Introduction
Selenium and selenoproteins importance
Selenium (Se) is an essential micronutrient for living beings, but toxic in large amounts. It has a very important role in essential human functions, since it takes part in antioxidant cellular processes, thyroid hormones metabolism and immunity (1). Our bodies contain about 14 mg of Se, and every cell in a human body contains more than a million selenium atoms (2). Its deficiency has been related to numerous physiopathological conditions as cardiovascular diseases, neuromuscular disorders, cancer or inflamation (3).
FIGURE 1. The 21st amino acid: Selenocysteine. Adapted from Lobanov AV et al, 2010.
A selenoprotein is any protein that includes a selenocysteine residue (U), which is composed by selenium. This amino acid is considered the 21st of the genetic code, because it is not among the usuals. Its coded by UGA, which may also be an stop codon (7).
Selenoproteins are very extended in all vertebrates, but they are not found in all living beings. For example, there is anything reported in yeasts and plants, which have similar proteins with cysteine. Consequently to this similarity, it can be explained why orthologues and paralogues with cysteine instead of selenocysteine can often be found(9). As SelenoDB reports, there are many selenoproteins classified: 37 different families in humans, 16 families in Drosophila melanogaster, and about 26 families in Zebrafish.
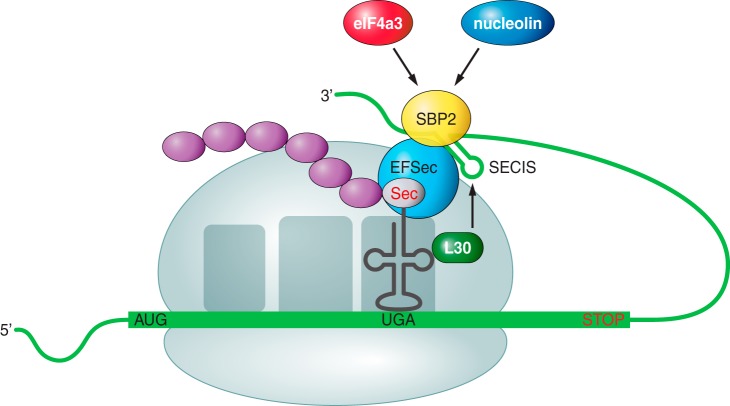
FIGURE 2. Sec incorporation to the growing polypeptide. Adapted from Labunskyy VM et al, 2014 (3).
As mentioned before, SECIS is an essential element to direct the process towards the selenocysteine (Sec) synthesis instead of stopping the protein translation. This SECIs element will be recognized for a SECIs Binding Protein (SBP2), and then eEFsec, a specific elongation selenoproteins factor, is recruited. After, tRNA together with a selenocysteine (Sec) will be called up and approximated to the UGA codon in the mRNA transcript, in order to incorporate the amino acid to the growing protein chain. The translation will continue normally until another stop codon is found by the translation machinery, which will end the process (5,11).
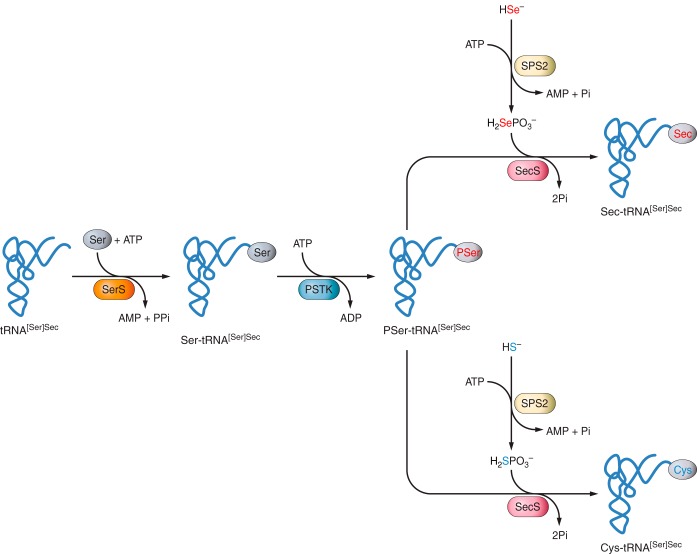
FIGURE 3. Sec synthesis. Adapted from Labunskyy VM et al, 2014.
This mechanism is very similar to the three life domains, suggesing that it appeared before the separation of these three domains, and just once during evolution. This ancestral capacity ability was lost in some lineages because they are not capable of incorporating a selenocysteine to a growing peptide (3).
The Selenoproteins research in genomes that have not been studied previously, like Xipophorus cochianus, starts by searching for already known selenoproteins from other species, found in SelenoDB databases. Then, analysis of SECIS elements should be performed. Is important to underline that these elements are located in the 3’ region, so when a possible candidate has a SECIS in the correct region and strand, it will probably be a selenoprotein in our specie.
Xiphophorus couchianus (The Monterrey Platyfish) is a type of freshwater fish belonging to the family Poeciliidae of the order Cyprinodontiformes. It is native to the vicinity of Monterrey, Mexico.
FIGURE 4. Xiphophorus Couchianus (see).
This specie has a short body, convex loin, small head and very large scales. It has a brown color, a dark lateral band and each flake has a large brown mole. Regarding to its dimensions, males can measure up to 5.5cm while females can reach up to 7cm, who have a rounded belly. Their fins are colorless and their body is slightly elongated, not as much as the related X.variatus but longer than the pervasive X.maculatus from the South. Males and females are similar in appearance, nevertheless males are thinner and females can exhibit a pronounced gravid spot which makes them easier to identify.
In relation to its functions, selenium is an essential component of selenoproteins,which have an antioxidant role in the cell by blocking free radicals, and strengthening the immune system. These are usually enzymes responsable for detoxification, such as glutathione peroxidase, thioredoxin reductases and so on, which can work by their own or in combination with Vitamin E (1).
Identification of patients with inborn errors in some of these proteins, reveals their significance for human health (4). As said before, its deficiency has been related to pathologic conditions like cancer, for example, and this is because in some instances, selenium can inhibit chromosomal damage and avoid mutations (5,6).
In order to read this codon as a selenocysteine and continue the translation instead of stopping it, an special structure at the 3’ side of the DNA is required: the SECIS element (SelenoCysteine Insertion Sequence) (8). Related to selenocysteine molecular structure, it is important to emphasize that it is a cysteine analogue, although cysteine’s sulfur atom has been replaced by a selenium one, which is more reactive (3,9). The main problem related to the annotation of these proteins is that, as most programs assume that UGA is a stop codon, they are not recognized as selenoproteins.
The aim of the project is to identify selenoproteins in the Xiphophorus cochianus genome in order to well-annotate its selenoproteome and provide accurate information about phylogenetic evolution.
Many of the known selenoproteins are ancestral, but after milions of years of evolution many of these may have evolved due to genetic duplication, genes loss or conversion to cysteine. Therefore, not all mammals have the same proteins in their genomes.
There are also some differences between aquatic species and terrestrial species, as selenoproteins evolution is defined by ambiental conditions and selenium availability. For example, selenoproteome in fishes is bigger than in other species as a result of their need of greater protection in front of oxygen concentrations and their high selenium backup (10,11).
In the process described before, it is important to learn how the selenocysteine (Sec) is added to the chain. Sec is the only amino acid that has its own tRNA called Sec-tRNA[Ser]Sec. First of all, a Seryl tRNA synthetase promotes the addition of a serine (Ser) to a Sec-tRNA, obtaining a Seryl-tRNA. Subsequently, the Phosphoseryl-tRNA kinase (PSTK) will introduce a phosphate to this Seryl-tRNA, generating a Phosphoseryl-tRNA. The next step is the action of the Selenocysteine synthase (SLA), which will replace this phosphate for a selenium donor (H2Se-P), previously activated with a Selenophosphate synthetase (SPS2). At the end, a selenocysteine-tRNA will be developed, and the Sec will be ready to be incorporated to the growing polypeptide (3,5).
Genomes prediction and annotation
Despite that, there can appear many problems during this analysis for different reasons. First of all, as UGA is a dual codon and it can result in a Sec or in a stop codon, sometimes the programmes are not accurate enough and some false positive can be obtained. Moreover, the conservation level between species is low, especially in fishes. Additionally, it has to be considered the fact that the selenoproteins in humans are very well annotated contrary to other species as fishes, in which some discrepancies between different databases could be found. All in all, this demonstrates the lack of knowledge feasible in the area.
Xiphophorus couchianus

It is considered an omnivorous specie, with special preference for vegetables, and in reference to its reproduction it is ovoviviparous. Females store the sperm inside them, which allows them to use it when they are physiologically prepared.
Finally, X. couchianus gender is determined by a standard XX or XY combination, unlike many other Xiphophorus whose sex determination is controlled either by a complex combination of genes or by a third sex chromosome.
For more information check the following web page: Viquipèdia