult the table
which sums up the most relevant information obtained in e!
Ensembl Human 4.4 Transcription
of the Cdk2 gene: its promoter
According to the
data previously reported in the literature, five transcription start sites
- spread over a 72-bp upstream region- were identified.
However, no consensus
TATA box was identified in the entire upstream sequence. Thus, this
promoter
falls into the category of TATA-less promoters similar to all other cell
cycle genes analyzed to date including: cdc2, cyclin A, cyclin D1, cyclin
D2 and cyclin D3, as well as Xenopus laevis Cdk2.
A YY1 box, which
in some TATA-less promoters is responsible for determining the transcription
start site, is present just upstream from the three start sites located
at positions +1, -5, and -9.
An Sp1 site was
identified upstream of each of the remaining transcription start sites
(-33 and -71), suggesting that these Sp1 regions may be responsible for
localizing the start of transcription at these sites . In addition, other
putative transcription factor binding sites were also identified, such
as a c-Myb binding site, or two putative p53 binding sites.
It is perplexing to assume that p53 would induce CDK2 since this induction
would most likely result in an accelerated cell cycle rather than a cell
cycle arrest.
4.5 Study
of the Cdk2 splicing variants As it can be observed,
the fragment missing in the second splicing variant corresponds to the
nucleotides 722 to 824 which codify for 34 aminoacid. This peptide sequence
was translated using ORF
Finder from NCBI to get the corresponding peptide chain:
H
H L S K D A A Q A
P D V H S C G I I F
A A Q E D F R S S
V P Q G H Once known the difference
in the aminoacid sequence, our intention was to locate these 34 aminoacid
within the 3-D structure of the protein ( human Cdk2) in order to know
in which domain was contained. Unfortunately, all the crystallography experiments
available in PDB have been done with the Variant2, and therefore our work
was frustrated.
However, we followed
this goal with news techniques. Below, a representation of the genomic
DNA and the isolated mRNAs overlapping regions are shown. This way, we
located this splicing variant in the 5th exon.
4.6 SNPs analysis Looking at these
results we conclude it could be possible that some genes - or pseudogenes
- belonging to this family haven't been identified yet. Only the progression
of the research on genes isolation and characterization will allow these
theoretical genes (or high homology sequences) to be accepted or refused. All NCBI SNPs in
CDK2
The SNPs distribution,
as expected shows a higher presence of single nucleotide polymorphisms
in the non coding / non translated regions than in the regions coding for
the protein. But we should consider the contig composition.
5.1 Structure
presentation
We compared,
manually, the regions of less similarity from the Clustalw run with all
the human Cdks and the the secondary structure from the protein. We could
appreciate that, as expected, the regions showing a greater divergence
level were located in the zones in between the alpha helix and the beta
sheets, were variations are allowed without representing a lose of activity.
5.2 Protein domains
and phosphorylation sites
This enzyme is formed
by two domains. A N-terminal beta sheet domain
where the phosphorylation activity is located and a Cterminal multi-helix
domain with kinase activity.
Cdk2 requires association
with cyclins and phosphorylation
by CAK at Thr-14 and Tyr-15 for being active. The inactivation
of the enzyme is done by the phosphorylation of the Thr-160
site. All these sites can be observed in an amplified
version of the previous picture.
[LIV]-G-{P}-G-{P}-[FYWMGSTNH]-[SGA]-{PW}-[LIVCAT]-{PD}-x-[GSTACLIVMFY]-x(5,18)-[LIVMFYWCSTAR]-[AIVP]-[LIVMFAGCKR]-K.
The position
of these consensus patterns in the tyrosine kinase domain of the human
Cdk2 was studied by manual direct observations and we could check their
presence in the aligned sequences of the hole human Cdk family.
Finally, we show
here the consensus of the phosphorylation sites of Cdk1 and Cdk2, which
are typically defined as S/T*-P-X-K/R, where 'S/T*' indicates the phosphorylated
serine or threonine, X represents any amino acid, and 'K/R' represents
the basic amino acids lysine or arginine.
NCBI allowed us
to find out the location of the Cdk2 human gene in the 12th chromosome.
Concretely in between the bands 12q13.11 and 12q13.12. SeeMaster
map genes in cr.12.
The FASTA files Variant1
and Variant2 from the mRNAs where used to perform
a Clustalw alignment in order to be able to see their common and the singular
fragments. Results are attached.
Small
nucleotide differences in exons 1,5,6 and 7 can be displayed at the
Evidence Viewer site.
Most
of the results obtained correspondent with DNA zones where no gen belonging
to the Cdk family was identified. Some homologous fragments to the Cdk2
sequence where found in fragments where no genes at all had been described.
Two genes (Cdk1 and Cdk4), already known to belong to the family did not
appear in the results when the search was performed with the standard options.
Two other genes appeared showing great homology, CdkL2 and CdkL3. This
made us realize there are 3 more members - Cdkl1, CdkL2 and CdkL3 - belonging
to the Cdk's family which we hadn't take in account. The hole of
this work will be done without including these in the searches.
* Lower case
letters indicate repetitive or low-complexity sequence
Genomic Data
Transcription
Data
SNP ID
Contig Accession
Position in Contig
Orientation
5' Flanking Sequence*
Nucleotide Change
3' Flanking Sequence*
Validation
Type
mRNA Accession
Protein Accession
ORF
Position in Protein
Amino Acid Change
rs2069413
NT_009458.6
1040319
reverse
TCTTCCAGGATGTGA
C/G
CAAGCCAGTACCCCA
+
coding-nonsynon
XM_049150
XP_049150
2
200
T/S
rs2069406
NT_009458.6
1042343
reverse
CTGCATCTTTGCTGA
G/A
ATGGTATGGAGGCTT
+
coding-synon
XM_049150
XP_049150
3
105
E/E
rs2069400
NT_009458.6
1044292
reverse
TTCACCTTCACCAGG
C/A
GGCTTTACTTACCTA
+
mrna-utr
XM_049150
--
--
--
--
rs2069401
NT_009458.6
1044233
reverse
TAATATATCCAAAAA
C/T
CACACCCTGACTACC
+
mrna-utr
XM_049150
--
--
--
--
rs2069397
NT_009458.6
1044952
reverse
GGGTTCCCAGGCCCC
C/A
GCTCCAGGGCCGGGC
+
mrna-utr
XM_049150
--
--
--
--
rs2069398
NT_009458.6
1044831
reverse
CAAGTTGACGGGAGA
G/A
GTGGTGGCGCTTAAG
+
mrna-utr
XM_049150
--
--
--
--
rs2069399
NT_009458.6
1044336
reverse
GGGGCTACTCCTGCA
T/C
TTTTTCCCCTCCATT
+
mrna-utr
XM_049150
--
--
--
--
rs2069402
NT_009458.6
1043355
reverse
TAACAAACATTTTTT
C/T
AATGCACAGGATGTA
+
intron
XM_049150
XP_049150
--
--
--
rs2069403
NT_009458.6
1042757
reverse
ttgggaggctgaggt
G/A
ggaggatcacttgag
+
intron
XM_049150
XP_049150
--
--
--
rs2069404
NT_009458.6
1042709
reverse
atcatgggcaacata
G/A
cgagaccccatctct
+
intron
XM_049150
XP_049150
--
--
--
SNPs
in
NT_009458.6
Length
SNPs presence
Normalized values
SNPs in introns
and utr mRNA
73,45%
80%
1.0891
SNPs in coding regions
26,55%
20%
0.7533
For further illustration
of the Cdk2 (variant2) protein structure, the secondary
and1B39 tertiary
( PDB) structure of the protein
can be observed. (The magnesium ion present in the structure
is due to the crystalization conditions of 2.5 mM ATPand 5 mM MgCl2).