
| Home ~ Intro ~ Project ~ Script ~ Results ~ Conclusions ~ Links |
SNPs distribution in genes Last Update: 21 March 2002 |

On February 2001 the sequence of human genome was completed and published in two of the most important and qualified scientific magazines of the world: Nature and Science. Despite of the fact's importance, we must say that now we only have, more or less, all the pieces of a huge jigsaw. The genome is a big mass of information waiting to be translated to a comprehensive language.
Scientists are working hard to achieve this grade of knowledge. It is required the understanding of the genetic code, composed of four nucleotides (adenine, guanine,cytosine and thymine) because they determine the functionality of the main pieces of life, proteins. Thus, sequence variants are a source of individuality: they can differentiate, at least at a genetic level, one human being from another and, for the same reason, they are also involved in differences in drug response, susceptibility to diseases, etc.
In our project we study one kind of variation: SNPs, an acronym that stands for single nucleotide polymorphisms.


Single nucleotide polymorphisms (SNPs) are stable, biallelic sequence variants that are distributed throughout the genome and are present at an appreciable frequency (>1%) in human population. With other types of polymorphism, like insertions or deletions, they cause part of genome variation among individuals. Nevertheless, the biggest part of this sequence variation is attributable to them.
There are three main reasons to identify SNPs:
The principal projects involved in this identification are the SNP Consortium (95% of the sequenced SNPs has been done by it) and the International Human Genome Sequencing Consortium.
 Medicine,
phisiology and population genetics
have found a new powerful tool in them.
Medicine,
phisiology and population genetics
have found a new powerful tool in them.
The SNP Consortium Ltd (TSC) is a non-profit foundation organised for the purpose of providing public genome data: the localisation of up to 300000 SNPs among human genome sequence.
It started in April 1999 with the founds of the Wellcome Trust and thanks to the collaboration of the most important pharmaceutical and technological companies of the world: APBiotech, AstraZeneca PLC, Aventis, Bayer AG, IBM, Motorola, Novartis...
For more information look at the SNP Consortium web site.
The Human Genome Project published the physical map of the human genome achieved with public founds. In that days it contained 90% of the whole sequence, so one of its objectives was ending it and, besides, locating 100000 SNPs by 2003. Nowadays they have mapped 971077, which can be consulted at the NCBI web site.


DNA, messenger RNA (mRNA) and proteins are the main characters of molecular life. DNA is transcripted into non-functional RNA that needs to be processed in mature RNA. This process is known as splicing and it happens in the nucleus, after capping, methylation and poly-adenilation.
RNA splicing is considered an essential regulatory mechanism to control gene expression. A single gene can give rise, by the combination of different exons, to several functionally different proteins. If the splicing machinery is altered, for example by an SNP in an acceptor site, it may lead to human disease.
There are consensus sequences implicated in the phenomenon at the end of each exon, beginning and end of introns and the initiation of the next exon. The AG/GT nucleotides on the boundaries of every intron are very important, as well as the branch site, positioned 18-40 nt from the 3' splicing site.

If you want to know more about the splicing mechanism click here.


We have focused our work on the relationship between SNPs and splicing sites, in order to get the hot points and study if they cause disease. This hot points are SNPs placed in acceptor or donor sites, because it is possible that a mutation there involves an alternative splicing and an error in the protein function. As commented before, nucleotide diversity is a sensitive indicator of biological and historical factors that affect human genome.
However, even if maybe at the first glimpse the possibility that SNPs cause a defective protein is the most attractive, illnesses caused by single mutations are not catching our eyes. The point is that the main use of the SNPs map will be in dissecting the contributions of individual genes to diseases that have a complex, multigenic basis. It will help in our comprehension of, for example, tissue and organ incompatibility, susceptibility (or protection, remember it) to Alzheimer's disease, AIDS...
But... why?
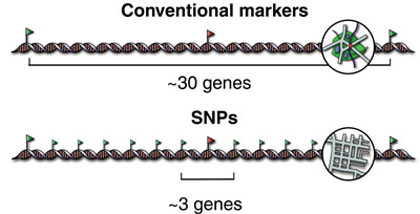
Because making an SNPs map will provide us an enough dense marker to identify all genome and make correlations between unknown mutations and mapped SNPs that are inherited together.
In our project we are locating SNPs among genomic features (exons, introns and splicing sites) and we are studying their distribution over there. Moreover we will try to identify genetic diseases caused by differential splicing due to SNPs.

