Introduction: Selenoproteins
SELENIUM
Selenium is an essencial dietary micro-nutrient, fundamental for human health. It is well known for its antioxidant activity, as well as, its chemopreventive, anti-inflammatory, and antiviral properties.(1) Insufficient selenium levels have been associated with several human diseases such as diabetes or cancer. On the other side, excessive levels of Selenium can be toxic for the organism. (2) Unlike other metal elements, selenium is incorporated into proteins by ionic and covalent associations with the selenocysteine amino acid (21st amino acid) (3).
SELENOCYSTEINE AND SELENOPROTEINS
Selenoproteins are selenocysteine-containing proteins found in eukaryotic, bacterial and archaeal cells.The selenocysteine amino acid (Sec) is an analog of the cysteine amino acid. The only difference between Sec and cysteine is the presence of a selenium atom instead of a sulfur which forms a selenol group. Sec is the only amino acid containing selenium as a constitutive component. Sec is encoded by the UGA triplet, which is a stop codon for non-selenoprotein genes and it is always synthesised in its tRNA (4).
Regarding eukaryotes and archaea, the insertion of this amino acid in selenoproteins takes place when a selenocysteine insertion sequence (SECIS or stem-loop structure) is located in the 3'-UTRs of selenoproteins genes. However, in regard to bacterial selenoproteins, the stem-loop structure has to be located immediately downstream of the UGA codon of the selenoprotein coding genes. (5)
Selenoproteins function depend on the presence of Sec. Although most of selenoproteins physiological roles remain still unknown, all of the functionally characterized selenoproteins share redox-active functions. (6)
BIOSYNTHESIS OF SELENOPROTEINS
The selenoprotein synthesis is a conserved process where some mechanism are different between prokaryotes, eukaryotes and archaea species. Other factors such as UGA-Sec codon, the specific tRNA and the SECIS element are conserved in every organism. (7)
In mammals, the biosynthesis of Sec depends on its own tRNA using serine as an intermediate (Ser-tRNASec).. For this process the enzyme SPS2 is required. It turns the seleno donor into its activated form: monoselenophosphatase (H2Se-P). Then Seryl-tRNA synthetase which binds the serine (Ser) to the tRNASec in order to form the Seryl-tRNA (tRNA[Ser]Sec). After that, Phosphoseryl-tRNA kinase (PSTK) phosphorylates the tRNA[ser]Sec to phosphoseryl-tRNASec which is then ready to be coupled to the activated donor of selenium called selenide. This donor, once activated by Selenophosphate synthetase 2 (SPS2) , replaces the phosphate of the phosphoseryl-tRNASec. This step is mediated by Selenocysteine syntase enzyme (SecS). At the end of this process from seryl-tRNASec (tRNA[ser]Sec) the selenocysteyl-tRNASec (tRNA[Sec]Sec) is obtained which delivers the Sec into the growing polypeptide chain. (7)
The Sec aminoacid is codified by UGA codon which normally codes for a translational termination. As the genetic code is redundant, the Sec codification by this codon is mediated by an trimensional non-coding structure found in a 3' extrem of selenoproteins called SECIS (SEleno CysteinInsertion Insertion Sequence). This structure recruits the so called SECIS Binding Protein (SBP2) which binds the specific elongation factor eEFsec. This factor will guide the Sec tRNA to the UGA codon and will allow its translation into a selenocysteine. (7)
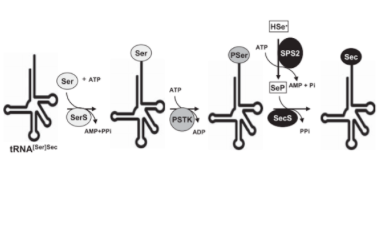
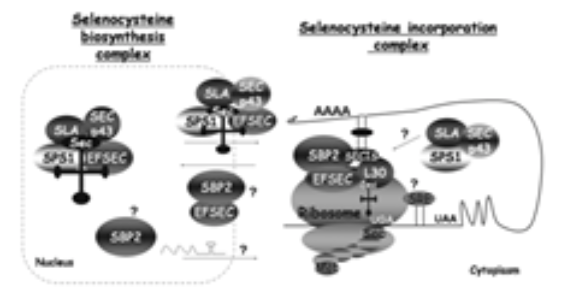
All the proteins and multiprotein complexes decribre until now in the literature as mediators and effectors of the biosintesis of selenoproteins.
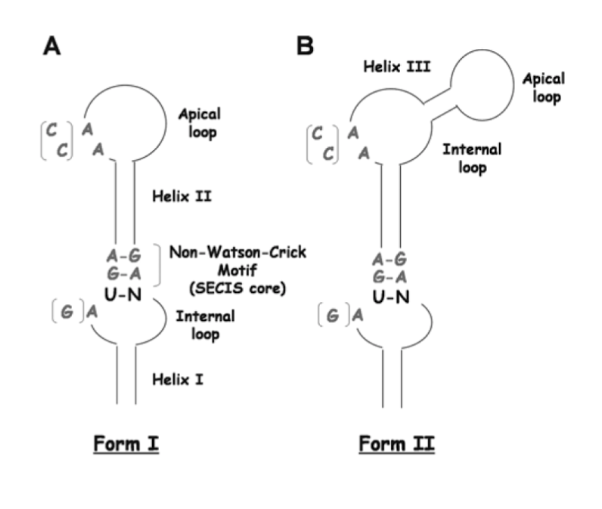
MAMMALIAN SELENOPROTEINS FAMILIES
25 mammalian selenoproteins have been discovered so far. Mammalian selenoproteins are divided in two main groups according to the Sec position: near the C-terminal part of the protein (TrxRs and selenoproteins S, R, I, O, K) and in the N-terminal part (GPxs, DIOs, Sep15, SPS2 and selenoproteins H, M, N, T, V, W). (5)
Glutathione peroxidase family in mammals consist of 8 GPx paralogs. However, only 5 of them contain a Sec in their active site (GPx1, GPx2, GPx3, GPx4, GPx6).The other three have a Cys instead of the Sec. Moreover, some of GPx6 homologs in mammals has also lost the Sec for a Cys. This family is involved in many physiological functions such as redox homeostasis, detoxification of hydroperoxides and hydrogen peroxide signaling.
GPx1, 2 and 3 are homotetrameric proteins that develope an intracellular antioxidant function by catalysing the reduction of hydrogen peroxide to water. Therefore, they are responsible for the regulation of H2O2 intracellular levels and, thus, for the regulation of biological processes where H2O2 plays an important signaling function, such as cell proliferation, apoptosis and stress response. GPx1 is the most abundant selenoprotein in mammals. It is found in all kinds of cells and tissues, specially in the liver and the kidney. GPx2 is specific epitellium of the gastrointestinal trac. GPx3 is primarly secreted from the kydney and is the major form of GPx found in the plasm.
GPx4 is a monomeric protein and can be found in many types of cells and tissues. GPx4 lacks a loop structure in the active site of the enzyme, as compared with GPx1-3. This particular configuration gives GPx4 specifity for phospholipid hydroperoxides such as colesterol hydroperoxide. GPx4 is responsible for the reduction of this type of molecules. GPx5 is not considered a selenoprotein since it does not have a Sec and GPx6 is found in embryonic tissues and olfactory epithelium. GPx7 and GPx8 are not considered selenoproteins either since they do not have a Sec.
There are three thioredoxin reductases in mammalian cells: TrxR1, TxrR2 and Thioredoxin glutathione reductase. All of them are considered to be part of the pyridine nucleotide-disulfide oxidoreductase family and comprise the major disulfide reduction system of the cell. TrxRs are three domain proteins consisting of one interface domain, a FAD-binding domain and a NADPH-binding domain. In regard to the thioredoxin glutathione reductase, it contains an additional glutaredoxin domain located in the NH2-terminal part of the protein. This suggests that this selenoprotein may be involved not only in thioredoxin functions, but also glutaredoxin functions. nevertheless, its real function remains unkwown.
TxR1 is found in the cytosol and nucleus of mammalian cells. It is responsible for the reduction of Tr1. TrxR2 is found in the mitochondria. Both TrxR1 and TrxR2 are found in all vertebrates.
All of them are homodimers integrated in membranes which have a transmembrane domain characterized by a thioredoxin fold. DIO1 and DIO3 are found in the cytoplasmic membrane, while DIO2 is found in the endoplasmic reticulum membrane.The Sec active site is located in the NH2-terminal part of the protein. Nevertheless, DIO2 has another Sec located in the COOH-terminal region. Its function remains unkwown.
DIO1 is mainly located in the kidney, liver and thyroid. DIO2 is located in the thyroid, skeletal muscle, brown adipose tissue, pituitary and brain. DIO3 is highly expressed in the pregnant uterus and placenta and located in the cerebral cortex and skin
DIOS are responsible for the regulation of the thyroid hormone activity by reductive deodination. The thyroid hormone is secreted its T4 form (inactivated). T4 can be transformed into T3 (active form) by a reaction which is catalysed by DIO1 and DIO2. Moreover, DIO3 can catalyse the reactions needed to inactivate T3 and T4 by transforming them to T2 and reverse T3, respectively.
Selenoproteins W, T, H and V belong to the Rdx family, a family of thiol-based oxireductases selenoproteins. They are characterized by a conserved Cys-x-x-Sec motif and a thioredoxin-like fold. However, they function remains unknown.
Selenoprotein W is mainly located in the citoplasm with small fractions found in the cell mambrane. It is commonly expressed in tissues and highly expressed in the muscles and the brain. SelW expression is highly regulated by the availability of Se in the diet, therefore, it belongs to the stress-related group of selenoproteins. However, its phisiological function remains unknown.
Selenoprotein T is mainly located in the endoplasmatic reticulum and the Golgi. It is ubiquitously expressed in embryonic and adult tissues. SelT has been associated with regulation of Ca2+ homeostasis and neuroendocrine function, as well as cellular adhesion and oxidoreductases genes expression.
Selenoprotein H is only located in the nucleoli. SelH has low levels of expression in adult tissue but is highly expressed in embryonic development. SelH has been associated with heat shock and stress response regulation. Moreover, it has glutathione peroxidase activity and is involved in the regulation of transcription of genes involved in glutathione synthesis.
Selenoprotein V is only found in placental mammals and only expressed in testes. SelV structure is very similar to SelW, but larger due to the incorporation of an additional Nh2.terminal domain. The physiological function of this domain and of SelV remain unknown. Sel V may be related to male reproduction.
Selenoprotein I is one of the latest discovered selenoproteins. It is only found in vertebrates. SelI is located in the endoplasmic reticulum membrane due to its 7 transmembrane domains. It may be associated with phospholipids synthesis.
Selenoprotein M (SELENOM) shares 31% of similarity with Sel15 indicative of functional and structural homology with this selenoprotein. It is found in all vertebrates and it is highly express in the brain. Its Sec is present as part of a -CXXU- a redox-active motif at its N-terminal end which suggest that this protein is involved in oxidative processes.
The 15-kDa selenoprotein is a selenorprotein located in the endoplasmatic reticulum and its related to disulfide bond formation and glycoprotein folding. Sep15 is highly expressed in prostate, liver, kidney, and testis.Two cDNA variants of this selenoprotein have been described, one of which is more efficient at incorporating Sec at the UGA codon. Moreover, loss of heterozigity have been associated with breast, head and neck cancer.
Selenoprotein P is a an unusual selenoprotein located in human plasma. It contains a total of 10 Sec residues in humans. SelP is responsible for the transport and delivery of selenium to body tissues from the liver, where it is secreted. It has also been associated to antioxidant functions.
Msr selenoproteins catalyze the reduction of methionine sulfoxide back to methionine. Methionine-R-sulfoxide reductase 1 (MsrB1) was initially defined as selenoprotein R and selenoprotein X. Is a zinc-containing selenoprotein which catalyzes the reparation of the R enantiomer of oxidized methionine residues in proteins. Moreover, methionine-R-sulfoxide reductase A (MsrA) is a selenoprotein that catalyzes the reduction of the other isomer.
These two selenoproteins have complementary functions. Nevertheless they do not have any sequence similarity and their structure is different. MsrB1 is located in the cytosol and nucleus, and it is mainly active in the liver and kidney.
Selenoprotein O is a widely distributed selenoprotein. It is the least characterized human selenoprotein. SelO has a Sec residue located at the COOH-terminal of the protein. Several homologs of human SelO have been found in animals, plants, bacteria ans yeast. However, most of them contain a Cys instead of the Sec residue. The function of this selenoprotein remains still unkown.
Selenoproteins K and S do not share any sequence similarity. However, they have structural similarities: a single transmembrane domain in the NH2-terminal sequence (transmembrane proteins III), positively charged amino acids, a high content of glycine and Sec residues located in the COOH-terminal of the protein.
SelK and SelS seem to be the most spread selenoproteins among individuals that incorporate selenium. They are both located in the endoplasmatic reticulum membrane and the cell membrane SelS and SelK are related to the ER-associated degradation of misfolded proteins.
Selenoprotein N is a transmembrane glycoprotein located in the endoplasmatic reticulum membrane. It is mainly expressed in embrionic tissues and less expressed in adult tissue, such as the skeletal muscle. It has been reported that SelN is mandatory for early muscle development and differentiation in zebrafish, but not in mice. Mutations in SelN gene (SEPN1) are associated to muscle disorders known as SEPN1-related myopathies.
After being converted to selenide, dietary selenium has to be converted again to selenophosphate by selenophosphate synthetase enzyme. Afterwards, selenophosphate is mandatory for the transformation of tRNA[ser]sec serine into selenocysteine, which will be inserted into the selenoprotein chain by the selenosome.




