Discussion
The Periophthalmodon schlosseri's selenoproteins are characterized by analyzing its homology with Danio rerio (zebrafish) and Homo sapiens (human) proteins. As P. scholsseri is phylogenetically closer with zebrafish than with human, the analysis is focused mainly on zebrafish, which has the largest selenoproteome found in bony fishes, with a maximum of 38 selenoproteins.[10]
Selenoproteins in Periophthalmodon schlosseri
Selenoproteins
Sel15
Sel15 selenoprotein has redox function and may be involved in the quality control of protein folding. [10] In P. schlosseri, after using the homology-based approach, only one hit was found for this protein. Thus, probably meaning that, in this fish Sel15 protein is encoded by the contig KN469434.1. The homology between the predicted one and the zebrafish sel15 protein is really high meaning it is conserved within species. As expected, a SECIS element was predicted in the 3’UTR of the transcript and also Seblastian could provide a selenoprotein structure form the mentioned contig.
Fep15 and SelM
Fep15 is a selenoprotein that has only been identified in fish. It is known that there was a lost of selenoproteins across vertebrates after their land colonization. As frogs do only have a cystein-homolog of this protein, it is known that Fep15 was in the ancestral vertebrate selenoproteome and its lost occur before the split of reptiles.[10,27] Fep15 resides in the endoplasmic reticulum and possibly in Golgi but its function remains unknown. It is homologous to mammalian Sep15. Fep15 evolved by duplication of SelM in animals, most likely in fish, followed by mutations. SelM is also located in the endoplasmic reticulum and its function is neither known.[10,27]
The results from this study predict a Fep15 and a SelM selenoprotein in P. schlosseri. The only hit obtained for SelM (in the contig KN483744.1) is the second best hit for Fep15 in the analysed organism. This fact could be explained for the similarity between the two proteins.
In both cases a SECIS element and a selenoprotein were predicted by using Seblastian but, for Fep15, the number of exons obtained from the exonerate results (3 exons) were not concordant with the one in Seblastian (4 exons). It could be due to: 1) an error in the annotation of P. schlosseri genome, 2) two different isotypes of the same protein are being considered or 3) the protein in zebrafish is shorter -or wrongly annotated- than the one found in P. schlosseri. It is also remarkable that the predicted selenoprotein in this organism has two selenocysteine residues instead of one.
Glutathione peroxidase (GPx)
Glutathione peroxidases, the largest selenoprotein family in vertebrates, play different roles but the main function of these enzymes is to protect the organism from oxidative damage.[10] In bony fishes, there are 9 glutathione peroxidases that can be explained by three GPx duplications:
- GPx1: generating GPx1a and GPx1b
- GPx3: generating GPx3a and GPx3b
- GPx4: generating GPx4a and GPx4b
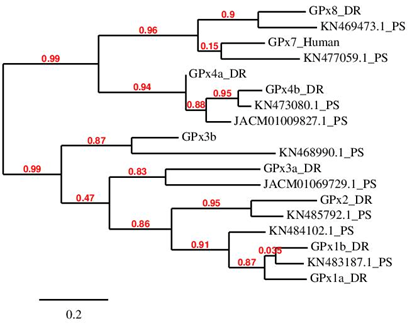
After analysing the obtained results, it is important to take into account that the first 3 obtained hits for GPx1a, GPx1b and GPx2 were the same. However, the best hit was different for each.
In addition, the identification of the best hit for GPx3b was only possible by the construction of the phylogenetic tree. However, it has to be considered that the identified GPx3b in P. schlosseri does not have aligned the selenocysteine residue with its zebrafish homologous.
It is known that Cys-containing GPx7 and GPx8 evolved from a GPx4-like selenoprotein ancestor, as it is observed in the phylogenetic tree. This event happened prior to separation of mammals and fishes. [10] Between the two cysteine homologues GPx7 and GPx8 there is a highly grade of similarity: the best result in blast for one is the second result for the other. However, there are differences between them. On one hand, neither SECISearch3 or Seblastian could predict a SECIS element for P. schlosseri GPx7. A possible explanation to this fact is that the SECIS element, as well as the selenocysteine residue, were lost during the evolution process. On the other hand, GPx8’s SECIS element could be predicted, supporting the idea that it was previously containing a sec residue that was substituted by a cysteine residue.
As expected, the other GPx proteins except these two had a SECIS element and a selenoprotein prediction obtained by using Seblastian.
Iodothyronine deiodinases (DI)
The iodothyronine deiodinases regulate the inactivation of thyroid hormones. It has been previously described that, the three enzymes DI1, 2 and 3a are found in frog, birds, mammals and fish whereas DI3b is only found in fish due to a duplication event.[10] When analysing the four members of the DI family from zebrafish or human (detailed in the results), the 3 obtained hits were overlapping between them. The fact that the hits were shared for the proteins that belong to iodothyronine deiodinase family is due to their significant intrafamily homology.[10] Using different parameters from the in silico analysis, the best hits were assigned to its corresponding homologous. However, DI2 did not have any assigned hit because the three that were originally proposed had better correspondence for other DI proteins: they showed higher homology and had better alienations. In conclusion, DI2 protein could not be predicted in the genome of P. schlosseri and that could be explained for: 1) the low quality of the assembly or 2) P. schlosseri has lost the DI2 protein.
In the three iodothyronine deiodinases that were found in this study, one (or more) SECIS element and a selenoprotein were obtained by using Seblastian. Thus, making the results more likely to be possible.
MsrA
MsrA is a widely distributed protein family, whose function is to repair methionine residues in proteins. MsrA catalyzes a stereospecific reduction of methionine-S-sulfoxides with thioredoxin. Only one selenoprotein MsrA was described, which is present in Chlamydomonas, green algae. All other known MsrAs are Cys-containing proteins.
After doing the homology-based approach with zebrafish and human MsrA proteins (MsrA1 and MsrA2) one of the obtained hits was not assigned to any of these, even so it has homology with MsrA1. A possible explanation could be that it is a partial duplication of MsrA1, since it has only 2 exons instead of the 9 exons that MsrA1 (I) contains.
The results obtained showed that the predicted MsrA proteins from P. schlosseri, MsrA1 (I), MsrA1 (II) and MsrA2, did not contain any selenocysteine amino acid, so that they are cysteine homologues. For this reason, any SECIS element was found in the genes which encode these two proteins.
SelH
Selenoprotein H is a 14 kDa thioredoxin fold-like protein with recently described redox functions. SelH is found in frogs, birds, mammals and fish and is part of the ancestral vertebrate selenoproteome.[10] The results allowed assigning the only hit obtained (corresponding to contig KN473035.1) with SelH protein due to its high homology.
The number of exons predicted by exonerate (4) does not match with the predicted by seblastian (3). This may be because both programs use two isoforms of the same protein to make predictions or simply due to the poor quality of the P. schlosseri genome annotation. The fact that both Seblastian and SECISearch3 were able to predict a SECIS element confirms the fact that SelH is a conserved selenoprotein.
SelI
Selenoprotein I is found in frogs, birds, mammals and fish and is part of the ancestral vertebrate selenoproteome. Regarding its function, it is only known that it has homologous sequence with enzimes involved in the phospholipid synthesis. In a recent work, human SelI reported a specific ethanolamine phosphotransferase (EPT) activity. However, a truncated form of SelI was expressed, lacking the Sec residue and the rest of the C-terminus. That is why the real molecular function of SelI has yet to be discovered. [10]
As for the results, 3 hits were obtained for SelI selenoprotein. Two of them were discarded due to the poor quality of the alignment. The third hit, corresponding to contig KN475358.1, was chosen to make the further comparisons. However, when analyzing the alignment, the selenocysteine residue present in the human protein was not aligned with the genome of P. schlosseri . In fact, the sequence of P. schlosseri was missing the last part of the protein (starting from the selenocysteine residue). Therefore, a manual blast without filtering by the e-value was performed to make sure that no other hits (aligning the selenocysteine residue) were discarted. Finally, a bad annotation of the P. schlosseri genome could be the explanation for the good genome-protein alignment but no selenocystein residue alignment.
SelJ1
Selenoprotein J is today found only in fishes, among vertebrates. The function of SelJ is not fully understood, although recent studies suggest it could have a structural role rather than a functional one.[10]
The results allowed to assign one of the obtained hits (corresponding to contig KN475703.1) with SelJ1 protein due to its high homology. Moreover, the fact that both Seblastian and SECISearch3 were able to predict a SECIS element confirms the fact that SelJ is a conserved selenoprotein.
SelK
Selenoprotein K is a small selenoprotein found in frogs, birds, mammals and fish and is part of the ancestral vertebrate selenoproteome. In fact, SelK is the most widespread selenoprotein, being present in nearly all eukaryotes that use Sec. The function of SelK has not been described yet in fishes, though several studies show that it has some well known functions in mice and humans, including its participation in the ERAD system and in T-cell proliferation (among others). [10]
Among the selenoproteins that have pseudogenes, SelK has been found to be the one that has more than any of them. Up until now, these pseudogenes have only been described in mammals, being rodents the ones that have the highest number.[10] Having this information in mind, the results suggested that fishes (or at least P. schlosseri ) do not present any of this pseudgenes as a single hit was obtained for this protein corresponding to contig JACM01060727.1. However, the selenocysteine residue present in the zebrafish protein was not aligned with the genome of P. schlosseri . In fact, the sequence of P. schlosseri was missing the last part of the protein (starting from the selenocysteine residue). Therefore, a manual blast without filtering by the e-value was performed in order to make sure that no other hits (aligning the selenocysteine residue) were discarted. Finally, a bad annotation of the P. schlosseri genome was considered a possible explanation for this situation.
SelL
Selenoprotein L is today found only in fishes, among vertebrates. Its function remains unknown until now. SelL is characterized for having multiple Sec residues[10]. In fishes, two Sec residues in SelL are only separated by two residues and are inserted with the help of a single SECIS element.
The results allowed to assign one of the two obtained hits (corresponding to contig KN476964.1) with protein SelL due to its high homology. Moreover, the classical SelL structure of 2 sec residues could be found and aligned in the two compared sequences. Finally, Seblastian program could not predict the selenoprotein but SECISearch3 program showed the presence of two SECIS elements (and not one, as expected).
SelN
Selenoprotein N can be found in frogs, birds, mammals and fish and is part of the ancestral vertebrate selenoproteome. SelN is a selenoprotein of unknown function although it was recently found to be implicated in the role of selenium in muscle function in mammals.[10] The results allowed to assign the only hit obtained (corresponding to contig KN472865.1) with protein SelN due to its high homology. Finally, Seblastian program could not predict the selenoprotein but SECISearch3 program was able to predict the presence of two SECIS elements.
SelO
SelO is a widely distributed protein that has homologs in animals, bacteria, yeast and plants, but the function of these proteins is not known. Only vertebrate homologs of SelO have Sec, which is located in the C-terminal before the penultimate position. However, in most of SelO homologs, Sec is replaced by Cys. Selenoprotein O can be found in frogs, birds, mammals and fish and is part of the ancestral vertebrate selenoproteome. In zebrafish only, we found an additional copy of SelO1 named SelO2.[10]
The results allowed to assign one of the obtained hits (corresponding to contig KN485144.1) with zebrafish protein SelO1 due to its high homology. Indeed, the Sec residue could be found (in both cases) before the penultimate position of the sequence. Moreover, the fact that both Seblastian and SECISearch3 were able to predict a SECIS element confirms the fact that SelH is a conserved selenoprotein.
As for SelO2 in P. schlosseri, the results showed it is encoded by two different contigs (KN481390.1 and KN476330.1) as it is shown in the results. However, neither the first nor the second contig contained a Sec residue. The selenocysteine residue present in the zebrafish protein was not aligned with the genome ofP. schlosseri. In fact, the sequence of P. schlosseri was missing the last part of the protein (starting from the selenocysteine residue). Therefore, a manual blast without filtering by the e-value was performed to make sure that no other hits (aligning the selenocysteine residue) were discarted. Finally, a bad annotation of the P. schlosseri genome was considered a possible explanation for this situation.
SelP
Selenoprotein P is found in frogs, birds, mammals and fish and is part of the ancestral vertebrate selenoproteome. It has a varying number of Sec residues (for example, human SelP has 10 Sec and zebrafish SelPa has 17 Sec residues) and is unique in that it contains two SECIS elements. This protein is in charge of the Se transport, which is important to maintain normal brain function.[10]
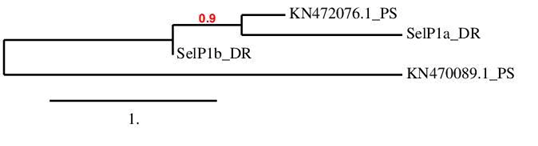
The results allowed assigning one of the obtained hits (corresponding to contig KN472076.1) with zebrafish protein SelP1A due to its good homology. Two Sec residues were found in this contig, one of them aligned with a sec residue of zebrafish protein. Unlike expected, a single SECIS elemet could be predicted for SelP1A, although the reason is not clear.
As for SelP1b, two hits were found. The first hit was already assigned to SelP1A (for which it had a greater homology and, as represented in the figure below, was closer phylogenetically). The method used in this study allowed indentifying a second hit, corresponding to the contig KN470089.1. The sequence of this hit contained a large number of selenocysteine residues (10) and, therefore, could encode a selenoprotein. However, the hit’s homology with SelP1b was too low to be considered valid. Therefore, an homologous protein of SelP1b could not be found in P. sclosseri genome.
The unmatched hit (corresponding to contig KN470089.1) was also analyzed, even it could not be assigned to any described zebrafish selenoprotein. Both Seblastian and SeciSearch3 were able to predict the selenoprotein and two SECIS elements, which indicates that this sequence probably encodes for a selenoprotein. Moreover, it may belong to SelP family as it presents its classical characteristics such as multiple Sec residues and two Secis elements. It was randomly named SelP~.
SelR
Selenoprotein R belongs to Msr protein family, being the only one that has a selenocysteine residue insted of a cystein residue in its active site. Its function remains unknown, though some studies suggest it could have a protection role against neurodegeneration and oxidative damage related to aging.[10]
The results allowed to assign one of the obtained hits (corresponding to contig KN475030.1) with zebrafish protein SelR2 due to its high homology. Neither the zebrafish nor theP. schlosseri sequence contained Sec residues, as they both converted them to cys (they presented 6 aligned cys residues). The fact that SECIS elements could not be found beyond this protein sequences confirms the absence of this amino acid. Similarly, contig JACM01052317.1 was asigned to zebrafish SelR3 protein. Sec residues could not be found in any of the sequences (query or contig), although they presented 5 cysteine residues aligned. None SECIS elements could not be predicted beyond the protein sequences either.
SelR1a and SelR1b in P. schlosseri were Sec-containing selenoproteins and were asigned to contigs KN472457.1 i KN481551.1 respectively. Furthermore, both Seblastian and SECISearch3 were able to predict a SECIS element, which confirms that SelR1a and SelR1b are conserved selenoproteins.
SelS
Selenoprotein S (SelS) is found in all vertebrates. In fishes, its function is not described, but in humans, this protein is related to various inflammatory diseases. Some studies suggest that it is involved in endoplasmic reticulum (ER) homeostasis regulation and antioxidative protection in a cell-type-dependent manner. The sequence of zebrafish SelS protein does not contain any Sec residue but has aligned cysteine residues with P. schlosseri SelS. However, SelS sequence of P. schlosseri has a Sec residue in its first exon known by genewise and t-coffee analysis but Seblastian did not provide any prediction. Finally, this protein in P. schlosseri has a SECIS element that strengthens the presence of the selenoprotein.
SelT
Selenoprotein T is a newly discovered thioredoxin-like protein, which is abundantly but transiently expressed in the neural lineage during brain ontogenesis. A recent study exposes that SelT deficiency leads to neurodevelopmental abnormalities and hyperactive behavior in mice. SelT is duplicated in bony fishes, probably owning to the whole genome duplication in the early evolution of ray-finned fishes. This event generated selenoproteins SelT1 and SelT2. In zebrafish SelT1 is also duplicated, and named SelT1b, but this protein is not found in P. schlosseri using the described methodology. In contrast, SelT1 and SelT2 are found in P. schlosseri, but in this last protein any SECIS element and selenoprotein are predicted with Seblastian, but using genewise or t-coffee it is possible to ensure that SelT2 has a selenoprotein in P. schlosseri.
SelU
SelU may regulate several biological processes through its redox function. SelU1 is found duplicated in bony fishes and named SelU1A and SelU1B.[10] Using the methododlogy described, in this study it is found two possible hits for SelU1A in P. schlosseri, called randomly SelU1A(I) and SelU1A(II). These 2 hits have high identity values and where located in different places of the genome, so they could be a duplication of the SelU1A protein. It does not seem unusual if we take into account that SelU1A is a duplication of SelU1. Referring to SelU1A(I), it is necessary to comment that 10 exons are found using exonerate, but the Seblastian prediction of the selenoprotein shows 5 exons. This could be because both programs are using two different protein isoforms or due to a bad quality annotation ofP. schlosseri ’s genome. On the other hand, SelU2 has a cysteine instead of a selenocysteine, as seen in mammals, although there is not any evidence that supports an early Sec-to-Cys conversion event for SelU2 and SelU3 proteins. SelU3, in addition, has a SECIS element and four Sec insertions among its sequence that are not found in zebrafish.
SelW
SelW, a protein containing a selenocysteine (Sec) in a conserved Cys-X-X-Sec motif, has been suggested to have an antioxidant role in cell metabolism. Several SelW homologs are observed across non-mammalian vertebrates[10]. SelW1 is found in all vertebrates, but it is lost in zebrafish and, using the methodology that has been described, no SelW1 could be predicted in P. schlosseri. However, SelW2 is only found in bony fishes, but also in frog and in elephant shark, due to part of the ancestral vertebrate selenoproteome was lost before the split of reptiles. In zebrafish SelW2 is duplicated, and named SelW2a and SelW2b. But in P. schlosseri, SelW2b could not be predicted by the steps followed in this study. Referring to SelW2a, Seblastian could not give a selenoprotein but using genewise or t-coffee it is possible to ensure that SelW2a has a selenoprotein in P. schlosseri . Finally, a SECIS element is found in SelW2a.
Thioredoxin reductase (TR)
Thioredoxin reductases control the redox state of thioredoxins, proteins that play an important role in redox regulation of cellular processes.[10]
TR2 and TR3 are selenoproteins whereas TR1 is not. This is why the encoding sequence for TR2 and TR3 were searched in the P. schlosseri genome and further analysed. The results showed that the contig KN477299.1 encodes two genes that compose the TR2 protein. In addition the predicted protein from P. schlosseri has 3 selenocysteine residues whereas the zebrafish TR2 does only have one. In this case, Seblastian could not predict any selenoprotein from the introduced sequence, maybe due to the fact that the genome is wrongly annotated. However, two SECIS structures were provided by SECISearch3. Thus, supporting the idea that P. schlosseri also has TR2 selenoprotein.
The obtained results describe that TR3 protein in P. schlosseri is codified by two diferent contigs: KN483500.1 and KN474115.1. In this case, Seblastian could not predict any selenoprotein. A possible explanation to this fact could be that the sequence introduced was obtained from one or the other contig so it was not the full-length sequence. However, two SECIS element were predicted in the 3’UTR by using SECISearch3.
Machinery
The machinery proteins are required for selenoprotein synthesis and, the results of this study reveal that are likely to be found inP. schlosseri
SPS and SPS2
P. schlosseri Selenophophate synthetase protein (SPS) is codified by the contig KN475839.1. The homology with the zebrafish SPS is high and therefore it could explain how important it is for the protein to conserve the primary structure in order to maintain its function. The SPS protein does not contain any selenocysteine residue (neither in zebrafish or in P. schlosseri) whereas it has 9 aligned cysteine residues. This is the reason why Seblastian could not predict a selenoprotein. However, a SECIS element was found by SECISearch3. Probably it is due to the evolution of the sequence and the change of the sec residue for a cys one.
The selenophosphate synthetase 2 (SPS2) is the enzyme that catalyses the generation of a Se donor (selenophosphate) which is required for Sec biosynthesis. In addition, it is a selenoprotein. In P. schlosseri, it is encoded by two different contigs (KN484547.1 and KN469560.1) as it is shown in the results. The first exon of the protein contains the selenocysteine residue.
In this case, Seblastian could not predict any selenoprotein. A possible explanation to this fact could be that the sequence introduced was obtained from one or the other contig so it was not the full-length sequence. However, a SECIS element was found in the 3’UTR by using SECISearch3.
eEFsec, Trnau1apa, C2H6orf52 and SecS
The approaches that have been used reveal that eEFsec, SECp43 proteins and SecS have a very high identity with the ones in zebrafish: the obtained alignments were almost perfect. It could be thought that, the conservation of this machinery proteins structure is really important for their function and they do not accept mutations in many sites.
It is important to consider that none of them are selenoproteins because there is not a sec residue among their sequences. Nevertheless, a SECIS element could be predicted in the 3’UTR for eEFsec, Trnau1apa and SecS. Thus, giving clues about how it could have evolved: probably changing the selenocysteine residue to a cysteine one.
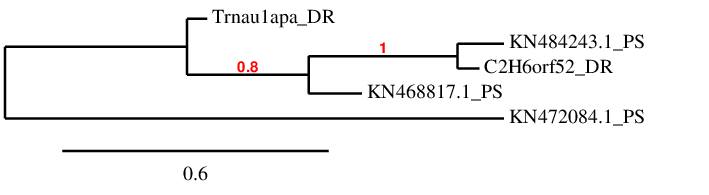
It is important to take into account that for SECp43 family, which is composed of two proteins, three hits were found. The first hit obtained for each protein was assigned, KN468817.1 to Trnau1apa and KN484243.1 to C2H6orf52, since they are closer phylogenetically and consequently they have higher homology.
The third hit that remained unassigned contained four selenocysteine residues in contrast to these zebrafish proteins which are Cys-containing proteins, meaning it could encode a selenoprotein. This finding could be supported by the prediction of a SECIS element in contig KN472084.1. However, the homology of the predicted protein with Trnau1apa is too low to be considered valid.
PSTK, SBP2, SBP2L
The alignments between these zebrafish machinery proteins and the genome of P. schlosseri were good enough to consider valid the obtained hit in each case. However, a deeper analysis of the alignments revealed how P. schlosseri sequences had some highly conserved parts whereas others were poorly conserved (unlike machinery proteins described above).
In this case, the more preserved regions could correspond to the protein active site or other domains extremely necessaries in order to maintain its functionality.