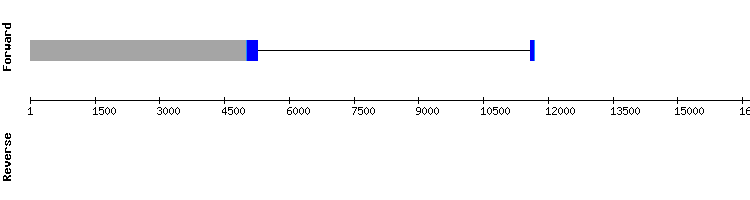Discussion
SELENOPROTEINS
15 kDa selenoprotein (Sel15) family
One scaffold has been found for Sel15: NW_018339842.1. This scaffold has identity percentage of 90. Regarding the t-coffee analysis, results showed a very good alignment between sequences with a score of 1000 and also a selenocystein residue was shown.
Regarding the SECIS, only one has been found at the relative position of 29421 - 29344 of the negative strand. The relative position of the SECIS is consistent with the relative position of the last exon (30000).
As the protein is not well annotated in Equus caballus in SelenoDB (as it is known that Sel15 is a very conserved selenoprotein and none selenocystein was shown), we compared our results with the information of Sel15 from Homo sapiens. However, the number of exons were not the same (there were five exons in the Seblastian prediction and three in SelenoDB) and the their relative positions did not match completely.
Nevertheless, we can conclude that Sel15 is a selenoprotein found in the Odocoileus virginanus texanus genome and it seems to be quite conserved among evolution.
Iodothyronine deiodinase (DIO) family
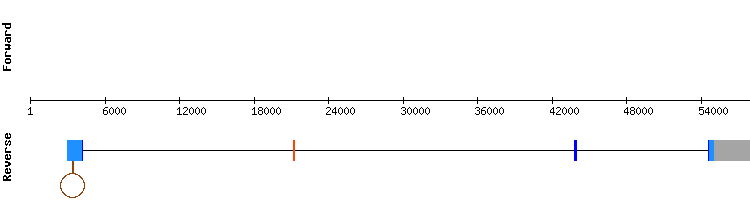
The protein DIO1 showed three scaffolds, being NW_018338175.1 the most significant as it had the higher identity percentage and length.The other two scaffolds were not analyzed since they were also present in other proteins of the same DIO family with a better identity percentages.
The results from t-coffee showed a very good alignment between sequences with a score of 996 and a gap of 5 amino acids in the middle.The alignment was divided in four exons and a selenocystein aligment was observed in the second one (relative position 37687 - 37830).
A SECIS element was predicted in the relative position 45275 - 45345, which matches with the 3'-UTR region of the protein.
Both the correct alignment and the predicted SECIS structure resolve DIO1 as a conserved selenoprotein present in Odocoileus virginianus texanus genome codified by four exon gene.
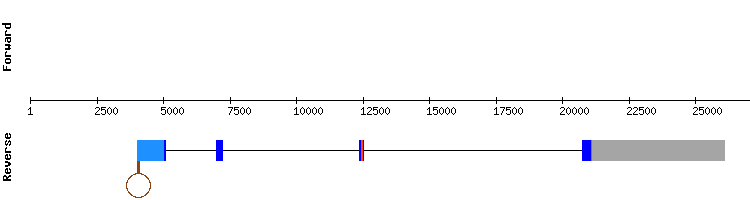
For DIO2 analysis we chose the scaffold NW_018337299.1, as the other scaffolds were least significant than those found in other selenoproteins from DIO family (DIO1 or DIO3). Its relative position in the chromosome is 30000 - 38922 and it is divided in two exons according to the exonerate output.
The results of the scaffold showed an almost perfect alignment with a score of 1000. A selenocysteine alignment was observed in the middle of the sequence.
The program Seblastian predicted three SECIS elements, but the position of the third one is too distant from the last nucleotide from the last coding exon of the protein (position 38701). The first and the second elements (relative positions 22963 - 22891 and 24736 - 24664, respectively) had the same infernal score. Thus, they were both likely to correspond to DIO2 protein. As both SECIS element were in a proper position, we can infer that it may have been a duplication of their sequence.
Since the protein shows a good alignment and a SECIS element, it is concluded that DIO2 is a conserved selenoprotein encoded by a two exon gene present in Odocoileus virginianus genome.
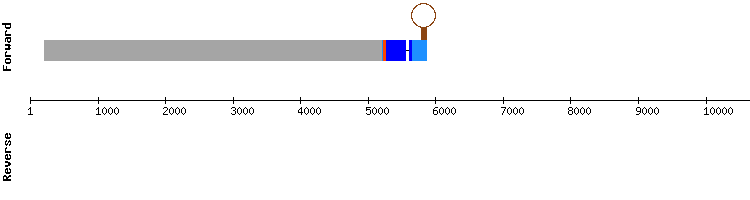
This protein is found in SelenoDB as DIO, since it is not well characterized yet. We named it DIOa to distinguish it from the other proteins of the same family. The scaffold we chose for this protein was NW_018336842.1, as it had the best identity percentage and length. The alignment showed in the results was good, with a score of 999.
Nevertheless, there was a gap of 12 amino acids in the initial positions of the sequence (relative position 30000).
We compared it to the alignment predicted by Seblastian and results were more consistent as the alignment was longer and it only contained a gap of three amino acids. A selenocysteine alignment was observed in both analysis.The SECIS element prediction was also consistent as it was in the 3'-UTR region of the protein (position 29341 - 29272).
However, according to Seblastian analysis, DIOa from Equus caballus was predicted as the thyroxine 5-deiodinase selenoprotein found in Bos mutus genome, which is DIO3 when talking about the specie Homo sapiens.
To verify this, we analyzed the results of this protein from Homo sapiens genome. The most significant scaffold was also NW_018336842.1, but it showed better results than for Equus caballus. Therefore, we conclude that DIOa is the equivalent for human DIO3 selenoprotein found in the genome of Odocoileus virginianus texanus, encoded by a gene with two exons.
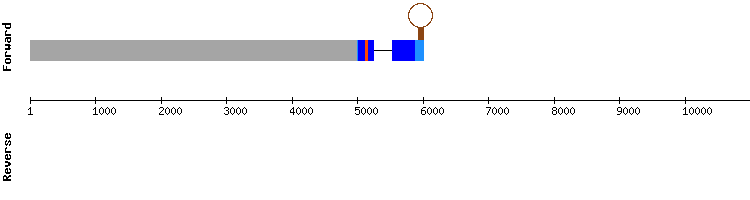
Glutathione peroxidase (GPx) family
In regard to GPx1, no significant scaffold was found when comparing this query protein of Equus caballus to Odocoileus virginianus texanus genome. Thus, we decided to compare it to a query from the species Homo sapiens. The chosen scaffold was NW_018337286.1 according to the identity percentage and the alignment length of its hits. The results from t-coffee showed a good alignment (score of 998) with a gap of five amino acids in the initial positions. A selenocysteine alignment was predicted in the second exon. However, Seblastian did not predict any SECIS element.
Since GPx1 is the most abundant selenoprotein in mammals and has a crucial role protecting the hemoglobin from oxidative breakdown in Homo sapiens (see introduction), it should be a well conserved selenoprotein. Besides, SelenoDB results show a SECIS element in the 3'-UTR region. Thus, we conclude that the reason why the SECIS prediction is missing is due to a short output of fastasubseq or because the program is not sensitive enough to detect the proper SECIS.
Finally, we conclude that GPx1 is a selenoprotein present in the genome of Odocoileus virginianus texanus and encoded by a gene with two exons, even though no correct SECIS could be found.
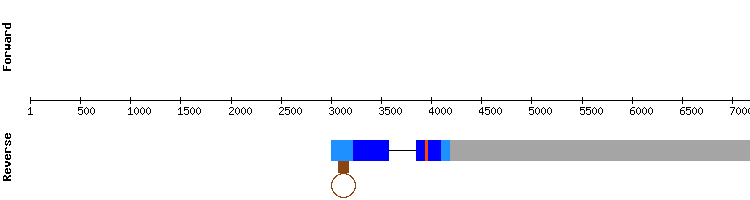
Even though the protein had multiple scaffolds, we discarded most of them since they were present in other proteins of the family with a better identity percentage. The scaffold selected was NW_018340754.1, which showed an almost perfect alignment with a score of 1000. A selenocysteine alignment was also predicted.
A SECIS element was predicted in Seblastian in the relative position 33317 - 33381, which is consistent as the gene encoding for the protein is located in the positive strand in the relative position 30000-33111.
Thus, it is concluded that GPx2 is a selenoprotein present in Odocoileus virginianus texanus genome encoded by a gene with two exons, differing from the GPx2 encoding gene from Equus caballus:, which has a single exon. This difference could be because the exonerate analysis was not exhaustive or because the annotation of the protein in SelenoDB is not correct, as it does not even have a selenocysteine in the gene structure:
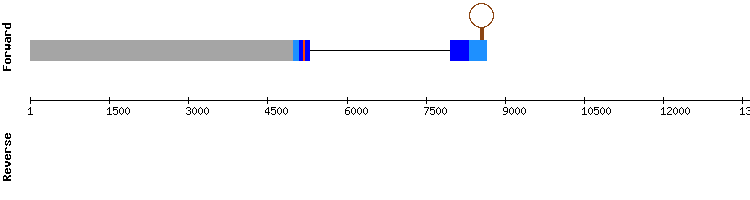
Significant hits were found in different scaffolds, but we only analyzed NW_018343298.1 as it had the best identity percentage and length. The alignment results from t-coffee confirmed this scaffold as the correct selenoprotein sequence, since the output showed the highest score (1000), no gaps and almost a perfectly conserved alignment.
The selenocysteine residue is conserved and was found in the second exon (relative positions 17803 - 17889), aligned with the same residue of the reference sequence.
Besides, Seblastian predicted a SECIS structure as well as a sequence alignment. It showed five exons (relative positions 17803-17889; 12692-12845; 11361-11478; 10818-10917; 10126-10344), whereas the sequence predicted by exonerate showed four (same exons except the one with relative position 17803-17889).
As Seblastian compared our specie Odocoileus virginianus texanus with Ovis aries musimon, it is concluded that the exonerate analysis was not exhaustive enough or that the annotation of this protein in SelenoDB is incomplete. The second option is more likely taking into account that the gene structure shown in this database does not have a SECIS element nor a selenocysteine:
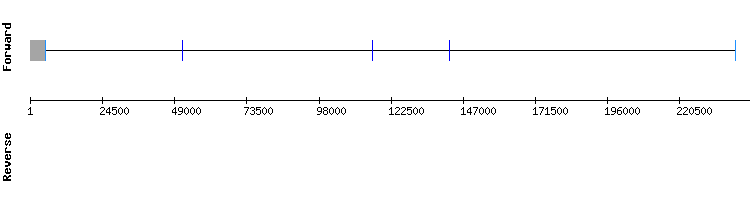
In conclusion, GPx3 is a selenoprotein present in Odocoileus virginianus texanus genome as it is conserved in Equus caballus and Homo sapiens.
As there were not hits predicted for this protein from Equus caballus, we used as query GPx4 from Homo sapiens. From the three scaffolds with good identity percentage and lenght, NW_018331773.1 was the one with the best alignment score (1000). A selenocysteine alignment was also shown.
Seblastian predicted a sequence alignment divided in five exons in the positive strand. Besides, it predicted a SECIS element in the 3'-UTR region of the gene (relative position 31172 - 31244).
These results are also consistent with the information in SelenoDB, even though the gene is divided in seven exons instead of five. This difference could be because the exonerate analysis was nos exhaustive.
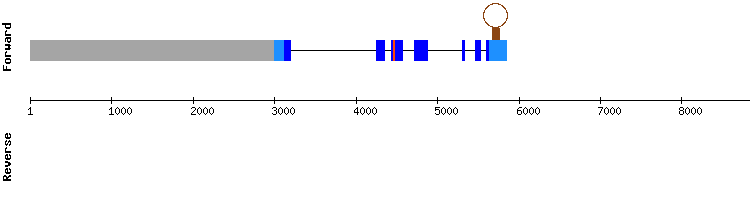
Thus, we conclude that GPx4 is a selenoprotein conserved in Odocoileus virginianus texanus. Besides, as according to the literature GPx4 is conserved among species, we could assume that the annotation of this protein is missing for Equus caballus in SelenoDB. (see introduction)
In regard to GPx5, the most significant hits belonged to the scaffolds NW_018327251.1, NW_018337286.1 and NW_018340754.1.The best alignment score (994) corresponded to the first scaffold, although there was a big gap in the reference sequence. Besides, Seblastian predicted the sequence of GPx5 from Equus caballus as the GPx6 of the species Capra hircus.
The scaffold NW_018327251.1 was also present in GPx6 from Equus caballus, with a better alignment and a score of 999. Nevertheless, the gap in the sequence of reference was still present.
Despite the gap, the protein started with the amino acid methionine in both GPx5 and GPx6, so we conclude that the gap could be due to the output from fastasubseq, which is longer than the protein so the programs align it to other sequences that do not correspond with the protein itself.
Even though there was a SECIS prediction for this scaffold, we could assume that GPx5 is an homologue protein of Gpx6 present in Odocoileus virginianus texanus genome, which conserves a part of the sequence but has lost the selenoprotein identity. Besides, the information found in the bibliography about this protein bear out our conclusion (see introdcutionlink).
From the multiple hits found for GPx6, we chose the hits from the scaffold NW_018327251.1, which had the higher identity percentage and lenght. Although the results from t-coffee showed a big gap in the alignment, the seblastian prediction showed a very good alignment and a single SECIS element. There was also a presence of a selenocysteine in the second exon. Regarding to the gap in the t-coffee results, it is discussed in the results of GPx5 (see above).
Both the alignment and the SECIS prediction determinate that GPx6 is a selenoprotein present in the genome of Odocoileus virginianus texanus, encoded by a gene with five exons, according to exonerate program. Nevertheless, in SelenoDB the gene encoding for the protein is divided in multiple exons, and it has the SECIS element missing. The difference in the number of exons could be because we did not perform an exhaustive exonerate, and the missing SECIS element in SelenoDB may be due to an incorrect annotation of the protein in the database:
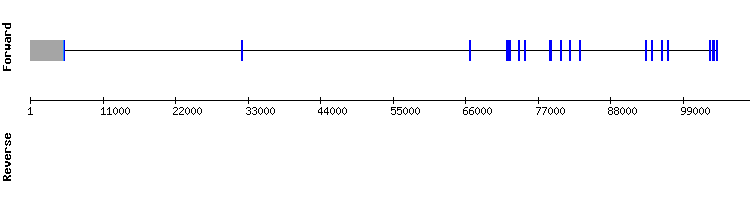
The hits with the best identity and length corresponded to the scaffolds NW_018338175.1 and NW_018334874.1, but only the first one showed an almost perfect alignment in t-coffee, with a score of 1000. It was divided in eight exons according to SelenoDB and it did not show a selenocysteine alignment. Moreover, Seblastian did not predict a sequence alignment nor a SECIS element.
We also compared the genome of Odocoileus virginianus texanus with the GPx7 query from the species Homo sapiens and a good sequence alignment was observed, with a score of 994. However, the gene encoding for this protein was divided in three exons (according to SelenoDB) instead of eight.
GPx7 gene from Equus caballus:
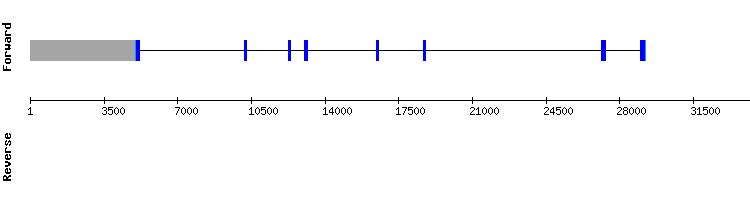
GPx7 gene from Homo sapiens:
From this information, we conclude that GPx7 is a partially conserved protein which has lost its selenoprotein identity in the species Odocoileus virginianus texanus, Equus caballus and Homo sapiens.This conclusion is consistent with the information we found in the bibliography (see intrductionlink).[a]
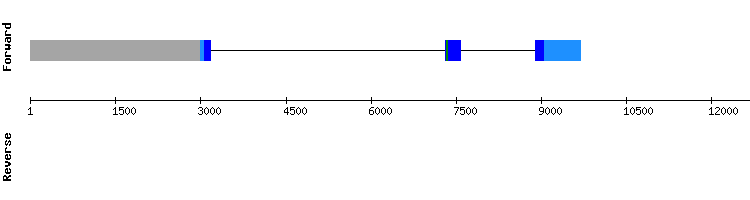
The scaffold with the best hits was NW_018334927.1, which showed an alignment score of 999 and a gap of three amino acids in the first three positions.
A SECIS element was predicted, but it was not consistent with the position of the protein (relative position of the SECIS 10541 - 10460 and relative position of the protein 9659 - 13480). Moreover, none selenocysteine alignment was shown.
From this information we conclude that GPx8 is a protein present in Odocoileus virginianus texanus genome, which has lost its selenoprotein identity but has conserved the sequence of the ancient SECIS element. This result is consistent with the information we found in the bibliography about this protein (see introduction).[b] Nevertheless, these results do not match with the information in SelenoDB, as the gene coding for GPx8 of Equus caballus appears to have a single exon and a SECIS element. However, the gene coding for the same protein in the specie Homo sapiens do not show the SECIS element and is divided in three exons. We attribute these differences to a possible incorrect annotation of the protein in the specie Equus caballus.
GPx8 gene from Equus caballus:
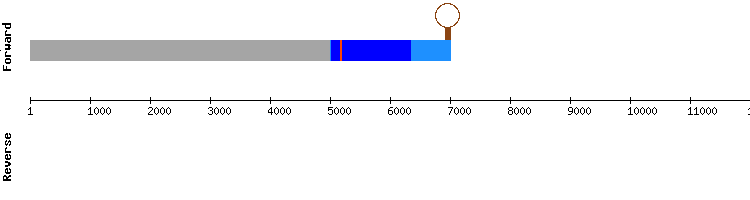
GPx8 gene from Homo sapiens:
GPxa, GPxb and GPxc are all proteins that were present in SelenoDB as GPx, thus we called them "e;a"e;, "e;b"e; and "e;c"e; following the order of appearence in the data base.
The protein GPxa had only two hits, both pertaining to the scaffold NW_018337286.1, which is the same scaffold we chose for the protein GPx1 of Homo sapiens and GPxb. The results did not show a very good sequence alignment (score of 974) and there was not a selenocysteine alignment although there were selenocysteine residues in both sequences. Besides, Seblastian did not predict any SECIS element for this protein.
It is not surprising to find the same scaffold for both GPxa and GPxb since the absolute positions are almost the same. Therefore, we thought that both sequences would be the same. Nevertheless, when comparing the query and the scaffolds sequences of GPxb and GPxa, they did only match in a few amino acids. This made us think that GPxa could be a duplication of GPxb found in the genome of Equus caballus, which has suffered multiple mutations losing its selenoprotein identity.
From the numerous hits found for GPxb, we chose the hits with best identity percentage and lenght, pertaining to the scaffolds NW_018337286.1 and NW_018336977.1.The first one is the same scaffold chosen for GPxa and GPx1 and is the one that got the best results, with an score of 999 and an almost identical alignment to the one of GPx1. However, it had a gap of three amino acids in the final positions of the sequence. A selenocysteine alignment was also observed, but Seblastian did not predict a SECIS element. The reason for the missing SECIS prediction could be that the output of fastasubseq was too short or the program in charge of the prediction was not sensitive enough to predict all of them.
Despite the sequences of GPxb and GPx1 are identical, the genes that encode for the proteins are formed by a different number of exons and are located in different strains, according to the information in SelenoDB. Nevertheless, we conclude that GPxb is the same selenoprotein as GPx1 from Homo sapiens and that it is present in Odocoileus virginianus texanus genome.
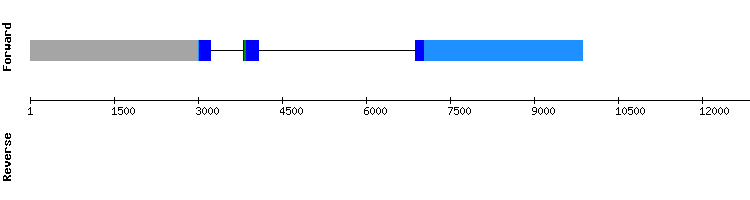
GPxb
From the multiple hits found for GPxc, we analyzed the ones corresponding to the scaffolds NW_018337286.1, NW_018340754.1, NW_018336664.1 and NW_018336977.1. The best alignment corresponded to the scaffolds NW_018337286.1 and NW_018340754.1, even though the alignment scores were only 981 and 974, respectively. Neither of both had a selenocysteine alignment when aligned with t-coffee and they had small gaps in the middle of the sequence. Besides, a SECIS element was only predicted in the scaffold NW_018340754.1 and Seblastian predicted the protein GPx2 for that sequence. Comparing the sequences of GPxc and GPx2, we observed that the sequence of GPxc is part of the sequence of GPx2, which is longer.
Regarding the gene structure in SelenoDB, it is observed that both proteins are encoded by genes with a different number of exons, but we conclude that it may be due to a bad annotation of the protein in the data base, as the SECIS element is missing too. So, GPxc could be either a duplication of GPx2 which has lost the selenoprotein identity or the GPx2 protein itself but more poorly annotated in the data base.
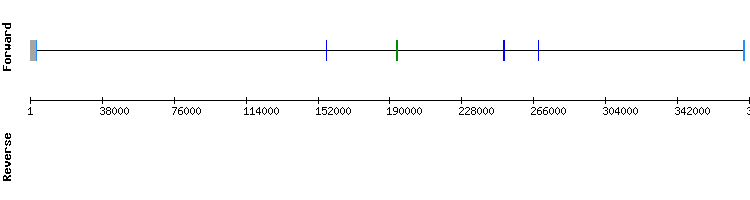
Methionine sulfoxide reductase A (MsrA) family
One scaffold has been found for MsrA: NW_018334876.1 as the most significant, with an identity percentage of 85. Regarding the t-coffee analysis, the results showed a good alignment between sequences with a score of 999, although there was a gap in the last amino acids. There was not a selenocysteine alignment.
In the Seblastian analysis, a single SECIS was found in the relative position 16340 - 16256 of the negative strand with an Infernal score of 10, which is consistent with the relative position of the first exon of the gene. This results do not match with the gene shown in SelenoDB, which has three exons in the positive strand and it contains a selenocysteine residue. Nevertheless, in the gene from Homo sapiens:, which is better characterized, a cysteine residue is found instead of a selenocysteine and there is not a SECIS element, so we could conclude that MsrA is a selenoprotein homologue that has conserved the cysteine residues among different species.
MsrA gene from Homo sapiens:
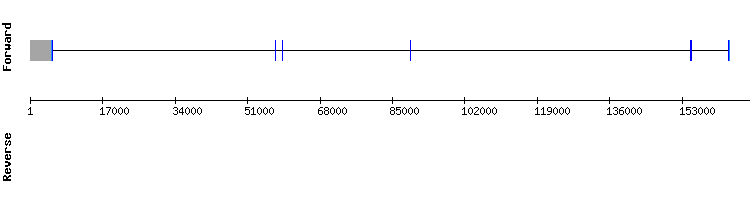
Selenoprotein H (SelH) family
Three scaffolds have been found for SelH. We did not discard any of them because the alignment observed in the t-coffee was good and had a selenocysteine alignment.
All of them showed SECIS. And all of the SECIS were properly located outside of the exons.Since it was surprising that three of the scaffolds had very good and similar results, we compared the three exonerate results.
In conclusion, it seems that selenoprotein H is present in the Odocoileus virginanus texanus genome and it is conserved when compared to the same protein for Equus caballus.
Moreover, the SelenoDB database also shows a four exon selenoprotein with its selenocysteine and SECIS.
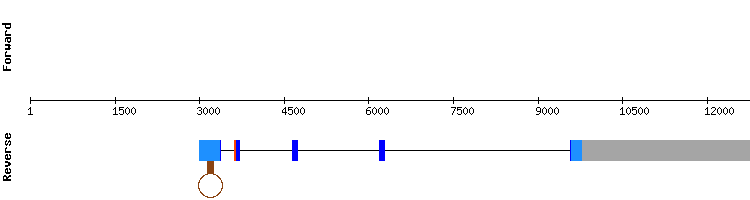
Selenoprotein I (SelI) family
One scaffold was found for SelI: NW_018336909.1. This scaffold had a high identity percentage in all of its hits. Regarding the t-coffee, results showed a very good alignment with a score of 1000. The sequences alignment of the t-coffee started with a methionine amino acid and a selenocyteine was also shown.
Regarding the Seblastian analysis, a single SECIS was found in the relative position 69477 - 69554. This is consistent with the fact that the last exon of the positive strand is at the relative position 68005.
Even though the gene encoding for this protein in Equus caballus does not show a SECIS element nor a selenocysteine in SelenoDB, we think that it can be due to an incorrect annotation of the protein in the database, as it is one of the latest discovered selenoproteins. So, we conclude that SelI is a selenoprotein present in Odocoileus virginianus texanus genome.
Selenoprotein K (SelK) family
Two different scaffolds were found for Selenoprotein K: NW_018335033.1 and NW_018341531.1. Both of them had proper e-values and high percentages of identity in their hits. However, when analyzing the t-coffee of NW_018335033.1, no alignment of a selenocysteine amino acid was found. Moreover, the alignment of these scaffold had a little gap at the beginning of the protein. Thus, this scaffold was discarded.
On the other side, the scaffold NW_018341531.1 showed a very good and complete alignment in the negative strand with a selenocysteine in the sequence.
In regard to the SECIS, three of them were found. Two of which were located in the negative strand, and one in the positive strand. Since our sequence is in the negative strand, we discarded the SECIS located in the positive strand. The other two SECIS were consistent options since they were located in positions before the closest exon of the 3'- UTR extreme.
Our results were not similar to the genome structure of Equus caballus for the same protein in SelenoDB as the gene is situated in the positive strand. Therefore we compared the query from Homo sapiens and results were consistent. Only one SECIS was found for this query in the negative strand at the relative position (28960-28873) with match with the location of the closest exon to the 3'-UTR extrem from the exonerate output (relative position 29336 - 29426).
This final result is consistent also with the structure found in SelenoDB, which is also located in the negative strand and has one SECIS. Therefore, we conclude that this is a conserved selenoprotein present in the Odocoileus virginianus texanus genome.
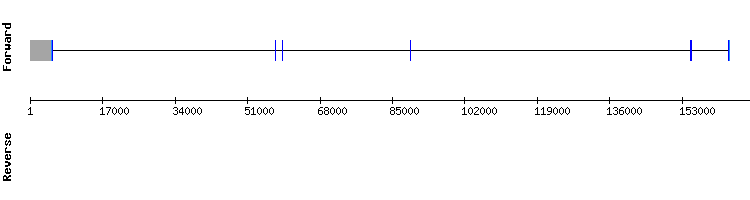
Selenoprotein M (SelM) family
One scaffold was found for selenoprotein M: NW_018338198.1. Even though it had a very low e-value, its percentage of identity only reached 65.71.
Regarding the t-coffee, results showed a very good alignment with a score of 1000. The sequences alignment of the t-coffee started, as expected, with a methionine amino acid. When analysing the Seblastian results, it was shown that the alignment for the selenocysteine residue was in the relative position 29357.
Only one SECIS was found in the relative position 30357 - 30429 of the positive strand. The position of the SECIS is consistent since the last position of the last exon of the positive strand is 30314. Therefore, we can conclude that Selenoprotein M looks very likely to the same protein found in the Equus caballus genome (with 5 exons), even though the gene annotated in SelenoDB does not show a SECIS element nor a selenocysteine.
Selenoprotein N (SelN)
A single scaffold was found for selenoprotein N: NW_018336176.1, which had a percentage of identity of almost 100.
In regard to the t-coffee, results showed a very good alignment with no gaps in the negative strand. However, the first amino acid of the alignment is not a methionine.
Regarding the Seblastian analysis two SECIS were found for this protein. However, only one of them was located in the negative strand as the our protein (from exonerate results). This SECIS, with an Infernal score of 40.73 was located in the relative position 28878 - 28808 of the negative strand.This position is consistent as the first position of the first amino acid of the negative strand is 30000.
Our results did not match with the SelenoDB database, neither for Equus caballus nor Homo sapiens, as the strand sense did not match.
Therefore, we can conclude that Selenoprotein N is present in Odocoileus virginianus texanus genome and it is conserved when compared to the other two species.
SelN gene from Equus caballus:
SelN gene from Homo sapiens:
Selenoprotein O (SelO) family
SelOa and SelOb from Equus caballus were present in SelenoDB as SelO. Thus, we called them "a" and "b" according to the order of appearence in the data base in order to differentiate them.
We chose one of three scaffolds as the most significant, the one with a highest identity percentage: NW_018332529.1. The alignment obtained by t-coffee was good, with a score of 997, but it did not start with a methionine. From Seblastian we did not obtain any SECIS nor protein prediction. When checking SelenoDB database, no SECIS nor selenocysteine are found either.
In conclusion, taking all the information into account, we can conclude that either a duplication happened losing the selenoprotein characterisation or that the annotation of SelenoDB may not be correct for Equus caballus.
From the second isoform, we chose NW_018337310.1 as the most significant scaffold. The alignment with t-coffee is good (score of 995) but only a short part of the protein was aligned. We did not obtain any alignment nor SECIS from Seblastian. We also checked SelenoDB database and no SECIS nor selenocysteine were found.
In conclusion, we think that either a duplication occurred losing the selenoprotein characterisation of the protein or that the annotation of SelenoDB may not be accurate for Equus caballus.
Three scaffolds were found for Selenoprotein O but only one was large enough to be selected: NW_018342064.1. Regarding the t-coffee analysis, it showed a very good alignment with only one gap of one amino acid at the beginning of the sequence. It had a score of 995 and showed an alignment of selenocysteine at the antepenultimate position.
In regard to the SECIS, a single one was predicted at the relative position 29933 - 29863 of the negative strand. This position was consistent as the first amino acid of the negative strand is located at relative position 30000.
Therefore, we can conclude that Selenoprotein O is well conserved among these species and is present in the genome of Odocoileus virginianus texanus in a nine exon gene. Nevertheless, according SelenoDB results there are only four exons for Equus caballus gene and it is located in the positive strand. Moreover, in regard to Homo sapiens genome, the number of exons match (nine) but they are also found in the positive strand.
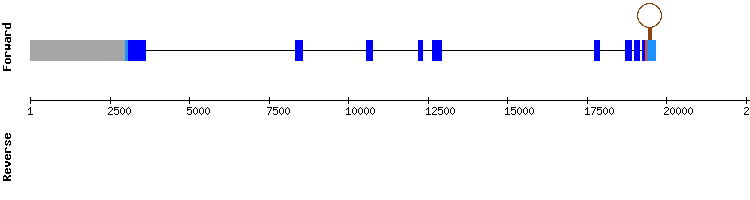
Selenoprotein P (SelP) family
One scaffold was found for selenoprotein P: NW_018330451.1, with a proper e-value, length and percentage of identity. In regard to t-coffee analysis, it had a good alignment with some small gaps of only one amino acid in the middle of the sequence and a score of 981. There is a selenocysteine alignment in the first of five exons.
Moreover, two SECIS were found in the relative position 1117 - 1046 and 668 - 603 of the negative strand. Both SECIS positions are consistent since the 3' UTR starts at the relative position 1366.
Therefore, we can conclude that SelP is found in the Odocoileus virginanus texanus genome and it seems to have been very conserved among evolution. Moreover, in SelenoDB we can also see that in Equus caballus this protein has five exons in which there is a selenocysteine but it is situated at the forward strand, which is not consistent with our results. Thus, we checked it in the SelenoDB 1.0 data base from Homo sapiens and the gene is also found in the reverse strand, as shown in our results.
SelP gene from Equus caballus:
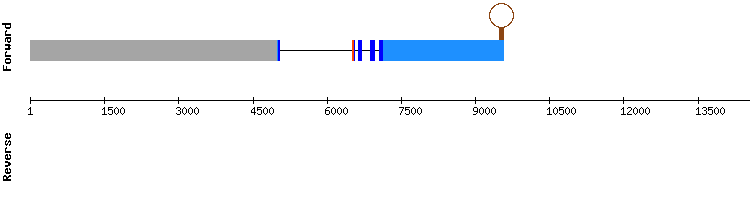
SelP gene from Homo sapiens:
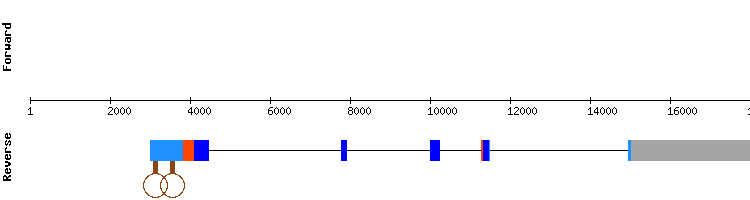
Selenoprotein R (MSRB) family
One scaffold has been found for MSRB1: NW_018343886.1 This scaffold has a very high rate of identity which is around 90%. Regarding the t-coffee analysis, results showed a very good but short alignment between sequences, with a score of 1000. A selenocysteine alignment was also predicted.
The SECIS prediction was consistent with the relative position of the last and third exon (SECIS relative position 28235 - 28165 and exon relative position 29976 - 30116), as they are located in the negative strand. Nevertheless, we have found that in SelenoDB the gene encoding for MSRB1 of Equus caballus is located in the positive strand. But, when looking at MSRB1 of Homo sapiens, the gene location is in the negative strand, matching our results.
As the selenocysteine and the SECIS element are conserved and the t-coffee and Seblastian alignment are coincident, we can conclude that MSRB1 found in Odocoileus virginanus texanus genome is a conserved selenoprotein among these species.
MSRB1 gene from Equus caballus:
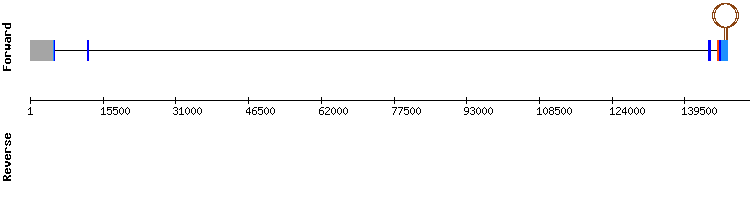
MSRB1 gene from Homo sapiens:

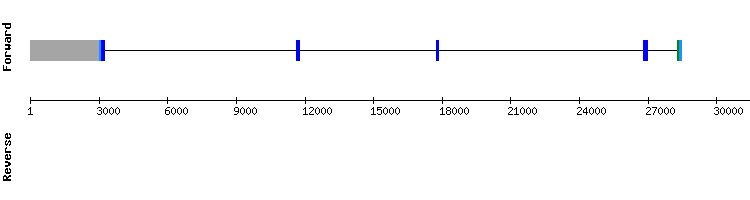
For this protein we obtained three scaffolds but we chose NW_018332573.1 as the most significant, as it had the higher identity percentage and length. The results from t-coffee analysis showed a good alignment between sequences with a score of 986, although the initial part of the protein showed a gap and none selenocysteine was shown.
Although Seblastian did not predict any alignment, three SECIS were shown for this protein. Two of them were found in opposite sense strand and one of them in the same strand (positive), but too far from the last exon of the coding gene (exon relative position: 30244-30402 and SECIS relative position: 228294 - 228362).
Therefore, we conclude that this protein is an homologous protein with four exons that has lost its selenoprotein identity in this species but still contains a conserved part of the original sequence. It could also be that we have not chosen an optimal length that include the genomic sequence as we did the fastasubseq for this protein.
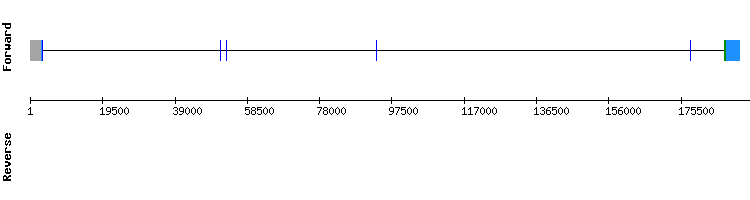
We chose the best of the three scaffolds obtained for this protein: NW_018329462, which was the one with a higher percentage of identity (100). Regarding the t-coffee results, we obtained a good alignment (score of 985) although in the beginning of the protein appeared a gap and none selenocysteine were found in the alignment, as it happened with MSRB2.
According to Seblastian, no protein prediction or SECIS were obtained.
Therefore, we conclude that either this protein is an homologous protein that has lost its selenoprotein identity or that we have not include an optimal length for this protein as we run the fastasubseq. This protein is found in Odocoileus virginianus texanus in a 5 exon gene as it was in Homo sapiens genome.
Selenoprotein S (SelS) family
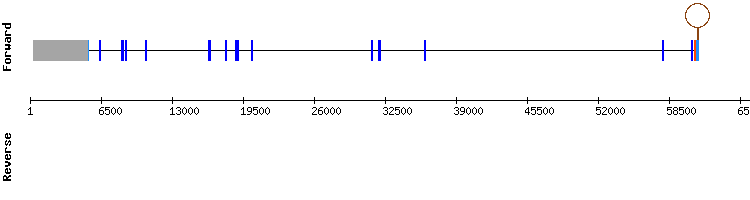
One scaffold has been found for SelS: NW_018334967.1. This scaffold has a very high percentatge of identity. Regarding the t-coffee analysis, results showed a good alignment between sequences with a score of 994. However, the initial part of our scaffold sequences had a gap. Also a selenocysteine was found. Therefore, the alignment is not good for the earliest positions. This can be due to a problem of sensitivity of the program.
The Seblastian alingment predicted a protein with 6 exons and found one SECIS structure between relative positions 27756 - 27677 in the negative strand with an infernal score of 27.05. This position of the SECIS is consistent with the relative position of the last exon (3'-UTR) of this protein (37789- 37879).
When compared with the genome structure for this protein in Equus caballus in SelenoDB the number of exons and the sense of the strand were not compatible. But when compared the output of our prediction with a Homo sapiens query and the structure in SelenoDB with the results were almost the same.
SelS gene from Equus caballus:
SelS gene from Homo sapiens:
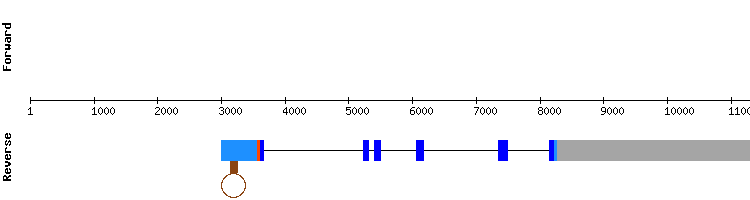
Therefore, we can conclude that Selenoprotein S found in the Odocoileus virginanus texanus genome, is very similar to a protein found in the horse genome, with 6 different exons.
Selenoprotein T (SelT) family
One scaffold has been found for SelT: NW_018336329.1. This scaffold had an identity percentage around 90. Regarding the t-coffee analysis, results showed a very good alignment between sequences with a score of 1000. However, the alignment of the selenoprotein did not start with a methionine, which was an unexpected result as most proteins start with this amino acid. This may be a result of a bad annotation of the protein SelT or it could be one of the exceptional proteins that do not start with a methionine.
According to the results from Seblastian, the selenocysteine residue was located at the relative position 30002.
A SECIS element was found in the relative positions 36052 - 36126 with an infernal score of 31.45. The position of the SECIS is consistent with the relative position of the exon in the 3' extreme (35314 -35435) which is located in the positive strand.
Since the results compared to the gene from Equus caballus: in SelenoDB were not consistent (it did not show a SECIS element nor a selenocysteine), we compared the specie of interest with the protein from Homo sapiens. The number of exons showed and the SECIS and selenocysteine residue prediction matched with our result.
Therefore, we conclude that Selenoprotein T is found in the Odocoileus virginanus texanus genome, encoded by a gene divided in five exons, as in the Homo sapiens species.
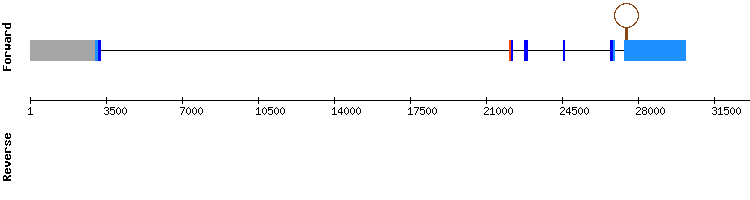
Selenoprotein U (SelU) family
Three scaffolds have been found for SelU1: NW_018343031.1, NW_018335057.1, NW_018338731.1.
Nevertheless, we discarded two of them because the alignment observed in the t-coffee was not good or they did not have a proper e-value and percentage of identity.
NW_018335057.1 is the scaffold we selected. Regarding its t-coffee results, it was well aligned and had a proper score of 1000. However, there is a gap in the earliest amino acids of the alignment. When looking at exonerate results, it is shown that the gene is in the positive strand and has 5 exons.
No selenocysteines were found along the sequence alignment. Finally, regarding the SECIS, none of them were found either. Moreover, we checked the results for this protein in SelenoDB data base for both Equus caballus and Homo sapien and both of them had also 5 exons situated in the positive strand, which is consistent with our results.
Therefore, we can conclude that Selenoprotein U1 is not a selenocysteine containing protein, but it is still a well conserved protein among this species.
SelU1 gene from Equus caballus:
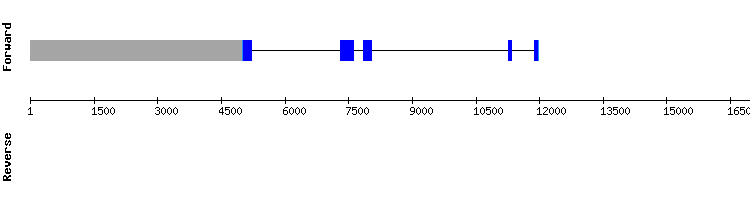
SelU1 gene from Homo sapiens:
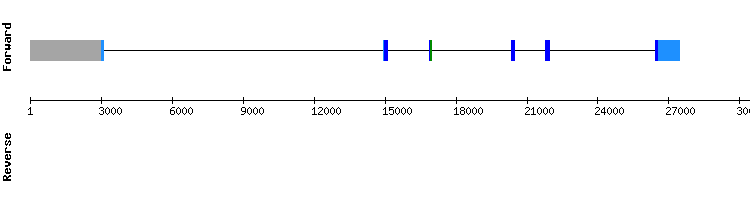
One scaffold has been found for SelU2: NW_018335050.1 with an average percentage of identity of 90. Moreover, regarding the t-coffee analysis, results show a very good alignment between sequences with a score of 1000. The sequences were located at the positive strand. However, the alignment of the selenoprotein does not start with a methionine, which is an unexpected result as most proteins start with this amino acid. No selenocysteine are found along the protein sequence and no SECIS could be found either in our scaffold. Therefore, we can conclude that SelU2 is not a selenoprotein since it does not have the mandatory amino acid to be.
We checked for the Homo sapiens genome. We did not find any selenocysteine in the t-coffee alignment either. Therefore, we could conclude that probably SelU2 is not a selenoprotein, but an homologue that has lost the selenocysteine residue.
Moreover, we checked the SelenoDB data base no selenocysteines nor SECIS were found either in Equus caballus and Odocoileus virginianus texanus. In regard to Homo sapiens, the number of exons matches since we found 6 exons in our protein. Nevertheless, the gene is located in the reverse strand according to SeleneDB. In regard to Equus caballus, SelenoDB shows 3 exons while we found 6 of them. However, the strand is located in the positive strand which is consistent with our results.
Selenoprotein W (SelW) family
SelWa and SelWb are proteins that were present in SelenoDB as SelW for Equus caballus. Thus, we called them "a" and "b" following the order of appearence in the data base.
Five different scaffolds were detected for selenoprotein Wa. We selected scaffolds NW_018338398.1 and NW_018342431.1 according to the e-value and the lenght.In regard to the t-coffee analysis, an almost perfect alignment with a score of 1000 was shown for both scaffolds, and a selenocysteine alignment was also shown. Moreover, both sequences were very similar but were located in very distant positions in the genome, according to the output of the tblastn (2462442 - 246223 and 368177 - 367968). Thus, we conclude that it may be due to a duplication of the sequence.
Seblastian predicted one SECIS element only for the scaffold NW_018338398.1, at the relative position 4903 - 4825 of the negative strand. This relative position is consistent with the relative position of the only exon as showed in exonerate (relative position of 4952 in the negative strand).
Therefore, we conclude that SelWa is a conserved protein in both Odocoileus virginianus texanus and Equus caballus species, since sequences are well aligned. Nevertheless, the protein of Equus caballus annotated in SelenoDB is shorter than the predicted in our results and the gene is divided in a different number of exons:
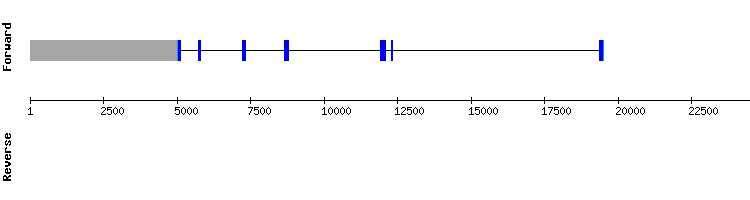
In regard to SelWb, we chose the scaffold with the best parameters, even though the identity percentage and the lenght were not very high: NW_018330951.1. The programs t-coffee and Seblastian did not show any sequence alignment, so we concluded that this protein may not be conserved in Odocoileus virginianus texanus, or that it is not well annotated for the species Equus caballus in SelenoDB.
Thioredoxin reductase (TXNRD) family
Five scaffolds have been found for TXNRD2. Nevertheless, we selected only one of them since the other showed a low identity percentage and a very bad alignment when the t-coffee was developed. We selected scaffold NW_018335521.1. This scaffold showed a good alignment with a score of 976. However, the first amino acids of the alignment are either incompleted or do not match.
Regarding the selenocysteine, an alignment of selenocysteine is found in the penultim position of the protein.
A single SECIS was found in the relative position 29114 - 29046 of the negative strand with an infernal score of 31.41. The relative position of the SECIS is consistent with the fact that the start of the first exon in the negative strand is 30000.
When checking the SelenoDB database, the number of exons and the strand sense for Equus caballus does not match with our results. However, when looking at Homo sapiens gene it has 17 exons and is also found in the positive strand.
In conclusion, TXNRD2 is a selenoprotein present in Odocoileus virginianus texanus genome, encoded by a gene with 17 exons.
TXNRD2 gene from human
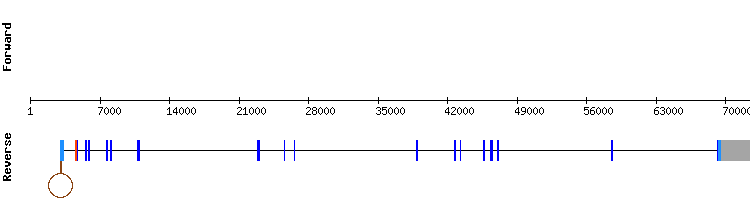
Seven scaffolds have been found for TXNRD3. Nevertheless, we selected only one of them since the other showed a low identity percentage. In some cases, we discarded them because the length of the hits were not long enough to be significant.
This scaffold showed a good alignment with a score of 1000 in a t-coffee alignment. However, the first amino acid is not a methionine, which is an unexpected result as most proteins start with this amino acid.
Interestingly, an alignment of selenocysteine is found in the penultim position of the protein, as it happened with TXNRD2.
Two SECIS were found. We selected the one with a higher infernal score of 17, which was in the relative positions 53168 - 53246 of the positive strand. The relative position of the SECIS is consistent with the fact that the start of the last exon in the negative strand is 53039.
In conclusion, TXNRD3 is a selenoprotein present in Odocoileus virginianus texanus,encoded by a gene divided in 17 exons. However, in SelenoDB we only observed 3 exons in the gene of Equus caballus codifying for this protein.
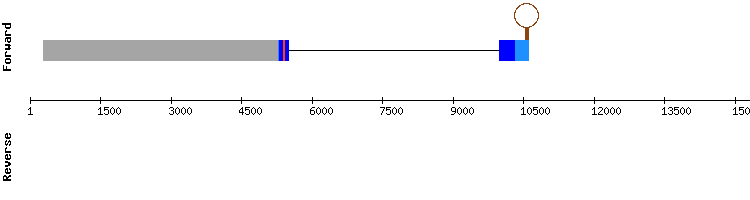
Six scaffolds were found this protein, but only one was selected for its t-coffee good alignment: NW_018335768.1. In regard to t-coffee results (with a score of 999), there were two big gaps at the beginning and end of the sequence which may be the reason why no selenocysteine alignment was found. No SECIS element was found using Seblastian.
Moreover, the results from SelenoDB of Equus caballus genome showed a selenocysteine and one SECIS in the forward sequence of a fifteen exon selenoprotein. Ths result does not match with our protein, since it had no SECIS and only 6 exons.
Therefore, it is difficult to draw conclusions, as the protein TXNRDa may not be well annotated in SelenoDB or our program may have not been sensitive enough to detect the proper SECIS.
MACHINERY
Eukaryotic elongation factor (eEFsec) family
The most significant scaffold for this protein NW_018329262.1. The alignment with t-coffee was good and had a score of 995, but no selenocysteine was shown. Also, none of the proteins aligned begun with methionine. No SECIS nor proteins were predicted by Seblastian.
Therefore, and according with the literature (where eEFsec is presented as a specific elongation factor which guides the Sec-tRNA to the UGA codon and will allow its translation into a selenocysteine (1,7), we deduce that this protein is part of the selenoproteome but may not be a selenoprotein itself because it does not have any selenocystein in its sequence.
This results are consistent with those showed by SelenoDB for the same protein from Homo sapiens, but not with the gene from Equus caballus:. However, this protein has not been yet well characterized in any species so we can not confirm whether it is a selenoprotein nor the number of exons of the gene.
eEFsec gene from Equus caballus:
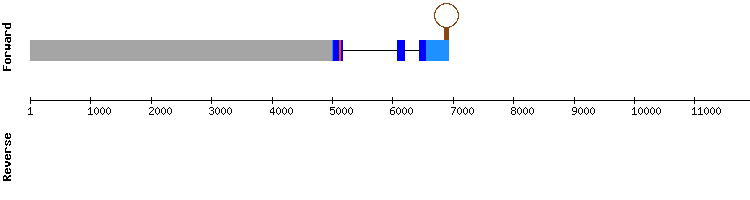
eEFsec gene from Homo sapiens:

Phosphoseryl-tRNASec kinase (PSTK) family
We only obtained one significant scaffold for this protein: NW_018335054.1. T-coffee showed a good alignment with a score of 998. However, none selenocysteine was shown in the protein sequence alignment. Besides, we did not obtain any alignment prediction or SECIS with Seblastian.
This is consistent with the information found in the bibliography (introdcutionlink), as it is known that this protein is involved in the selenoprotein biosynthesis and is not a selenoprotein itself. This is why we conclude that this protein is part of the selenoproteome prenset in Odocoileus virginianus texanus and encoded by a gene with two exons.
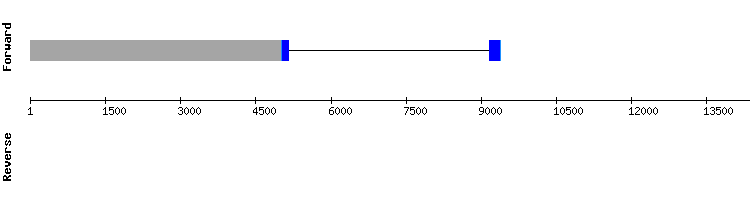
SECIS binding protein (SBP2) family
SBP2a and SBP2b from Equus caballus were present in SelenoDB as SBP2, as the external ID was missing. Thus, we called them "a" and "b" according to the order of appearance in the data base so we could differentiate them.
The hits with best identity percentage and length of this protein pertained to the scaffold NW_018332373.1, which showed a good sequence alignment in t-coffee, with a score of 997. It did not show a selenocysteine alignment and Seblastian could not predict a SECIS element. However, in SelenoDB the gene show a selenocysteine and a SECIS element:
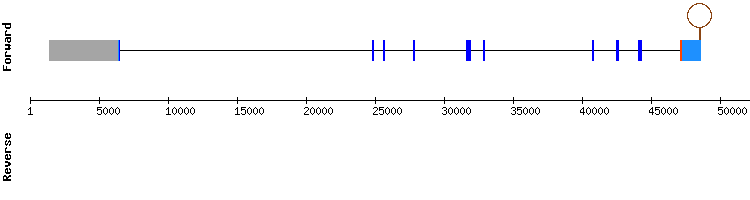
The information from SelenoDB is not consistent with out results nor the information found in the bibliography (see introduction), as SBP2 is not a selenoprotein itself but a protein involved in the synthesis of selenoproteins.
Two scaffolds were found for SBP2b but only one of them was selected: NW_018343786.1. The other one was discarded because it was also found for SBP2a with a higher percentage of identity. In regard to t-coffee results, they showed a good and large alignment. However, two long gaps appear at the beginning of the sequence. It had a score of 994.
Moreover, there was no selenocysteine alignment and no SECIS could be predicted with Seblastian program. This is consistent with the fact that this protein is not considered a seleprotein as it its only involved in their synthesis. However, as it happens with SBP2a, SelenoDB shows the presence of both SECIS and selenocysteine, which is not a consistent result with the literature about selenoprotein synthesis machinery.
Moreover, the scaffold selected for this protein matches with the scaffold used in the same protein for Homo sapiens. In regard to the Homo sapiens protein, no SECIS nor selenocysteine was found either.
In conclusion, SBP2b in Equus caballus seems to be the homologous protein of SBP2 in Homo sapiens, even though the results from SelenoDB are not exactly the same. In addition, it is present in Odocoileus virginianus texanus genome.
SBP2b gene from Equus caballus:
SBP2 gene from Homo sapiens:
Selenocysteine synthase (SecS) family
We chose NW_018334953.1 as the most significant scaffold for this protein as it had the best identity percentage. The results from t-coffee showed a good alignment (score of 1000) and both prediction started with a methionine. None selenocysteine was found in our results.
About the Seblastian results, we did not obtain an alignment prediction with this program but two SECIS were found. None of them were consistent as they were not well situated in the 3' UTR region.
According to the literature (1), this protein is known to be involved in the selenoprotein biosynthesis but is not a selenoprotein itself. With this results we conclude that this protein is part of the selenoproteome present in Odocoileus virginianus texanus encoded by eleven exons, the same as in the gene from Homo sapiens:.
Selenophosphate synthetase (SEPHS) family
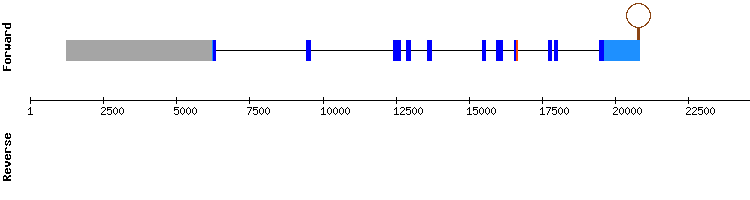
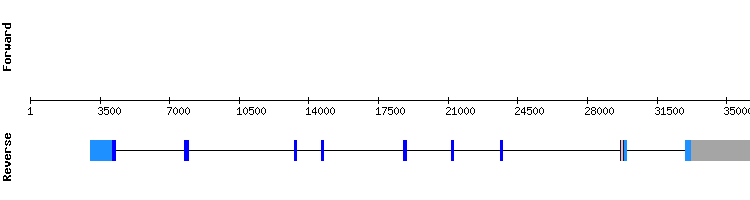
We chose NW_018341730.1 as the best scaffold because it was the longest of the four scaffolds obtained. The results from t-coffee showed a good alignment (score of 1000) although none selenocysteine was shown.
We did not obtain any protein alignment nor SECIS prediction in Seblastian. However, in SelenoDB the gene showed a SECIS element in the 3'-UTR and a selenocysteine. This discrepance does not exist when we compare our results to the gene of this protein from the species Homo sapiens.
As found in the literature (1), SEPHS1 is one of the two Selenophosphate synthetase enzimes (SEPHS1 and SEPHS2) involved in the biosynthesis of selenoproteins, but it differs from its functional homologous, as SEPHS1 does not work as a direct factor.
Therefore, we conclude that SEPHS1 may be a protein that has lost its selenoprotein identity in Odocoileus virginianus texanus, among other species.
SEPHS1 gene from Equus cabalus
SEPHS1 gene from Homo sapiens:
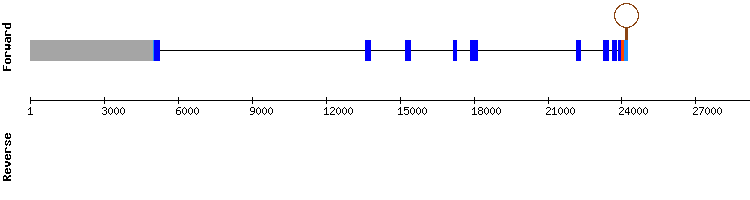
NW_018343187.1 was the most significant scaffold found and the only one which did not show a t-coffee alignment with a gap. The results found in the t-coffee showed a good alignment (score of 996) with the protein beginning with methionine and the presence of an selenocysteine.
We did a Seblastian and we obtained the prediction of a protein and one SECIS in the relative position 31810 - 31883 with an Infernal score of 27,25. This SECIS result is consistent with the exon relative position, as the closest exon to the 3' extreme is found in the position 29898 - 31244. This results indicate that the protein might be a selenoprotein as we also found a selenocysteine alignment.
According to the literature consulted, SPSH2 is a selenoprotein itself involved in the biosynthesis of selenoproteins (1,7). Therefore, we conclude that is a selenoprotein present in Odocoileus virginianus texanus genome, encoded by a gene with nine exons.
tRNA Sec 1 associated protein 1 (SECp43) family
SECp43a and SECp43b from Equus caballus were present in SelenoDB as SECp43. Thus, we called them "a" and "b" according to the order of appearence in the data base so we could differentiate them.
Both of them showed the same scaffold as the most significant: NW_018337598.1, but SECp43a showed too short hits. For this one we did a t-coffee and obtained a good alignment (score of 1000) but the protein was only aligned in a very small part (only by 30 nucleotide). Therefore, we chose the isoform SECp43b which gave a scaffold with a longer length. We did a t-coffee and obtained a good alignment (score of 1000) but none selenocysteine was shown in the sequence.
We did not obtain SECIS element or alignment predictions as we did the Seblastian analysis.
According to the literature (1), this protein is known to be involved in the selenoprotein biosynthesis but is not a selenoprotein itself. So we could conclude that SECp43b is a protein from the selenoproteome which is present in the Odocoileus virginianus texanus genome.