Discussion
The Xiphophorus cochianus selenoproteins are characterized by analyzing their homology with Zebrafish selenoproteins, because is the specie with whom is closer phylogenetically. Two of them, SELENOL and SELENOM were annotated using another fish specie, due to the results with Zebrafish were not satisfying enough.
From this point, we will describe and discuss the results and limitations of our analysis, correlating them with the literature found upon both Xiphophorus cochianus and Zebrafish selenoproteins.
Selenoproteins
Sel15 function is unknown but some evidences revealed that its levels change depending on selenium supplementation. Some studies also suggest that this selenoprotein has redox functions and may be involved in protein folding.
This protein was obtained from SelenoDB database. Although the annotation was not entirely correct as a methionine was not present at the beginning of the sequence, a selenoprotein could be predicted. The conting used was KQ557225.1 and a high homology could be seen by comparing the predicted protein and Zebrafish Sel15, meaning a high identity between them and a good preservation within these species. While using seblastian analysis, a selenoprotein was predicted, containing a SECIs element in 3’UTR position, near the last exon.
Along evolution, change from aquatic habitat to terrestrial was challenging for both plants and animals, being forced to adapt to new environments, like the exposure to higher oxygen levels. As a result, many terrestrial organisms lost selenoproteins or replaced them with cysteine-containing homologs. Some proteins like SelE protein showed a narrow distribution among aquatic eukaryotes and this protein in particular was only detected in fish (13).
In the alignment, only one scaffold (KQ557204.1) could be found. From there, high identity between the predicted protein and the compared from Zebrafish could observed, as well as the presence of a Sec residue aligned with a stop codon. For this reason, a selenoprotein could be predicted with seblastian analysis, giving a SECIs element in 3’UTR region. Apart from that, more interesting result could be found in Xiphophorus’ genome. While doing the blast, the same query was aligned in two different parts of the studied genome, being 10100000 nucleotides away from each other. This event could be interpreted as a possible duplication of this gene in Xiphophorus’ genome. Fishes have a high degree of gene fluctuation and evolutionary, so events like duplications and deletions are constantly happening.
Glutathione peroxidase (GPx) family is a widespread protein superfamily found in many organisms throughout all kingdoms of life. Selenium-containing GPx proteins reduce H2O2 and organic hydroperoxides by employing glutathione (GSH) as an electron donor (14). A total of eight GPx families have been described in mammals.
A common ancestral origin for this diverse glutathione peroxidase clusters has (not) been tried to identify but it has not been possible. This might mean that they have complex relationships and evolutionary rates. This can suggest that they have originated from independent evolutionary events such as gene duplication, gene losses, lateral gene transfer among invertebrates and vertebrates or plants (15). The differentiated enzymatic properties which can be found along this family might be acquired by the evolutionary relaxation of selection pressure and/or biochemical adaptation to the acting environments, being the most abundant in almost all aerobic organisms.
Reactive oxygen is accumulated in aerobic reactions, what can be toxic for the cells. For this reason, aerobic organisms have developed several enzymatic systems to neutralize these compounds. These systems include a set of gene products, such as superoxide dismutases, catalases, ascorbate peroxidases and glutathione peroxidases (GPx), whose principal function is to protect organism from oxidative damage.
By doing the comparison between Zebrafish and Xiphophorus’ proteins, which are two close species with similar evolutionary characteristics, many discrepancies and unique occurrences could be seen.
First of all, not all Zebrafish subfamilies could be seen in Xiphophorus’ genome. By doing a phylogeny analysis, the identity of each subfamily could be determined and classified along subfamilies.
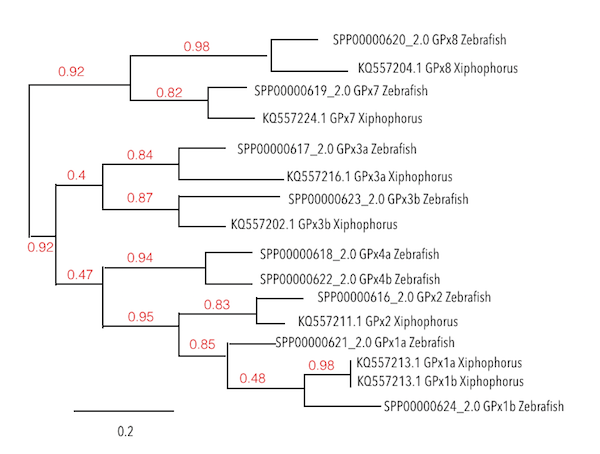
For example, GPx1b has been identified in some particular species, like Zebrafish, as it was created from a duplication event from GPx1a. Thus, just one of these proteins could be found in the analysed genome. Surprisingly, GPx1b should have derived from GPx1a, even though, in Xiphophorus’ genome, the protein found is located closer to GPx1b, reaffirming the presence of this one in the genome of interest.
Continuing with the analysis, in GPx3 subfamily, a selenoprotein could be predicted for GPx3a, having a high homology degree. For GPx3b, a protein could be predicted but SECIs elements could not be found. So, after seeing that this protein was also duplicated in Zebrafish, some doubts regarding the possible selenoprotein identity came out. This leads to two possible options: it can be understood as a seblastian error due to poor sources; or as a protein, not strictly implying to be a selenoprotein.
For the GPx4 subfamily, none of its members could be found in Xiphophorus’ genome.
Finally, while analysing GPx7 and GPx8, a high degree of conservation was seen. These subtypes are Cys containing homologs, which means that they have lost their Sec residue due to evolution. Then, as expected, this homologs without Sec residue were also found in Xiphophorus. Interestingly, a SECIs element was predicted in GPx7 protein but its characteristics were not the best ones as it was located far from the 3’UTR region. It could be thought that this protein was still containing the SECIs element present some time before the loss of this Sec residue which was converted to cysteine.
The distinct Iodothyronine deiodinases catalyze the biotransformation of TH in practically every tissue of the organism. Each DIO1, DIO2 and DIO3 isotypes, which are ubiquitous reductive dehalogenases, have different catalytic properties and specific tissue and developmental expressions. From an evolutionary perspective, DIOs may be considered pivotal players in the emergence and functional diversification of both TS and their iodinated messengers (16).
Even though mammals and fishes’ DIO1 and DIO2 are coded by single genes, the majority of the studied fishes have two genes coding for both DIO3 isoforms (DIO3a and DIO3b) whereas vertebrates have only a single gene for DIO3. (17, 18).
Phylogenesis of Zebrafish Iodothyronine deiodinases shows DIO1 as the oldest deiodinase because it is the gene with the highest number of mutations, so it has had more time to variate during evolution.
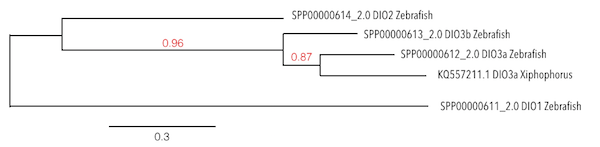
Considering the evolution of these proteins, results were confusing. It was found the same and only significant scaffold (KQ557211.1) for all of DIO proteins and the same hits were located in the same position inside the genome. After doing the analysis with the human genome, which was better annotated than Zebrafish genome, the same results were found. Considering that DIO3b was the protein with the best query score and length, the analysis was made based on this protein.
As it can be observed in the phylogeny analysis, it could be revealed that DIO3a from Xiphophorus was showing more homology with DIO3a from Zebrafish than DIO3b. This could mean that DIO3b was a duplication exclusively from Zebrafish and that the protein which was being predicted was the homologue of DIO3a in Xiphophorus, as it was the first being created.
Moreover, the seblastian analysis predicted a SECIs element located less than 6000 nucleotides after the last exon in 3’UTR position. Finally, a selenoprotein was predicted, which was consistent with results obtained in translate and T-Coffee analysis, where the Sec residue was aligned with a stop codon.
In conclusion, it has been predicted one DIO protein in Xiphophorus. Probably more research is needed regarding this protein family and more analysis can be done by using other database sources.
This is an ancestral selenoprotein that is widely distributed in all vertebrates. It is located in cell nucleus and it is suggested to play an antioxidant role. It was also seen that it is involved in gene regulation, affecting the synthesis of glutathione.
Using Zebrafish genome, KQ557216.1 was the only possible scaffold fulfilling the requirements in terms of score and E-value. Positive similarities between the compared genomes were presented in T-Coffee analysis. Moreover, a Sec residue, typical from selenoproteins, could be observed in Xiphophorus’ genome. Moreover, with the seblastian analysis, a SECIs element was found in the 3’UTR region, so a selenoprotein could be predicted.
This is a multi-pass transmembrane protein contained in the ancestral vertebrate proteasome. It is encoded by a gene belonging to CDP-alcohol phosphatidyltransferase family so it has a phosphotransferase activity and it is also found in phospholipid synthesis (19).
In the analysis, three scaffolds were predicted, but KQ557207.1 was the one selected as it is the one with the highest score and the best E-value. While doing the alignment using T-Coffee, a high homology between sequences could be seen. Unexpectedly, the seblastian analysis showed no output, so SECIs element could not be predicted. These results were unexpected as a Sec residue aligned with a codon stop could be identified in the predicted protein. Thus, it could be concluded that, in this case, seblastian was not accurate enough or that the predicted protein was not classified as a selenoprotein so this subfamily could not be found in Xiphophorus’ genome.
This protein has a strict phylogenetic distribution as it is missing in mammalian genomes. It is assumed to play a structural role although most of the proteins have enzymatic functions.
This proteins constitutes a distinct subfamily within the large family of ADP-ribosylation enzymes, finding a close relationship between SelJ (with a Sec residue) and J1-crystallins (with a Cys residue), suggesting the possibility of SelJ also being a crystallin. SelJ and J1-crystallins may have been derived from the same ancestral enzymes, so selenoproteins may have acquired specialized functions from them (20)
Analysis between Zebrafish protein and Xiphophorus was done, obtaining a selenoprotein prediction as a result. KQ557214.1 was the scaffold taken as the query to confront with the studied genome. In exonerate analysis, an extra exon was found, and a SECIs element was predicted between two of the exons. This last exon was located really far away, probably forming part of another protein.
Although all the analysis was consistent and a high homology could be observed, some remarkable results were obtained. Fistly, SelJ in Zebrafish was not containing a SECIs element while it did in Xiphophorus. Secondly, more than one stop codons were present in the protein prediction even if only one them was well-aligned, meaning that it was the only Sec residue in the sequence. So, as a result of the compiled data, a selenoprotein could be predicted. However, more research would be needed in order to determine whether those first stops codon were affecting the protein translation or not.
Selenoprotein K (SelK) is an ancestral selenoprotein found in all vertebrates. It is a small selenoprotein found in the endoplasmic reticulum (ER) with an unknown function. Comparative genomics analysis indicate that this family is the most widespread eukaryotic selenoprotein family. A biochemical research for proteins that interact with SelK revealed ER-associated degradation (ERAD) components (p97 ATPase, Derlins, and SelS). In this complex, SelK showed higher affinity for Derlin-1, whereas SelS had higher affinity for Derlin-2, suggesting that these selenoproteins could determine the nature of the substrate translocated through the Derlin channel. SelK co-precipitated with soluble glycosylated ERAD substrates and was involved in their degradation. Its gene contained a functional ER stress response element, and its expression was up-regulated by conditions that induce the accumulation of misfolded proteins in the ER. Components of the oligosaccharyltransferase complex (ribophorins, OST48, and STT3A) and an ER chaperone, calnexin, were found to bind SelK (21).
The contig used in SelK in order to find it in the studied genome was KQ557201.1. Although seblastian output did not predict any selenoprotein, a Sec residue was found in the Xiphophorus’ sequence, as well as a SECIs element in 3’UTR position. Moreover, high homology between sequences could be seen in the T-Coffee analysis and, as previously mentioned, this selenoprotein was largely present in all vertebrates. For all these evidences, a selenoprotein could be predicted.
SelL is a specific type of selenoproteins present in fishes but not in mammals, whose function remains unknown. SelL contains two selenocysteine, which form the first known diselenide bond in proteins. This selenide bons corresponds to the disulfide bond between cysteines at the active site of mammalian thioredoxin residues, suggesting a redox role for SelL in fish. Even though, the two Sec residues are only separated by two residues, so it will not be needed more than one SECIs element.
In this case, sequence from Zebrafish was unable to be aligned with the sequence from Xiphophorus, so the alignment was done with Gasterosteus aculeatus SelL sequence. In the blast, one possible hit (KQ557207.1) was found, showing an E-value of 5e-35. During the alignment, although a short region of 30 amino acids was not aligned with Gasterosteus aculeatus’ query, high homology between these species could be observed. Finally, only one Sec residue (rather than two) could be predicted in Xiphophorus’ genome, and it was aligned with a stop codon in the T-Coffee analysis.
These results were consistent with the ones from seblastian, which predicted a SECIs elements in 3’UTR region and a selenoprotein, as it was expected.
Selenoprotein M (SelM) takes part in the formation of disulfide bonds. It presents a redox motive with a Sec residue that may function as a redox regulator. This protein is closely related to Sep15, SepM and Fep15, which are predicted to reside in the ER (22).
As well as SelenoL, the alignment was unable to be performed with the sequence from Zebrafish. In this case, the query was taken from Gadus morthua genome, as Gasterosteus aculeatus sequence was not useful neither.
Results from the study predicted a SECIs elements and a selenoprotein in Xiphophorus’genome. Three hits were obtained for SelM, but KQ557204.1 was the only significant. Despite the short length of the studied sequence, high homology could be seen in T-Coffee analysis. Moreover, a Sec residue was found in the predicted sequence and it was aligned with a stop codon, as supposed.
SelN is a protein found in fishes, frogs, birds and mammals, and it is known to take part in the ancestral vertebrate selenoproteome. It is highly active in tissues before birth and it is suggested to be involved in myogenesis. SelN is also essential for muscle regeneration and satellite cell maintenance.
Recent data suggested that it participates in oxidative and calcium homeostasis (triggering muscle contractions), with a potential role in the regulation of the ryanodine receptor activity. This is why it can be ubiquitously expressed in the membrane of the endoplasmic reticulum (23,24).
Scaffold KQ557211.1 was picked for the subsequent analysis. In T-Coffee analysis, high homology between sequences was shown and a Sec residue was aligned with a stop codon. Even in the predicted protein three stop codons could be found, after doing a new exonerate without the egrep, just one of the residues was aligned with a stop codon.
Moreover, exon information from seblastian and exonerate was not the same, as an extra exon was predicted in exonerate output, located far from the other exons.
This could happen as the protein being tested was analysed with the extension of some nucleotides upstream and downstream the query. Then, a bigger sequence could be browsed, so the prediction of the protein of interest in Xiphophorus’ genome would be more likely to be found. Is for this reason that while performing exonerate analysis, two different proteins could have been predicted in the same hit.
Even that, a selenoprotein could be well predicted as it was containing all the essential components which characterize this kind of proteins.
In Zebrafish two subfamilies of SelO can be found. Both SelO1 and SelO2 take part in the ancestral selenoproteome. SelO is widely distributed in animals (frogs, birds, fishes and mammals), bacteria, yeast and plants. These genes have Sec residues in their sequences, located on the active sites, but their functions are still unknown. However is the largest mammalian selenoprotein with orthologs found in a wide range of organisms, including bacteria and yeast (25).
For SelO1 three hits were obtained, but two of them were significant. Considering the E-value of each hit, scaffold KQ557202.1 was assigned for SelO1. For SelO2, the same three hits were obtained and analysed, in which Sec residues could not be found, confirming what it was supposed. SelO2 was a duplication of SelO1 as similar hits and scaffolds were obtained in the same positions of the genome. This could be understood as a specific feature of Zebrafish, and this duplication event was not required to be found in other species.
For SelO1, high sequence homology between species could be seen, and a Sec residues was aligned with a stop codon. These results were consistent with the ones obtained from seblastian, which predicted a SECIs element and a selenoprotein.
Evidence support that Selenoprotein P has antioxidant properties and functions in selenium homeostasis. Interestingly, it has been found that plasma selenoprotein P is the best indication for human selenium nutritional status (26). This protein also stands out from the others as 17 Sec residues can be found in Zebrafish.
While analysing SelP1 inXiphophorus genome some homology could be seen, even though some gaps were present because of the multiple Sec residues.
From all scaffolds, KQ557204.1 was the one selected as it was the one containing more Sec residues. Then, unexpected results were found as one single Sec residue could be seen in Xiphophorus’ protein, but two SECIs elements, equally valid, were predicted, as it was happening in the Zebrafish protein.
The presence of these multiple Sec residues in Zebrafish genome could be explained as fishes are constantly suffering duplication events and these residues can mutate and evolve through the years.
Then, a single Sec residue was found in SelP2 . The contig used was the KQ557221.1 and poor homology could be seen between genomes, as matches were found in a small part of the alignment, probably in the conserved part which were giving functionality to this protein. Even this low homology, a selenoprotein could be predicted with seblastian analysis and one SECIs element was found, contrary to Zebrafish in which SECIs element could not be identified.
This proteins belong to the methionine sulfoxide reductase (Msr) family, which includes repairing enzymes that reduce oxidized methionine residues. Cysteine homologs of SelR are present in all organisms except certain parasites and hyperthermophiles. In several genomes, SelR and MsrA genes are fused or clustered, and their expression patterns suggest a role of both proteins in protection against oxidative stress. Methionine residues in proteins are susceptible to oxidation by reactive oxygen species, but can be repaired via reduction of the resulting methionine sulfoxides by methionine-S-sulfoxide reductases.
The most significant scaffold for MSR1 was KQ557208.1 in which a selenoprotein could be predicted. High homology between the expected protein and the one taken from Zebrafish was seen. Also, a potential SECIs element could be predicted by seblastian output. Unexpectedly, while analysing the alignment, a duplication could be hypothesized as same queries were having different locations in Xiphophorus’ genome.
MSRB1b is considered a specific protein of bony fishes. The most significant scaffold was KQ557202.1, but no selenoproteins or SECIs elements could be predicted, even the high homology with the sequence from Zebrafish. This does not match with the results from T-Coffee, as a prediction of a SECIs element would have been expected.
Results for MSRB2 and MSRB3 were similar. For MSRB2 the most significant scaffold was KQ557214.1. No cysteines could be found either in Zebrafish genome or Xiphophorus’ genome, but a SECIs element could be predicted in the seblastian analysis. Then, considering the results and that SECIs element was predicted more than 6.000 bp from the last exon, it could be considered a false positive result. For MSRB3 the most significant scaffold was KQ557209.1. No selenocysteine could be found either, but a grade B SECIs element could be found in the seblastian output. Again, a false positive could be expected.
This event could be explained as these two proteins were classified as cysteine homologues. These proteins had lost their Sec residue due to evolutionary processes, converting this residue for a cysteine. This is why a SECIs element could still be predicted.
Selenoprotein S is found in all vertebrates, but the function in fishes is unknown, even though it is known that in humans is related to inflammatory diseases. This enzyme is in the ER and the cell membrane. It is known that is involved in ER homeostasis regulation and antioxidative protection in a cell-type-dependent manner. Other studies suggest that participates in intracellular membrane transport and maintenance of protein complexes by anchoring them to the ER membrane.
High homology between sequences could be seen, meaning that a selenoprotein could be predicted. In Xiphophorus' protein, one Sec residue was identified. By doing seblastian analysis, three SECIs elements could be found in 3’UTR region, avoiding the STOP codon signal. Then, only one of them was chosen depending on its grade and its strand location.
SelT1 is known to be one of the most conserved proteins and is found in all vertebrates. It is transiently expressed in the neural lineage during brain ontogenesis. In most fishes SelT1 was found duplicated generating SelT2. Additionally, some gene duplications were observed only in specific lineages as Zebrafish, where SelT1a (the original one, SelT1 is found in all vertebrates) and SelT2 were found (9).
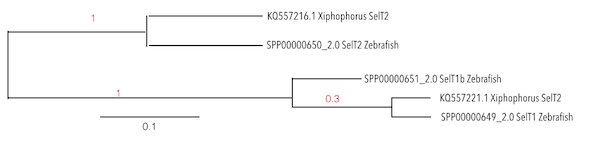
Taking into account the phylogeny, it was observed that T1b is a protein exclusively from Zebrafish, so the analysis was done for SelT1 and SelT2.
Three scaffolds were found for SelT1 and SelT2, but just two of them were significant. Considering the E-value we assigned the KQ557221.1 for SelT1 and KQ557216.1 for SelT2. SECIs elements were obtained and selenoproteins were predicted, meaning that Xiphophorus had SelT1 and SelT2 in its genome, except for SelT1b because as previously mentioned, it could only be found in some species due to duplication events.
Selenoprotein U was firstly found in fish and also reported in birds and unicellular eukaryotes. It may regulate a myriad of biological processes through its redox function. SelU1 takes part of the ancestral selenoproteome. Fishes have Sec residues rather than Cys, present in mammals. This last ones can probably have evolved from earlier Sec residues. There are also two more proteins known as SelU2 and SelU3, but in this case their sequences do not contain Sec residues. However, there is no evidence that supports an early Sec-to-Cys conversions event for these proteins, but a high fluctuation between these different residues can be seen (9).
By analysing each particular protein, some interesting result could be extracted. Firstly, an homologous selenoprotein of SelU1a could be predicted as a high homology between genomes could be seen. Moreover, a Sec residue was found in the predicted protein as well as a SECIs element in 3’UTR position.
On the contrary, while analysing SelU2 and SelU3, no selenoprotein could be predicted. This is due to the absence of Sec residues in Zebrafish, as these were replaced by Cys. When seblastian analysis was performed, SECIs elements could be predicted but their respective positions were far from the end of the last exon and on the opposite strand of the taken queries. This prediction could be considered as a false positive or as previously reported, these SECIs elements could have been preserved since a Sec residue was present in their origins.
Selenoprotein W is a small selenoprotein first identified in sheep suffering from selenium deficiency. It is mostly expressed in muscle, heart, spleen and brain. The biological function of SelW has not been identified. Evidences show that it can serve as an antioxidant that responds to stress and it can be involved in cell immunity, it can be a specific target for methylmercury, and it also has thioredoxin-like function (27).
After performing some analysis, SelW was predicted in Xiphophorus’ genome. A difficulty was encountered while studying this family as no classification between subfamilies could be found in SelenoDB database. For this reason, distinction between subfamilies was not possible to do.
Analysis of the Selenoprotein W was done exclusively for the first protein from Zebrafish (which was named as SelW1). However, the predicted sequence of SelW in Xiphophorus was compared with all Zebrafish SelW in a phylogenetic tree and the predicted SelW was presenting more homology with SelW2 than with SelW1, from which the query was taken. Finally, looking to the phylogeny, it was observed that SelW3 was located at the same level as SelW2 of Zebrafish, so it could be thought that SelW3 was a duplication from SelW2, named as SelW2b in the literature.
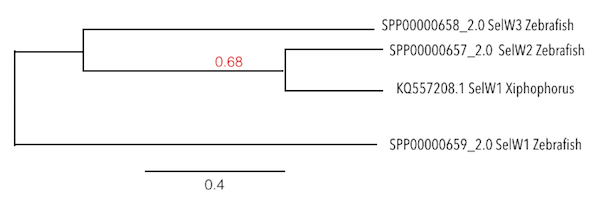
These results were consistent with the ones obtained by Gregory et al, where it was said that Zebrafish has SelW1, SelW2a and SelW2b (which was considered as SelW3 during the analysis), and that SelW2b was an exclusive duplication of Zebrafish.
Moreover, interesting results were found as for SelW2, two selenoproteins could be predicted in the same sequence while using seblastian analysis. This would mean that a unique duplication had occurred in Xiphophorus’ genome. It could also be understood as a pseudogene from the first predicted gene.
The only concern about these results would be that SelW1 was reported to be a member of the ancestral selenoproteome and it could not be identified in Xiphophorus’ genome. To ensure the absence of SelW1, the same analysis was carried out using the protein from Human’s genome. However, alignment could not be done as poor homology was observed.
On the contrary, SelW2 was reported to be characteristic from bony fishes, frogs, elephants and sharks and if the suppositions were right, it could be identified in the studied genome. Nevertheless, as no classification is made in SelenoDB, this result cannot be absolutely concluent.
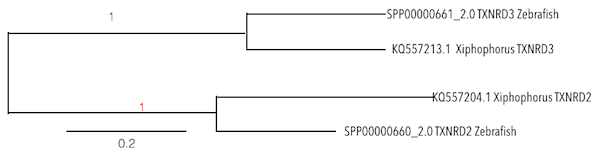
This family of genes encode for members of class I pyridine nucleotide-disulfide oxidoreductase family. The encoded protein is a selenocysteine-containing flavoenzyme that maintains thioredoxins in a reduced state, so the cellular redox environment can be regulated. Mammals have three related thioredoxin reductases but just two of them can be found in Zebrafish genome. This gene encodes a mitochondrial form important to avoid reactive oxygen species in mitochondria (28).
The sec residue in this family is essential for enzyme activity. It is located in the penultimate C-terminal position and is encoded by a UGA codon. Sec differs from cys in that it substitutes selenium, a better nucleophile, for sulfur (29).
In the analysis of both genes, a Selenoprotein could be predicted and thus, a SECIs element could be found in the sequence by performing seblastian analysis.
To obtain the predicted proteins, scaffolds KQ557213.1 and KQ557204.1 were taken. By contrasting Zebrafish sequences withXiphophorus sequences, a high conservation between species could be seen. As both proteins were similar from each other, a phylogenetic analysis was done in order to ensure the obtained results.
Machinery proteins
Due to the important function of machinery proteins in the organism's survival, its structure is highly conserved and mutations are not well accepted.
Considering the phylogenetic proximity between species, Zebrafish genome was chosen as a reference to align Xiphophorus’ genome. It has been observed that the majority of these proteins have not Sec residues in their sequence. Considering that machinery proteins from Xiphophorus should be as similar as possible from the ones in Zebrafish due to the high conservation, no SECIs elements should be predicted.
These proteins are involved in the early steps of selenocysteine biosynthesis and tRNA (Sec) charging resulting in the incorporation of selenocysteine into selenoproteins. Binding tRNA[ser]sec (SECp43) has been identified as a key factor in orchestrating the interactions and localizations of the other factors involved in selenoprotein biosynthesis, as selenocysteyl-tRNA[Ser]Sec-EFsec (30,31).
By comparing eEFsec, SecP43 and SecS with the sequences from Zebrafish, and with the T-Coffe alignment, a high identity was revealed. Moreover, as high conservation between these groups and a substitution for a cysteine residue was present, any of them were containing a SECIs elements so selenoproteins were not predicted.
Two protein members were included in SECp43 family. For SECp431, two different scaffolds were found, but only KQ577221.1 was working. In SecP432, none of the scaffolds were working, meaning the possible absence of this protein in Xiphophorus’ genome.
When SecP431 was analyzed with seblastian, a SECIs element was predicted. Considering that there were not selenocysteine residues and that the residues aligned were four cysteines, it could be thought that Sec residues were substituted by cysteines but SECIs element were still present in the sequence.
However, in this case, SecP43 proteins were not well-differentiated as both were included in the same family but the descriptive of the subfamily appeared as “NONE”. On the other hand, both eEFsec and SecS could be predicted in Xiphophorus’ genome, being homologues with cysteine.
Phosphoseryl-TRNA Kinase (PSTK) is a Protein Coding gene. SECIs binding proteins (SBP) are required, as well as SECIs elements, to decode UGA codon as selenocysteine.
SBP family was showing different homology degrees depending on the aligned part of the protein. As explained in the results, most of the protein was aligned with Zebrafish sequence but surprisingly, a gap could be seen in the middle of the predicted protein. This event could be understood as random acquisition, due to the high gene fluctuation in fishes. Although this extra genomic material was not contained in the analysed query, the protein remained with the same function.
In general lines, it is known that parts showing greater homology were supposed to correspond to those coding for the active site of the protein or having important domains necessary to perform its function.
Moreover, poor homology of the predicted PSTK in Xiphophorus could be seen compared with the one from Zebrafish. This results were unexpected as machinery genes and proteins are characterized for having high conservation.
Despite these results, it should take into account that sequences from SelenoDB did not have a methionine residue at the beginning so this could be disrupting the analysis.
MsrA gene encodes an ubiquitous and highly conserved protein that carries out the enzymatic reduction of methionine sulfoxide to methionine, so its function is to repair proteins damaged by oxidative processes to restore its biological activity.
Two proteins were included in MsrA family. These protein were obtained from SelenoDB database even they weren’t correctly annotated, as a methionine was not present at the beginning of the sequences. This was an important fact to consider while analyzing the results.
KQ557207.1 and KQ557211.1. were the contigs used for the analysis. Then, high homology could be found in both proteins, always comparing with a reference sequence from Zebrafish.
Results revealed no selenocysteine residue in the predicted MsrA sequences, and for this reason, no SECIs elements or selenoproteins were found. Thus, these genes could be described as machinery proteins. These proteins are known for being essential in selenoproteins’ assembly.
These genes encode an enzyme that synthesizes selenophosphate from selenide and ATP. Their function is to provide the selenium used to synthesize selenocysteine, which is co-translationally incorporated into selenoproteins. It is conserved in all prokaryotic and eukaryotic genomes and SPS is itself a selenoprotein in many species (32).
For both member of this family, three hits were obtained, but only two of them were significant (as a threshold of 0,05 was applied). Contigs KQ557212.1 for SEPHS1 and KQ557201.1 for SEPHS2 were the ones chosen. Results from T-Coffee revealed a high homology between Zebrafish and the studied organism, meaning high conservation between sequences.
Sec residues were not found in SEPHS1, so SECIs elements were not predicted. Based on the homology, this machinery protein could be predicted in Xiphophorus' genome.
Interestingly, SEPHS2 was containing a Sec residue in the sequence and it was aligned with a stop codon while doing T-Coffee analysis. However, in the seblastian output, a SECIs element could be predicted but a selenoprotein was not found. This result could probably be due to the lack of information in public sources or because the contig chosen was not having the full-length of the sequence. This was reasoned as information from previous data suggested that this protein was classified as a machinery protein but also as a selenoprotein.