Selenoproteins
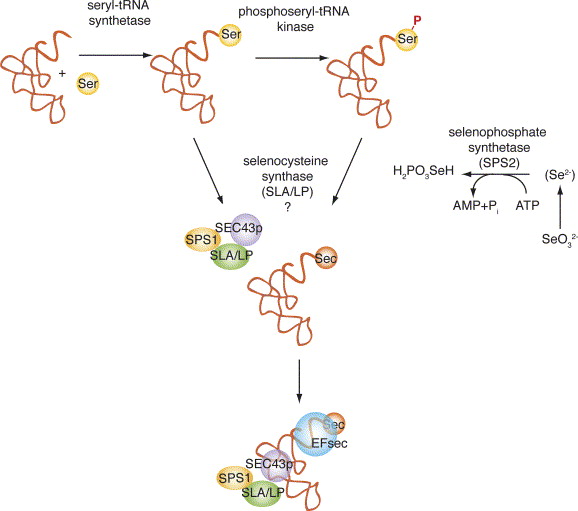
The selenium is a non-metal element considered a micronutrient with several health benefits (1). It is an essential component of several metabolic pathways, such as: thyroid hormona metabolism, antioxidant defence system or immune function (2), as a component of selenium-containing proteins. This element is incorporated to the organism through water and food as selenite and selenate. These compounds need to be transformed to selenide, because is the selenium source used for selenocysteine (Sec) biosythesis, the transformation can be done by glutathione-glutaredoxin or thioredoxin systems. Besides, the selenide can be also obtained from dietary selenomethionine and selenocysteine via lyase system. The selenoprotein selenophosphate synthetase 2 (SPS2) transforms the selenide into monoselenophosphate (3), the active selenium donor. The Sec is synthesized on its own tRNA by using serine (Ser) as an intermediate. The synthesis is initiated with the attachment of Ser to the tRNASec by seryl tRNA synthetase. Then, the phosphoseryl tRNA kinase (PSTK) phosphorylates the complex and the Sec synthase (SecS) produces Sec-tRNASec, due to the incorporation of monoselenophosphate (3). Finally, when charged this tRNASec can carry the amino acid Sec (4).
Allmang C, Krol A. Selenoprotein synthesis: UGA does not end the story. Biochimie. 2006;88(11):1561-1571.
The Sec is the 21st amino acid (5) and its structure is nearly identical to cysteine (Cys), as exception of a selenium instead of a sulfur atom (6). This amino acid is enconded by UGA codon, which signalize the termination of protein synthesis in nonselenoprotein genes (1, 7), but a specific structural signal in the mRNA leads to the incorporation of Sec and thus, the evasion of the STOP codon (5). Besides, is inserted in the polypeptide chains of selenoproteins by the tRNASec, which has an anti-codon complementary to UGA and therefore controls the expression of the entire selenoprotein family (7). The Sec-containing proteins have oxidoreductase functions in a variety of organisms in all three domains of life, in which Sec is found in the active site (1).
Selenoproteins biosynthesis
Selenoprotein synthesis is an evolutionary conserved process. Nevertheless, some major differences are found in this process between the three domains (Bacteria, Archaea and Eukarya). The common features to all organisms are: the specific tRNA codon, the Sec insertion sequence (SECIS elements) and several protein factors (7). The SECIS elements are conserved secondary structures found in 3' untranslated region of selenoproteins mRNAs and are required for recognition of UGA codons as Sec (8). Furthermore, Sec insertion depends on two types of factors: cis-acting elements and trans-acting elements.
On one hand, the selenoprotein mRNA has to contain cis-acting elements such as UGA codon and a stem–loop structure (SECIS element) in the 3'-UTR region. Because this elements allows the UGA decoding and Sec insertion (9).
On the other hand, the trans-acting elements are the structures that are not found directly bind mRNA sequence. The biosynthesis of selenoproteins requires the SECIS binding protein 2 (SBP2) binding to the stem region of SECIS element and the recruitment of selenoprotein-specific elongation factor (eEFSec). The eEFSec recruits tRNASec and mediates its ribosome incorporation (6, 9). In conclusion, the combination of these factors on selenoprotein mRNAs in the nucleus leads to the UGA decoding and insertion of Sec (7).
Selenoproteins families
Selenoproteins can be grouped in different families depending on their function and structure.
Although selenoproteins have been found in bacteria, archea and eukarya, not all the species of this three domains contain selenoproteins (1).
Sel15 Family
Sel15 is a 15-kDA thioredoxin like selenoprotein. It is located in endoplasmic reticulum (ER) and it’s involved in the quality control of glycoprotein folding through its interactions with UDP-glucose-glycoprotein glucosyltransferase (UGT). Sel15 may assist UGT function and control folding or secretion of certain glycoproteins.
It has an oxireductase domain and the selenocysteine is located in the catalytic position.(10).
Glutathione Peroxidases (GPxs)
The GPxs were the first selenoproteins described.
GPxs are selenoenzymes that have a key role in protecting the organism against the oxidative damage of hydroperoxides. It is in charged of catalyzing the reduction of hydrogen peroxide, therefor its elimination producing glutathione disulfide and water (2, 11, 12).
GPx2 is located in gastrointestinal tract. It protects mammals from the toxicity of ingested lipid hydroperoxides. It is the important selenoprotein antioxidant in the colon.
GPx3 is found in the extracellular space. It has a specific antioxidant function in renal tubules. It is also found in plasma.
GPx4 is found in multiple tissues and cell types. This protein is responsible for the reductive destruction of lipid hydroperoxides. It has also been shown that it is implicated in the regulation of protein tyrosine phosphatases.
GPx7 and GPx8 evolved from a GPx4-like selenoprotein prior to the separation of mammals and fishes.
Deiodinases (DIO)
The deiodinases are responsible for the activation/inactivation of thyroid hormones (12). There are 3 different selenoproteins and are found in all vertebrates.
DIO1 is expressed predominantly in the liver and kidney and provides most of the circulating T3. The activation reaction involves the conversion of the prohormone T4, secreted by the thyroid gland, to the active one,T3.
DIO2is found in the ER and activates the thyroid hormone by deiodination of the outer tyrosyl ring. It has in its mRNA a second in-frame UGA codon. This can insert Sec when the first UGA codon is mutated.
DIO3 irreversibly inactivates the thyroid hormone by deiodination of the inner tyrosyl ring (13).
Methionine Sulfoxide Reductase A (MsrA)
This gene encodes an ubiquitous protein that carries out the enzymatic reduction of methionine sulfoxide to methionine. There is higher expression in kidney and nervous system (13).
Selenoprotein H
This protein is located in the nucleus. It is an oxidoreductase and it protects neurons against UVB-induced damage by inhibiting apoptotic cell death pathways, promote mithocondrial biogenensis and mitochondrial function and supress cellular senescence through genome maintenance and redox regulation (13).
Selenoprotein I
Selenoprotein I is a transmembrane protein that belongs to the CDP-alcohol phosphatidyltransferase class I family. It catalyzes the transfer of phosphoethanolamine from CDP-ethanolamine to diacylglycerol to produce phosphatidylethanolamine, which is involved in the formation and maintenance of vesicular membranes, regulation of lipid metabolism, and protein folding (13).
Selenoprotein K
Selenoprotein K is a transmembrane protein localized in the ER. It is involved in ER-associated degradation of mislfolded glycosylated proteins. It has a role in the protection of cells from ER stress-induced apoptosis too (13).
Selenoprotein M
It belongs to the selenoprotein M/Sel 15 family. It may have a role in maintaning redox homeostasis, but the exact function is not known. It is localized in the perinuclear region and it’s highly expressed in brain (13).
Selenoprotein N
This protein is important for cell protection against oxidative stress and in the regulaton of redox-related calcium homeostasis. it is located in the endoplasmic reticulum (13).
Selenoprotein O
The exact function of this protein is not known, but it is related with redox family. It is predicted to have kinase activity. It is a mitochondrial protein and its deficiency plays roles in osteoarthropathy (14).
We found two selenoproteins from this family: SELENOO 1 and SELENOO 2.
Selenoprotein P
Selenoprotein P it’s located in plasma. Its function is related with selenium transport and oxidant defense.
When selenium supply is limited, selenoprotein P synthesis have priority over glutathione peroxidase synthesis (15).
Selenoprotein R
No known function, but it has been related with methionine sulfoxide reduction pathway. It has also been said that have a role in protection against oxidative stress and redox regulation of cellular processes. This protein is related with MsrA. It has zinc (16).
Selenoprotein S
This protein is a transmembrane protein localized in the endoplasmic reticulum. It is involved in the degradation process of misfolded proteins in the ER, and may also have some function related with inflammation control (13).
Selenoprotein T
This protein is localized in the endoplasmic reticulum. It belongs to the SelWTH family that possesses a thioredoxin-like fold and a conserved CxxU (C is cysteine, U is Sec) motif, suggesting a redox function for this gene (13).
Selenoprotein U
The protein encoded by this gene belongs to a subfamily of FAM213A/selenoprotein U (SelU), within a peroxiredoxin-like FAM213 superfamily. SELENOU is unusual because it has restricted phylogenetic distribution in vertebrates, such as in chicken and fish. Other vertebrate members of this family, including mammals, contain cysteine-containing homologues. SelU proteins contain UxxC (U for selenocysteine and C for cysteine) motif in place of the catalytic CxxC motif found in cysteine-containing homologues. The latter function as redox regulatory proteins, suggesting a similar role for SelU proteins (13).
TRXR
TRXR plays an important role in a variety of biological functions such as DNA synthesis. (Ref 2) = Thioredoxin reductase (TR) is a recently identified seleno-cysteine containing
enzyme which catalyzes the NADPH dependent reduction of thioredoxin and
therefore plays a regulatory role in its metabolic activity (15).
Selenoproteins Machinery families
Eukaryotic Elongation Factor (eEFSec)
EEFsec is responsible for recruiting tRNA [Ser]Sec and inserts Sec into nascent protein chains in response to UGA codons.
This protein has GTPas activity and it has high specificity for aminoacylated tRNA[Ser]Sec and does not bind phosphoseryl-tRNA[Ser]Sec or other aminoacylated tRNAs.
It has been shown to form a compex with SBP2 (17).
Phosphoseryl-tRNA Kinase (PSTK)
This protein phosphorylatess the seryl.tRNA [Ser]Sec. (selenoprot: molecualr pathways)
It seems to be resoposible for the tRNA discrimination, as it phosphorylates Ser-tRNASec but not Ser-tRNASer. Thus, the mechanism by which PSTK discriminates Ser-tRNASec from Ser-tRNASer still remained elusive.
PSTK may recognize the length of the long acceptor stem of tRNA sec.
This protein is formed by two independent subunits (18).
SECIS binding protein 2 (SBP2)
It is one of the essential components of the machinery involved in co-translational insertion of selenocysteine into selenoproteins. Mutations on this gene have showed a reduced activity of DIO 2, and consequently, abnormal thyroid hormone metabolism. (13)
Selenocysteine synthase (SecS)
The amino acid selenocysteine is the only amino acid that does not have its own tRNA synthetase. Instead, this amino acid is synthesized on its cognate tRNA in a three step process. The protein encoded by this gene catalyzes the third step in the process, the conversion of O-phosphoseryl-tRNA(Sec) to selenocysteinyl-tRNA(Sec) (13).
Selenophosphate synthetase (SEPHS)
Selenophosphate synthetase is necessary for the incorporation of selenocysteine into selenoproteins. This enzyme catalyzes the reaction that produces monoselenium phosphate, a selenium donor in biological reactions.
Two forms have been identified, SEPHS1 and SEPHS2 but only SEPHS2 is a selenoprotein (14).
tRNA Sec 1 associated protein 1 (SECp43)
SECp43 has two ribonucleoprotein-binding domains (RNPs) that constitute an RNA-recognition motif (RRM). SECp43 appears to regulate the levels of methylated Sec tRNA[Ser]Sec, which in turn is known to control the synthesis of stress-related selenoproteins (3).
Nannopterum harrisi
| Nannopterum harrisi phylogeny | |
|---|---|
| Domain | Eukaryota |
| Kingdom | Animalia |
| Phylum | Chordata |
| Class | Aves |
| Order | Suliformes |
| Family | Phalacrocoracidae |
| Genus | Phalacrocorax |
| Species | Nannopterum harrisi |
Nannopterum harrisi (or Phalacrocorax harrisi) also known as Galapagos cormorant and flightless cormorant, is a bird from the genus Phalacrocorax. Nevertheless, before was classified with its own genus, Nannopterum but now is classified with the other cormorants. It lives in two islands of the Galapagos islands in Ecuador, Isabela and Fernandina, usually they are found in the west coast of Fernandina and east coast of Isabela (19, 20).
This cormorant is the only specie that has lost the ability to fly, probably because it was isolated in islands without terrestrial predators and it is easier to them for forage for food, like octopuses, eels and fish. Although it is an aquatic predator, its feathers are not waterproof, so it spends a lot of time drying the feathers ,by opening their wings, after being in the water (21, 22).
They are between 89 and 100 cm high and weight 2.5-5 kg. Its wings are about one-third the size that would be required for a bird of its proportions to fly. The keel on the breastbone, where birds attach the large muscles needed for flight, is also greatly reduced. They have their toes joined by webbed skin, the upper parts are blackish and the underparts are brown. Their eyes are turquoise and the long beak is hooked at the tip (21).
Male and female look similar, but the male is larger. Juveniles are generally similar to adults, but differ in that they are glossy black in colour with a dark eye. Besides, the Galapagos cormorant are polyandric. In one season male and female have one brood, and after 70-90 day of both taking care of them, female desert and finds another male to have a second brood and the male stays taking care and feeding the first chick (21, 23).
More information can be found at Viquipèdia.