Selenoproteins
All of the following selenoproteins were predicted in an homology-based approach with both the T. guttata and G. gallus proteome annotated in SelenoDB2.0 except for MSRB1, which was only found in the G. gallus genome.
SEL15
Sel15 is a selenoprotein, whose specific function is unknown. However, It has been seen that its levels change in selenium supplementation. Some studies in mouse suggest that this selenoprotein may have redox function and may be involved in the quality control of protein folding.
This protein’s gene was found in the scaffold NEVG01000627.1 in the positions 10721124-10732764 in the reverse strand. The protein is encoded in 4 exons. T-Coffee results show an almost perfectly conserved alignment between the model proteomes and the query, with only a few punctual amino acid changes.
A selenocysteine element was found in the sequence. SECIS element were found in the 3’ UTR.
Both the Seblastian predicted protein and our predicted protein contain the same exons as well as SECIS elements in the same positions. Both sequences are conserved, however the protein predicted by Seblastian contains some extra residues in the beginning of the sequence. None of the sequences start with methionine residue. This two limitations could be occured due to the poor annotation of selenoDB protein, because protein we use as query didn’t contain methionine as the first residue.
GP FAMILY
The following proteins belong to the glutathione peroxidase family, which catalyze the reduction of organic hydroperoxides and hydrogen peroxide (H2O2) by glutathione, and thereby protect cells against oxidative damage. Several isozymes of this gene family exist in vertebrates, which vary in cellular location and substrate specificity. Several isoforms have been reported to have lost the selenocysteine residue to cysteine.
In selenoDB2.0 8 different isoforms from this family were annotated in T. guttata, however, after having analyzed all of them we can conclude that only the following can be properly annotated as selenoproteins in N. harrisi.
In order to analyze if the selection of every scaffold for each protein was correctly done in this family, we realized a phylogenetic tree using clustal omega from EMBL-EBI in order to see if every predicted protein was close to its query. In the following phylogenetic tree we can see that for every GPx selenoprotein, both the T. guttata (Zebra finch) and the G. gallus selenoproteins are presented as well as the predicted protein using the homology-based approach from both the T. guttata and G. gallus genome. We then conclude that indeed, for every isoform the proteins (both predicted and reference proteins) are closer within each isoform.

GPx2
GPx2 does majority of the glutathione-dependent hydrogen peroxide-reducing activity in the epithelium of the gastrointestinal tract.
As for this gene, it is encoded in the contig NEVG01000034.1 between positions 126214-126865 in the reverse strain. It contains two exons. T-Coffee output shows the highest score (100), no gaps and almost a perfectly conserved alignment.
The selenocysteine residue is conserved, aligned with the same residue of the reference sequence. SECIS structure were found in the 3’UTR.
As for the Seblastian prediction, both the exon and SECIS elements are the same. However, the sequence of the predicted protein contains 4 more residues at the beginning, in which a Methionine is included resulting in a Methionine as the first aminoacid. Thus, we can conclude that the prediction made by Seblastian is, in this case, more complete and informative than our predicted protein so we would annotate this as GPx2 in N. harrisi.
GPx3
This protein is involved in the detoxification of hydrogen peroxide, and is one of the most important antioxidant enzymes in humans. Secreted to the plasma.
It is encoded in the contig NEVG01000044.1 between the positions 5628926-5629999 in the forward strand. As observed in exonerate it is encoded in 4 exons. T coffee output file shows an almost perfectly conserved alignment.
A selenocysteine residue is found in the coding sequence, aligned with the same residue of the reference sequence. As for the SECIS structures, a SECI element was found in the 3’UTR.
As for the Seblastian prediction, both the exon and SECIS elements are the same as in our predicted protein. As for the sequence, both of them lack of methionine as the first aminoacid.
GPx4
This protein catalyzes functions in the protection of cells against oxidative damage. It is highly expressed in the sperm.
A hit for this protein is found in the contig NEVG01000622.1 between the positions 726555-727573 in the forward strand. T-Coffee output file shows the highest score (100), no gaps and almost a perfectly conserved alignment.
A selenocystein residue is found in the coding sequence, aligned with the same residue of the reference sequence. A SECIS element is found in the 3’UTR.
For this protein, even though we obtained a fasta subseq output file, we could not obtain the following steps, as in the terminal appeared an storage error. Thus, as we did not have a predicted protein for this one, we used the Seblastian predicted protein and did the T-Coffee with this protein. We are not sure why this error appeared, since it worked with the rest of files.
GPx7
This protein is involved in the detoxification of hydrogen peroxide, and is one of the most important antioxidant enzymes in humans.
A hit for this protein is found in the contig NEVG01000627.1 between positions 580705-582996 in the reverse strain. The protein is encoded in three exons. T-Coffee output file shows an almost perfectly alignment.
No selenocysteine residue was found in the predicted protein, nor in the G. gallus or T. guttata protein. Therefore, as previously mentioned we would conclude that this isoform is only present in the cys form in N. harrisi. No SECIS elements were found in this protein.
Seblastian has not predicted this protein. Therefore, due to lack of evidences, we cannot definitely conclude that N. harrisi has GPx7 in its genome.
GPx8
A hit for this protein is found in the contig NEVG01000229.1 between positions 1154507-1157404 in the forward strain. The protein is encoded in two exons. T-Coffee output file shows an almost perfectly alignment between our predicted protein and both T. guttata and G. gallus.
No selenocysteine residue was found in the predicted protein, nor in the G. gallus or T. guttata protein. Consistent with the literature (24, 25, 26), this protein amongst this family is one of the cases in which Sec residue has been lost. No SECIS elements were found for this protein.
Seblastian has not predicted this protein. Therefore, due to lack of evidences, we cannot definitely conclude that N. harrisi has GPx8 in its genome.
DIO FAMILY
The Iodothyronine deiodinase family regulates the activation and inactivation of thyroid hormones.
In order to analyze if the selection of every scaffold for each protein was correctly done in this family, we realized a phylogenetic tree in order to see if every predicted protein was close to its query. In the following phylogenetic tree we can see the contigs for each protein are correctly selected, since the predicted proteins for both T. guttata and G. gallus are closer to its isoform than the rest.

DIO1
For this proteins, two statistically significant hits in the same contig and in the same positions were found for different proteins (SPP00002518_2.0 and SPP00002519_2.0), however only one of them was taken into account as the other was significantly smaller than the first and with worse alignment.
The selected hit for this protein is found in the contig NEVG01000575.1 between positions 277803-281421 in the forward strain. The protein is encoded in four exons. T-Coffee output file shows an almost perfectly alignment between our predicted protein and both T. guttata and G. gallus.
A selenocysteine residue was found in the predicted protein. SECIS element was found in the 3’UTR.
As for the Seblastian prediction, both the exon and SECIS elements are the same as in our predicted protein. As for the sequence, both of them contain methionine as the first aminoacid.
Therefore, we can definetly annotate DIO1 as a selenoprotein.
DIO2
DIO2 transforms T4 into T3, activating it. It is an ER-resident protein which activates the thyroid hormone by deiodination of the outer tyrosyl ring.
A hit for this protein is found in the contig NEVG01000059.1 between positions 3654602-3666715 in the reverse strain. The protein is encoded in two exons. T-Coffee output file shows an almost perfectly alignment between our predicted protein and both T. guttata and G. gallus.
A selenocysteine residue was found in the predicted protein. SECIS element is found in the 3’UTR. Seblastian has not predicted this protein. Therefore, due to lack of evidences, we cannot definitely conclude that N. harrisi has DIO2 in its genome.
DIO3
DIO3 is the last member of Iodothyronine deiodinase family. It catalyzes the inactivation of thyroid hormone by inner ring deiodination of T4 and T3. It is highly expressed in the pregnant uterus, placenta, fetal and neonatal tissues, suggesting that it plays an essential role in the regulation of thyroid hormone inactivation during embryological development.
A hit for this protein is found in the contig NEVG01000413.1 between positions 1842474-1843088 in the reverse strain. The protein is encoded in one exon. T-Coffee output file shows an almost perfectly alignment between our predicted protein and both T. guttata and G. gallus.
A selenocysteine residue was found in the predicted protein. SECIS elements were found in the 3’UTR.
As we observe in the protein predicted by Seblastian, our protein contains the same exons as the protein predicted by Seblastian and SECIS element are located in both proteins in 49595 - 49515 positions in respect to the fastasubseq file. About the sequences, we observed that predicted protein by Seblastian starts with methionine due to the addition of some residues at the beginning in comparison of our predicted protein. We hypothesised that, the lack of this residues in our query are the due to the wrong annotation of selenoDB proteins. For this reason, we think that protein predicted by Seblastian is better than our predicted protein.
MsrA
MrsA is implicated in the enzymatic reduction of methionine sulfoxide to methionine, thus it is highly conserved amongst species. Human and animal studies have shown the highest levels of expression in kidney and nervous tissue.
For this protein, two contigs obtained an statistically significants hits (NEVG01000097.1 and NEVG01000260.1). Therefore, with the possibility of two isoforms in this proteins we analyzed them, one of them was rapidly discarded as the length was almost two-fold the other and the T-Coffee alignment showed very bad results.
So, taking into account the selected contig, a hit for this protein is found in the contig NEVG01000260.1 between positions 267686-369281 in the reverse strain. The protein is encoded in 7 exons. T-Coffee output file shows a good alignment between our predicted protein and both T. guttata and G. gallus.
As read in the literature (24, 25, 26), all MsrA found in vertebrates changed to the Cys form. Thus, as expected, no selenocysteine residue was found in the predicted protein, nor in the G. gallus or T. guttata protein. No SECIS elements were found for this protein.
As a cys-containing homologous protein, it does not have a Sec residue and thus, Seblastian has not predicted any selenoprotein and any SECIS element.
SELENOH
This selenoprotein known as SelH, resides in the nucleus. The function of this protein may plays a role as an antioxidant.
This protein’s gene was found in the scaffold NEVG01000369.1 in the positions 527821-528137 in the forward strand. The protein is encoded in 3 exons. T-Coffee results show an almost perfectly conserved alignment between the model proteomes and the query.
A selenocysteine element was found in the sequence. SECIS element were found, located in 3’UTR.
Seblastian predicts a protein with the same exons as our protein. SECIS elements are located in the same positions in both sequences. But, as in other proteins, the protein predicted by Seblastian contains some residues at the beginning of the protein which our protein did not contain, resulting in a lack of methionine in our predicted protein. We therefore think that, the Seblastian predicted protein is more complete and informative and we would annotate it as a selenoprotein in Nannopterum harrisi.
SELENOI
SELENOI is a membrane protein whose function is not fully understood.
For this protein, the T. guttata and G. gallus were considerably different; the one from T. guttata was much smaller (55 aminoacids) and did not contain Met as the first aminoacid in contrast to the one from G. gallus, which contain Met as the first aminoacid and contained 400 residues. Therefore, we decided to only use the protein from G. gallus as it was better annotated.
For this protein in G. gallus, two statistically significant hits were found, one in contig NEVG01000381.1 and the other in contig NEVG01000087.1. However, the alingment for the second hit was very bad so be discarded it.
This protein’s gene was found in the scaffold NEVG01000381.1 in the positions 477694-489594 in the reverse strand. The protein is encoded in 10 exons. T-Coffee results show an almost perfectly conserved alignment between the model proteomes and the query.
A selenocysteine element was found in the sequence. SECIS element were found located in 3’UTR.As for Seblastian predicted protein, it contains the SECIS element in the same positions, however the protein predicted is smaller than ours with only 5 exons in contrast to the 10 exons protein we have predicted. Seblastian predicted protein does not start with Met nor Met is included in the first 10 residues, making us doubt of the reliability of this protein.
Overall, we would conclude that SELENOI is definitely in N. harrisi proteome, however, the fact that Seblastian doesn’t predict the same protein as ours makes us doubt of the reliability of our results.
SELENOK
This selenoprotein is localized to the endoplasmic reticulum and is highly expressed in the heart, where it may plays a role as an antioxidant.
The gene that encodes for this protein is located in the scaffold NEVG01000427.1 between the positions 1193202 and 1193107 in the reverse strand. This gene contains 4 exons. T-Coffee results show a perfect alignment between our predict protein and the reference sequence.
A selenocysteine residue is found in the sequence. The SECIS elements were found in 3’UTR.
As for Seblastian prediction, both proteins contain the same exons as well as the same length and the SECIS element of both proteins are located at the same location. Both protein contain Met as the first aminoacid.
Therefore, due to our favorable results, we can annotate this protein as a selenoprotein of Nannopterum harrisi.
SELENOM
This gene is expressed in a variety of tissues, and the protein is localized in the endoplasmic reticulum.
The gene that encodes for this protein is located in the scaffold NEVG01000398.1 between the positions 3760340 and 360647 in the forward strand. This gene contains 5 exons. T-Coffee results show a perfect alignment between our predict protein and T. guttata genome.
A selenocysteine residue is found in the sequence. The SECIS elements were found and located in 3’ UTR.
Protein predicted by sebastian and our predicted protein contain the same number of exons and SECIS element are located in the same positions. As for the sequences, the Seblastian predicted protein is longer than ours, however none of neither the Seblastian one nor our predicted protein contain Met as the first aminoacid.
Therefore, due to our favorable results, we can annotate this protein as a selenoprotein of Nannopterum harrisi.
SELENON
The function of this protein remains unknown. Mutations in this gene cause the classical phenotype of multiminicore disease and congenital muscular dystrophy with spinal rigidity and restrictive respiratory syndrome.
The gene that encodes for this protein is located in the scaffold NEVG01000540.1 between the positions 1525313 and 1536366 in the forward strand. This gene contains 12 exons. T-Coffee results show a perfect alignment between our predict protein and reference genome.

A selenocysteine residue is found in the sequence. The SECIS elements were found and located in 3’ UTR.
As for the Seblastian prediction, both the exon and SECIS elements were located in the same positions. However, our protein contains extra aminoacids in the beginning of the protein.
Therefore, due to our favorable results, we can annotate this protein as a selenoprotein of Nannopterum harrisi.
SELENOO
This gene encodes a selenoprotein that is localized to the mitochondria. The exact function of this selenoprotein is unknown, but it is thought to have redox activity. Amongst this family of selenoproteins, 4 selenoproteins are annotated in SelenoDB2.0 in T. guttata, however after our prediction, only two of these can be properly annotated as selenoproteins, as the other two are too short and within the coding sequence of the others.
As read in the literature (24, 25, 26), usually SELENOO family contains a Sec residue in the C-terminal region. However, in some cases homologs have been found in which the Sec residue is lost and a Cys residue can be found.
In order to analyze if the selection of every scaffold for each protein was correctly done in this family, we realized a phylogenetic tree. As we can see, most of the proteins are predicted correctly. We see a missmatch in some proteins, for example in the case of the shorter versions of the proteins. However, this can be due to the fact that, because this protein is very small its not correctly sorted phylogenetically. Moreover, this shorted versions where rapidly discarded so this missannotation does not affect our results.
SELENOO 1
The gene that encodes for this protein is located in the scaffold NEVG01000221.1 between the positions 4558726 and 4581633 in the reverse strand. This gene contains 7 exons. T-Coffee results show a perfect alignment between our predict protein and the reference genome.
No selenocysteine residue was found in the predicted protein, nor in the G. gallus or T. guttata protein. No SECIS elements were found for this protein. No selenoprotein has been predicted by Seblastian as it is a Cys-containing protein.
SELENOO 2
The gene that encodes for this protein is located in the scaffold NEVG01000289.1 between the positions 4418838 and 4431861 in the reverse strand. This gene contains 9 exons. T-Coffee results show a perfect alignment between our predict protein and reference genome.
A selenocysteine residue is found at the end of the sequence, this residue is located in the C-terminal region, agreeing with the literature (24, 25, 26). The SECIS elements were found and located in 3’ UTR.
As for the Seblastian prediction, our protein contains one extra exon than Seblastian predicted protein. SECIS element of both proteins are located at the same positions. Due to the addition of this extra exon at the beginnning, our protein does not start with methionine, in contrast to Seblastian predicted protein. For this reason, we conclude that Seblastian protein is better than our protein, we would annotate Seblastian’s protein as a selenoprotein of Nannopterum harrisi.
SELENOP
This selenoprotein is an extracellular glycoprotein. It is the selenoprotein family that contains the most Sec residues. It is considered to play an important role in the preservation and transport of selenium due to the abundance of Sec residues.
In order to analyze if the selection of every scaffold for each protein was correctly done in this family, we realized a phylogenetic tree. As we can see, our predicted proteins are close enough to its reference proteomes. Interestingly, SELENOP1 in G. gallus is far from the one in T. guttata, therefore we decided to check its alignment and, due to its poor results, we concluded that probably N. harrisi SELENOP1 is closer to T. guttata and at the same time poorly conserved to the one from G. gallus. As for SELENOP2, both in T. guttata and G. gallus as well as N. harrisi the protein has been conserved, with few residues changed.
SELENOP 1
The gene that encodes for this protein is located in the scaffold NEVG01000627.1 between the positions 4837210 and 4837968 in the forward strand. This gene contains 3 exons. T-Coffee output file shows a perfect alignment between our predict protein and T. guttata genome as well as the G. gallus.
This protein contains only one selenocysteine residue in contrast to what we would expect, this result is unexpected, since as read in the literature (24, 25, 26), SELENOP contains multiple selenocysteins. However, nor in T. guttata or G. gallus, this gene contained multiple selenocysteins. Seblastian predicts SECIS elements located in 3’ UTR.
Seblastian predicts a protein with the same exons as our protein. SECIS elements are located in the same positions in both sequences. But, as in other proteins, the protein predicted by Seblastian contains some residues at the beginning of the protein which our protein did not contain, resulting in a lack of methionine in our predicted protein. We therefore think that, the Seblastian predicted protein is more complete and informative and we would annotate it as a selenoprotein in Nannopterum harrisi.
SELENOP 2
The gene that encodes for this protein is located in the scaffold NEVG01000053.1 between the positions 159078 and 163868 in the forward strand. This gene contains 4 exons. T-Coffee results show a good alignment between our predicted protein and the reference genome, with some aminoacids not being conserved.
This protein contains 14 selenocysteins, as expected. As read in the literature (24, 25, 26), this selenoprotein is characterized by multiple Sec residues located in the N-terminal region which is consistent with our results. Seblastian predicts SECIS element, located in 3’ UTR.
Seblastian predicts a protein with the same exons as our protein. SECIS elements are located in the same positions in both sequences. But, as in SELENOP1, in our protein a few more aminoacids are added at the beginning, leading to a lack of Met as a first aminoacid in our protein in contrast to Seblastian predicted protein. We would attribute this to a poor annotation in SelenoDB2.0 database.
We therefore think that, the Seblastian predicted protein is more complete and informative and we would annotate it as a selenoprotein in Nannopterum harrisi.
MSRB
This family Selenoprotein R (SelR) plays an important role in maintaining intracellular redox balance by reducing the R-form of methionine sulfoxide to methionine.
For this family, three proteins were annotated in T. guttata proteome and two in G. gallus proteome. Interestingly, MSRB1 was only annotated in G. gallus, even though we managed to find this protein in N. harrisi. As for the T. guttata annotation, as explained, contained three selenoproteins, one of which was rapidly discarded as it was contained within the sequence of another one (same contig and same position) which a larger sequence and better alignment. Therefore, the following proteins are the ones we selected and consequently hypothesize that are found in N. harrisi proteome.
In order to analyze if the selection of every scaffold for each protein was correctly done in this family, we realized a phylogenetic tree in which we can see that, in the case of MSRB2 and MSRB3 the contigs were correctly chosen. However, for the protein we believed to be MSRB1 in T. guttata (Zebra Finch) was much closer to MSRB2 than to MSRB1 of Gallus gallus, leading us to think that this hit was not referring to MSRB1 but to MRSB2. We later check this protein and, as explained below we could see that its sequence was contained within the sequence of MSRB2.
MSRB1
This protein belongs to the methionine sulfoxide reductase (Msr) protein family. This gene encodes for a protein that localizes to the cell nucleus and cytosol and is expressed in a variety of adult and fetal tissues.
The gene that encodes for this protein is located in the scaffold NEVG01000124.1 between the positions 32.028 and 33.039 in the forward strand. This gene contains 3 exons. Exonerate predicts 4 exons, however one of them we considered is far from the others so we have ignored it. T-Coffee results shows a perfect alignment between the G. gallus query and our genome.
A selenocysteine residue is found in the sequence. SECIS elements were found in 3’UTR.
Protein predicted by Seblastian contains the same number of exons comparing with our protein. SECIS element are located in the same position in both proteins. Sequences are very similar and both contain methionine as the first residue of the protein. For this reason, we would use our protein to annotate this protein as a selenoprotein in Nannopterum harrisi.
For this protein, we could only find a hit with the G. gallus genome, not with the T. guttata. Thus, we can conclude that G. gallus is probably better annotated than T. guttata.
MSRB2
This protein plays an important role in maintaining intracellular redox balance by reducing the R-form of methionine sulfoxide to methionine.
The gene that encodes for this protein is located in the scaffold NEVG01000221.1 between the positions 3051806 and 3053430 in the forward strand. This gene contains 2 exons. T-Coffee results show a perfect alignment between our predict protein and the reference genome.
A selenocysteine residue was found in the coding sequence. No SECIS elements could be found.
MSRB3
This protein catalyzes the reduction of methionine sulfoxide to methionine. This enzyme acts as a monomer and requires zinc as a cofactor.
The gene that encodes for this protein is located in the scaffold NEVG01000107.1 between the positions 11056176 and 11106529 in the forward strand. This gene contains 6 exons. T-Coffee results show a perfect alignment between our predict protein and the reference genome.
This protein doesn’t contain any selenocysteine, but it contains 6 cysteins, probably due to a loss in the selenocysteine residue in evolution. No SECIS element were found.
Since it does not has a Sec residue, Seblastian has not predicted any selenoprotein nor SECIS elements. Since the alignment of the query with the genome is good, we can conclude that N. harrisi contains MSRB3 in its genome.
As read in the literature (24, 25, 26), even though MRSB1 usually contains Sec residue in metazoans, two additional MSRB homologous (MSRB2 and MSRB3) contain a Cys residue instead of the Sec residue in the active site. In the case of MRSB2 our results are not consistent with this, as for MSRB3 our predicted protein was consistent with the literature (24, 25, 26).
SELENOS
This protein regulates the production of the cytokine, for this reason, is a key protein on the control of the inflammatory response.
The gene that encodes for this protein is located in the scaffold NEVG01000025.1 between the positions 10804869 and 10804813 in the reverse strand. This gene contains 4 exons. T-Coffee results shows a perfect alignment between our predict protein and T. guttata genome.
A selenocysteine residue was found in the coding sequence. SECIS elements are located in 3’ UTR.
Protein predicted by Seblastian contains two more extra exons that our predicted protein. As for, SECIS element this is located in the same positions. Due to this addition of extra exons in the beginning of the protein our protein doesn’t start with Met in contrast to Seblastian’s one. We would attribute this to the fact that there is probably a wrong annotation of T. guttata and G. gallus in SelenoDB2.0.
For this reason, we think that protein predicted by Seblastian is better than our protein and we would annotate this protein as a selenoprotein of Nannopterum harrisi.
SELENOT
The gene that encodes for this protein is located in the scaffold NEVG01000066.1 between the positions 2103991 and 2108537 in the forward strand. This gene contains 5 exons. T-Coffee results shows a perfect alignment between our predict protein and reference genome.
A selenocysteine residue was found in the coding sequence. SECIS elements are located in 3’ UTR.
Our predicted protein and protein predicted by Seblastian contain the same number of exons as well as SECIS element at the same positions. As for the sequences, our protein contains some further residues at the beginning, which if they are removed results in the same predicted protein as in Seblastian and with Met as the first aminoacid. We attribute this to the bad annotation of Selenodb.
Therefore, we can conclude that the Seblastian predicted protein is more complex and more informative than ours. We can annotate this protein as a selenoprotein in Nannopterum harrisi.
SELENOU
The protein encoded by this gene belongs to a peroxiredoxin-like FAM213 superfamily. The function of SelU1 remains unclear. SelenoU(1) is known to be expressed in bone, brain, liver and kidney. In high mammalian species, such as humans and mice, all SelU proteins exist in Cys form.
In order to analyze if the selection of every scaffold for each protein was correctly done in this family, we realized a phylogenetic tree. Interestingly, in this phylogenetic tree we can observe that in G. gallus there is the only one SELENOU protein annotated, which corresponds to SELENOU1. We attributed this to the fact that a hit in the same positions has been found in the same contig as SELENOU1 from T. guttata and also due to phylogenetic distance.

SELENOU 1
The gene that encodes for this protein is located in the scaffold NEVG01000168.1 between the positions 5298587 and 5304182 in the forward strand. This gene contains 5 exons. T-Coffee results shows a perfect alignment between our predict protein and T. guttata genome as well as the G. gallus genome.
A selenocysteine residue is found in the coding sequence, this information is consistent with the literature (24, 25, 26), as the Sec to Cys event in this protein is considered to have occurred during the primitive mammalian stage. SECIS elements were found in 3’ UTR.
Protein predicted by Seblastian contain the same number of exons as our predicted protein and SECIS element is located in the same positions. About the sequences, we observed that protein predicted by Seblastian contain some more extra residues between our sequences, but in contrast, our sequences starts with a methionine as a residue. For this reason we think that our protein is better to annotate this protein as a protein of Nannopterum harrisi.
SELENOU 2
The gene that encodes for this protein is located in the scaffold NEVG01000140.1 between the positions 1483855 and 1487777 in the reverse strand. This gene contains 6 exons. T-Coffee results shows a perfect alignment between our predict protein and the reference genome.
No selenocysteins were found in the coding sequence nor any SECIS element.
Due to the favorable results in both proteins, we can annotate them as selenoproteins. As mentioned, in high mammals this proteins presents a loss in the selenocysteine residue. As our model is a bird, we have observed both cases: in SELENOU(1) the selenocysteine residue is conserved while in the SELENOU(2) the selenocysteine residue has been lost.
TXNRD
This family protein are thiredoxin reductase proteins (TR), which control the redox state of thioredoxins, key proteins involved in redox regulation of cellular processes. there has been described multiple isoforms of TXRND1 as well as TXRN3D. As read in the literature (24, 25, 26), TXNRD1 and TXNRD3 are present in all vertebrates, thus N. harrisi is expected to have them.
For this family, 5 selenoproteins were annotated in Selenodb2.0 for T. guttata, however, after having done our analysis, we can conclude that only 3 of them can be properly annotated as selenoproteins in N. harrisi genome. Two of this proteins were discarded because they were found in a contig with the same positions which had already found a hit for another one with larger length and better alignment. The following selenoproteins are the one we can conclude can be annotated as selenoproteins in N. harrisi genome.
In order to analyze if the selection of every scaffold for each protein was correctly done in this family, we realized a phylogenetic tree. In this case, we have observed that there is a missannotation between TXNRD1 and TXNRD3. In our case, we can see that our predicted TXNRD3 from T. guttata is much closer with TXNRD1 from G. gallus than with the TXNRD3 from T. guttata itself, the same happens with TXNRD1. We would attribute this fact to a wrong annotation in selenoDB2.0.
TXNRD1
The gene that encodes for this protein is located in the scaffold NEVG01000091.1 between the positions 12619013 and 12637850 in the reverse strand. This gene contains 16 exons. T-Coffee results shows a perfect alignment between our predict protein and the reference genome.
A selenocystein residue was found in the predicted protein. SECIS element was found in the 3’UTR.
As for Seblastian, both proteins predicted contain the same number of exons as well as SECIS element located in the same positions. About the sequences, these are very similar between them, however the protein predicted by Seblastian contains methionine as the first residue in contrast of ours. For this reason we think that Seblastian’s protein is better to annotated as a selenoprotein of Nannopterum harrisi.
TXNRD2
The gene that encodes for this protein is located in the scaffold NEVG01000088.1 between the positions 9574014 and 9607800 in the forward strand. This gene contains 16 exons. T-Coffee results shows a good alignment between our predict protein and the reference genome.
A selenocystein residue was found in the predicted protein. SECIS element was found in the 3’UTR.
Our predicted protein contains 6 more extra exons than protein predicted by Seblastian and SECIS elements is located at the same positions. We observed that 6 extra exons located in the beginning in our predicted protein. Due to this extra residues, our protein starts with Met in contrast to the Seblastian predicted protein. Therefore, we conclude that our protein is more complex and informative than the protein predicted by Seblastian, so we would annotate it as a selenoprotein of Nannopterum harrisi.
TXNRD3
The gene that encodes for this protein is located in the scaffold NEVG01000033.1 between the positions 9310697 and 9428083 in the forward strand. This gene contains 13 exons. T-Coffee results shows a perfect alignment between our predict protein and the reference genome.
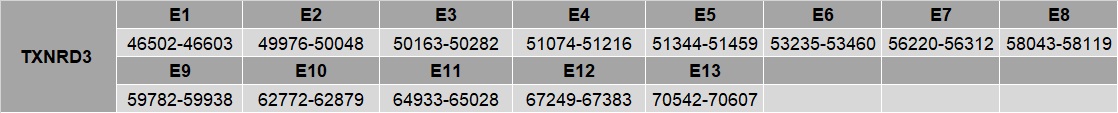
A selenocysteine residue was found in the predicted protein. SECIS element was found in the 3’UTR.
Our predicted protein contains 4 more extra exons than the protein predicted by Seblastian. SECIS element is located at the same positions. About the sequences are similar between them but our predicted protein contains extra aminoacids at the beginning. None of the sequences start with Met, however in our predicted protein this residue is within the first 5 aminoacids of the sequence. Therefore, we would conclude that our protein is better predicted than the protein predicted by Seblastian and we can use our protein to annotate it as a selenoprotein of Nannopterum harrisi.
Selenoproteins machinery
eEFsec
eEFsec is an essential translation factor needed to incorporate selenocysteine into proteins. Its function is to recruit Sec-tRNASec due to its specifically binding to Sec-tRNASec and deliver it to the ribosome. Thus, this protein is implicated in the selenoprotein biosynthesis.
In selenoDB2.0 two eEFsec proteins (SPP00002557_2.0 and SPP00002558_2.0) were annotated both for G. gallus and T. guttata, both with statistically significant hits in the same contig approximately for the same positions. Thus, we checked the alignment for both hits and concluded that only one of them was actually reliable, since one of them was much larger than the other and with much worse T-Coffee alignment.
Therefore, we concluded that eEFsec was found in the scaffold NEVG01000091.1 between positions 13740775-13741233 in the forward strain. The protein is encoded in one exon. T-Coffee output shows a good alignment between the predicted protein and the annotated one.
No selenocysteine residues are found in the predicted protein, being consistent with the available information about eEFsec in mammals since this protein belongs to the transcription machinery and it is not a selenoprotein. As for the SECIS elements, a SECIS element 3’ UTR.
Due to the favorable results, we can conclude that eEFsec can be annotated as a selenoprotein in N. harrisi.
PSTK
Phosphoseryl-tRNA kinase is found to be highly conserved in evolution, hence suggesting that it plays an important role in selenoprotein biosynthesis and/or regulation.
A hit for this protein is found in the contig NEVG01000299.1 between positions 3634132-3636771 in the reverse strain. The protein is encoded in 7 exons. T-Coffee output file shows a good alignment between our predicted protein and both T. guttata and G. gallus.
No selenocysteine residue was found in the predicted protein, nor in the G. gallus or T. guttata protein. No SECIS elements were found for this protein.
SBP2
SBP2 is a nuclear protein that binds to SECIS elements. Mutations in this gene have been associated with a reduction in activity of a specific thyroxine deiodinase, a selenocysteine-containing enzyme, and abnormal thyroid hormone metabolism.
For this gene, two proteins were annotated in selenoDB2.0. In one of them we observed a hit in the contig NEVG01000098.1 and the other a hit in the contig NEVG01000025.1, both of them with similar length. However, the protein of NEVG01000098.1 contig was discarded since it obtained a bad alignment with the predicted protein and the reference genome.
In order to analyze if the selection of every scaffold for each protein was correctly done in this family, we realized a phylogenetic tree. As seen in the following image, the selection has been done correctly.

A hit for this protein is found in the contig NEVG01000025.1 between positions 2309919-2326162 in the reverse strain. The protein is encoded in 16 exons. T-Coffee output file shows a perfectly conserved alignment between our predicted protein and both T. guttata and G. gallus with some aminoacids not being conserved.
No selenocysteine residue was found in the predicted protein, nor in the G. gallus or T. guttata protein. No SECIS elements could be predicted.
SecS
The amino acid selenocysteine is the only amino acid that does not have its own tRNA synthetase. Instead, this amino acid is synthesized on its cognate tRNA in a three step process. The protein encoded by this gene catalyzes the third step in the process, the conversion of O-phosphoseryl-tRNA(Sec) to selenocysteinyl-tRNA(Sec).
For this protein, in SelenoDB2.0 of T. guttata as well as G. gallus two proteins were annotated (SPP00002531_2.0 and SPP00002532_2.0). Thus, after having analyzed both of them, we concluded that one of them was actually non existent, since it encoded for a peptide sequence which its size was much smaller and encoded for an exon of the other protein. Therefore, for our analysis, we took into consideration the hit corresponding with SPP00002532_2.0 protein.
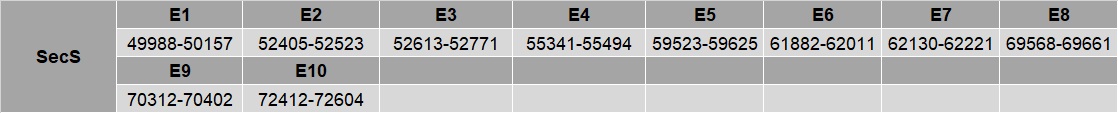
A hit for this protein is found in the contig NEVG01000256.1 between positions 1188823-1212771 in the reverse strain. The protein is encoded in ten exons. T-Coffee output file shows an almost perfectly alignment with only a few aminoacids not conserved.
No selenocysteine residue was found in the predicted protein, nor in the G. gallus or T. guttata protein. No SECIS elements were found in this protein.
SEPHS
The Selenophosphate synthetase encodes an enzyme that synthesizes selenophosphate from selenide and ATP. Selenophosphate is the Selenocysteine donor compound necessary for Sec biosynthesis, and interestingly it is itself a selenoprotein.
This gene was found in two contigs: One hit was found in the contig NEVG01000144.1 between positions 1569776-1591144 in the reverse strain and the other hit was find in the contig NEVG01000519.1 between positions 187873-193215 in the forward strain. Because both hits had a significant e-value (2,00E-37 and 7,00E-33 respectively), and after having analyzed both of them, we can consider that this protein experienced a duplication in the N. harrisi proteome.
As for the protein found in the contig NEVG01000144.1, it is encoded by 8 exons and T-Coffee output file shows an almost perfectly alignment. The predicted protein has a length of 392 aminoacids and no selenocysteine residue was found in the sequence, nor in the G. gallus or T. guttata protein. SECIS elements were found in this protein in the 3’UTR.
As for the protein found in the contig NEVG01000519.1, it is also encoded by 8 exons and T-Coffee output file shows a good alignment, even though some of the aminoacids are not conserved. The predicted protein has a length of 374 aminoacids and no selenocysteine residue was found in the sequence, nor in the G. gallus or T. guttata protein. No SECIS element could be predicted. The fact that this protein lacks of selenocysteine and SECIS elements makes us doubt of the reliability of our results.
Thus, we can conclude that this protein has been duplicated in as only SEPHS protein is annotated in T. guttata as well as in G. gallus in SelenoDB2.0. Moreover, the sequence of both proteins have been analyzed, they contain similar length and most of the aminoacids but not all are conserved (leading us to think this is a duplication and not an alignment error). However, for the second protein described (contig NEVG01000519.1) no SECIS element could be predicted, decreasing the reliability of our results.
SECp43
This family protein is responsible to control selenoprotein expression. This protein is only annotated once both in the T. guttata and G. gallus genome, however in N. harrisi genome we have found a duplication, as two significant hits were found in different contigs encoding for proteins of the same size (8,00E-21 for NEVG01000260.1 and 3,00E-12 for NEVG01000067.1).
In order to analyze if the selection of every scaffold for each protein was correctly done in this family, we realized a phylogenetic tree in which we can see that there is only one SECp43 statistically significant in G. gallus and that this one is probably closer to SECp43(2) that to SECp43(1). However, we decided to dismiss the result from the phylogenetic tree as we observed that SECp43 from G. gallus was contained in the same contig and same positions with a good alingment as SECp43(1) from T. guttata.

SECp43 (1)
This protein is encoded by a gene which is located in the scaffold NEVG01000260.1. The positions of the gene are from 397151 to 408923 and is encoded in the reverse strand. This gene contains 7 exons and T-Coffee shows a perfect alignment between the predict protein and reference genome. The predicted protein has a length of 245 aminoacids.
This protein doesn’t contain any selenocysteine residue but contains multiple cysteines, probably due to the evolution and the loss of selenocysteine residue. Seblastian results show us the prediction of SECIS elements located in 5’ UTR.
SECp43 (2)
This protein is encoded by a gene which is located in the scaffold NEVG01000067.1. The positions of the gene are from 4621537 to 4727096 and is encoded in the reverse strand. This gene contains 8 exons and T-Coffee shows a good alignment between the predict protein and T. guttata genome, even though it is not as conserved as in the first case. The predicted protein has a length of 227 aminoacids.
This protein doesn’t contain any selenocysteine residue but contains multiple cysteines, probably due to the evolution and the loss of selenocysteine residue. Seblastian results show us the prediction of SECIS elements located in 3’ UTR.
To conclude, after our analysis and given that both proteins have similar aminoacid length, have good alignments and are conserved within them, we would say that SECp43 has two isoforms in N. harrisi, however, the fact that one of them doesn’t contain any selenocysteine and that SECIS elements are found in 5’UTR make us doubt of the reliability of our results.