Conclusions
As it has been already described, selenoprotein detection in genomes is a big challenge: many handicaps have to be faced. First of all, selenocysteine is encoded by the UGA codon, which is usually a STOP signal. Secondly, it is important to take into account that, the sec residue is frequently substituted by a cysteine one. Moreover, selenoproteins play many different roles in the organism even though their functions are usually unknown. However, the study of selenoproteomes is important to have a wider knowledge of the genome. Furthermore, the elucidation of selenoproteins phylogenetic distribution could be a key point to discover evolutionary relationships between organisms.
The aim of this study was to describe selenoproteins in P. schlosseri genome. For this purpose, an homology-based comparison method was used as this proteins are generaly highly conserved between close species.
The results of this study allowed characterizing a total of 28 selenoproteins and 11 cys-containing selenoprotein homologues, while 6 selenoproteins could not be predicted. In addition, 9 machinery proteins were found and one of them (SPS2) contained sec residues. Interestingly, during the analysis of the results 2 new protein duplications (not decribed in zebrafish) were found: SelU1A and MsrA1.
The following figure tries to clarify and cluster the results obtained in this study:
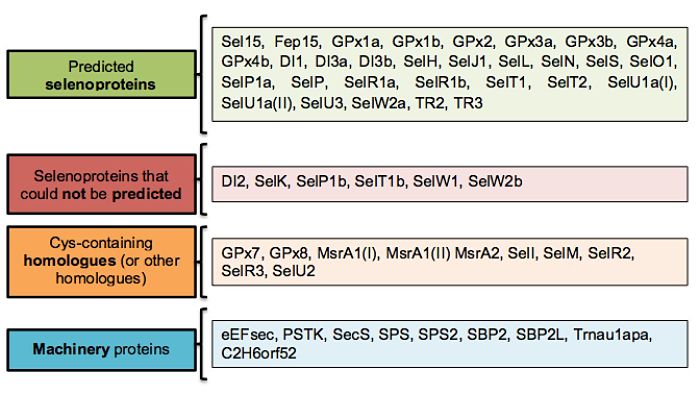
In general, a good homolgy can be found betweeen zebrafish and P. schlosseri selenoproteome.
The main limitation in this study is that only the selenoproteins already described in zebrafish are searched in P. schlosseri’s genome, meaning that could be some selenoproteins not identified in P. schlosseri. In further studies, non-homologous selenoproteins should be searched in the genome of this animal. Another limitation is that not all the proteins in SelenoDB or UniProt are well annotated and in some cases, instead of using zebrafish proteins to do the comparison, humans ones were used. This weakness could be improved with future research studies. Finally, it is important to consider that, in the performed analysis, the existence of SECIS elements was only used as a “confirmatory” tool. However, the reason why in some predicted selenoproteins could not be found, is not clear. Possible explanations are: 1) the genome is not perfectly annotated, 2) the SECIS element is not in the introduced sequence that is scanned or 3) it does not exist.
In conclusion, this study represents a small contribution to the selenoprotein current knowledge by the description of the selenoproteome in an animal that until now, had never been analysed.