 |
|||||||
| Abstract | Introduction | Materials & Methods | Results | Discussion | Conclusions | References | Acknowledgments |
|
|
IntroductionSelenium is an essential nutrient for microorganisms, animals and other eukaryotic organisms. However, it also has a high toxicity in its free form. Therefore, both excess and defficiency of selenium will cause pathology. Selenium defficiency results in Keshan's disease which produces myocardium necrosis and heart weakening. Currently, new studies show that there may be an association between selenium consumption in our diet and prostate cancer prevention proving once again the importance of this nutrient. An excess will be toxic because it will surpass the organism's capacity of selenium chelation and, as said before, its free form has high toxicity. As such, we will find selenium in selenocysteines, an aminoacid that is represented by Sec and U. 1 SelenocysteineSelenocysteine is not part of the 20 main aminoacids but is considered as the 21st aminoacid. Structurally, selenocysteine is the same as a cysteine but there is a selenium atom in place of sulphur atoms in the cysteine. The proteins in which we find selenocysteines are the selenoproteins. Due to the similarity described, we can find homologues where a selenocysteine has been substituted by a cysteine during evolution, these are cysteine homologues.2 3 SelenoproteinsAs mentioned before, selenoproteins have selenocysteines in their sequence. They are redox enzymes and some studies show that they could have antioxidant, antitumoral and immunologic capacity related to selenium chelation.
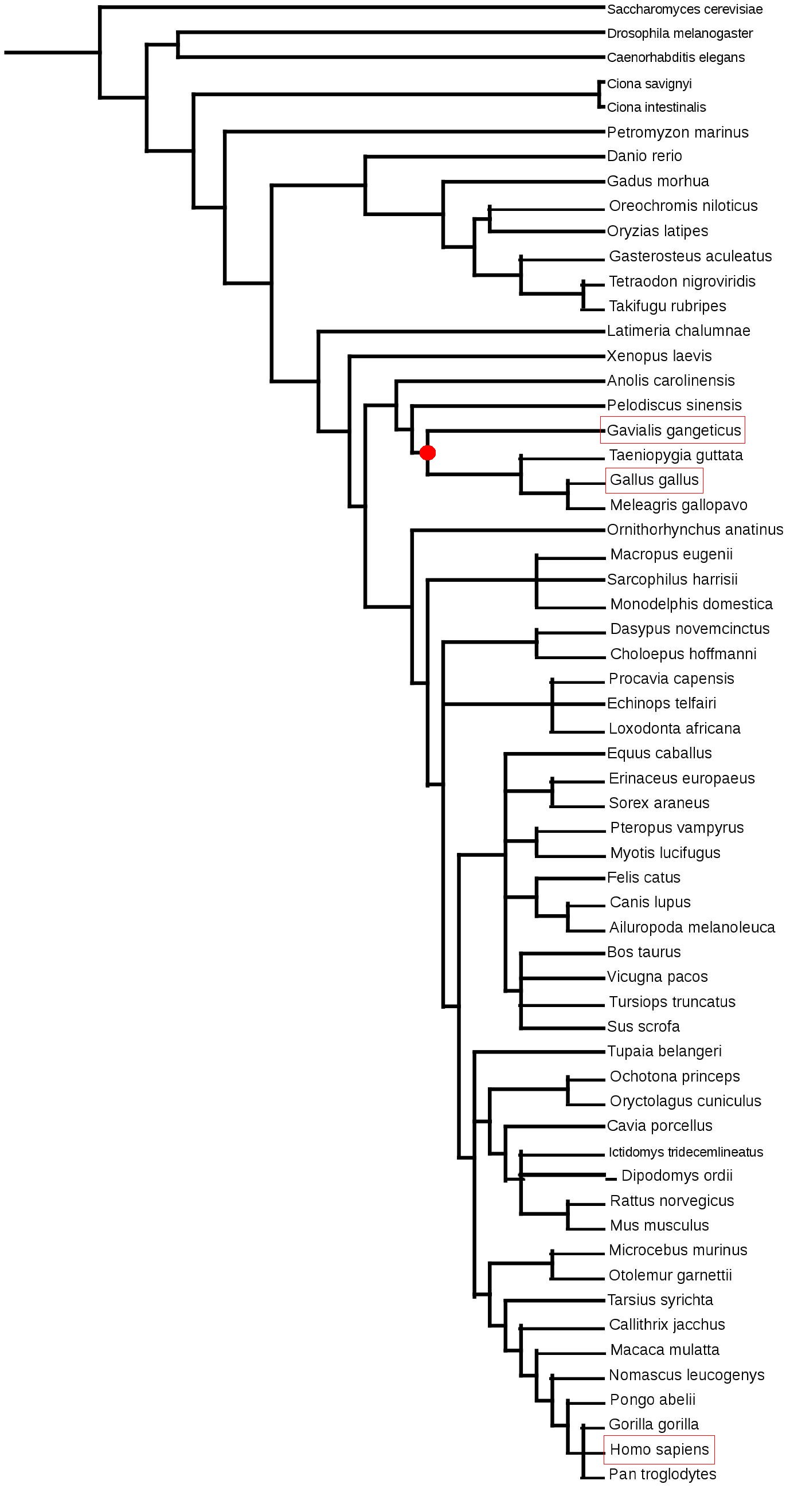
4 It is possible to find selenoproteins in 3 domains: bacteria, archaea and most eukaryotes, but not in plants. We can find a lot of diversity in organisms with selenoproteins. For example, orthologues may contain cysteine instead of selenocysteine (cysteine homologues), there are some species which do not present selenoproteins, they can be different and found in different number among different species.5 Selenoprotein synthesisGenerally, selenoprotein synthesis is similar to other protein synthesis. The difference is that they include an additional aminoacid. We will explain this aminoacid synthesis and its incorporation into the protein during translation explanation. In order to synthesize selenocysteine we can only use selenium in its selenophosphate form because it is the only one we can incorporate. We can obtain it from the dietary selenium thanks to Selenophosphate Synthetase (SPS2). The responsible structure for introducing selenocysteine into the protein sequence is selenocysteinil-tRNA. The synthesis of selenocysteinil-tRNA is as follows: The enzyme Seryl-tRNA synthetase binds serine to the tRNA which will be the precursor for selenocysteine. Then, phosphoseryl-tRNA kinasa (Pstk) will phosphorilate the serine in tRNA and, finally, selenocysteine synthase (SLA/LP) incorporates the selenophosphate to the aminoacid. Eventually, this form will lead to the formation of selenocysteinil-tRNA.6 Once we have synthesized selenocysteinil-tRNA we have to incorporate it in the protein during its translation. The codon assigned to selenocysteine is UGA which usually is a stop codon. The translational machinery should stop when it detects that codon, however, in selenoprotein codifying sequences, the translational machinery does not interpret some of the codons UGA as an STOP, so introduces a selenocysteine and continues until the next STOP codon.7 8 In eukaryotes, the responsible structure for this different interpretation is SECIS (Selenocysteine Insertion Sequence) which is an RNA loop located in the 3'-UTR extreme. The translational machinery introduces a selenocysteine in UGA codons when there's a SECIS element due to two proteins: The protein SBP2 (SECIS Binding Protein 2) which is linked to the three-dimensional structure of SECIS, when SBP2 is attached to SECIS, eEFsec binds to the translational machinery and recruits Selenocysteinil-tRNA. The incorportaion of Sec and the function of eEFsec as elongation factor enables the translation to continue instead of stopping it. 6 7 9 Gavialis gangeticusGavialis gangeticus also known as gavial is a native crocodilian of the northern part of the Indian subcontinent. The global population is estimated in 235 individuals so, it is a critically endangered species. It is one of the longest crocodilians measuring up to 6,25 m and, with 110 sharp interdigitated teeth in its long and thin snout, it is very adapted to catching fish. Gavials once inhabited all the major river systems of the Indian Subcontinent but, nowadays, they only inhabit 2% of their former range.10 Domain: Eukarya Kingom: Animalia Philum: Chordata Class: Reptilia Order: Crocodilia Family: Gavialidae Genus: Gavialis Species: G. gangeticus Choose reference genomeThe phylogenetically closest genomes for which we have selenoprotein notations in SelenoDB are birds. More specifically, we have used Gallus gallus genome because we consider that it will be the best studied due to its role as a model animal.11 In addition, we used the human genome as reference because is the best characterized genome. 
|