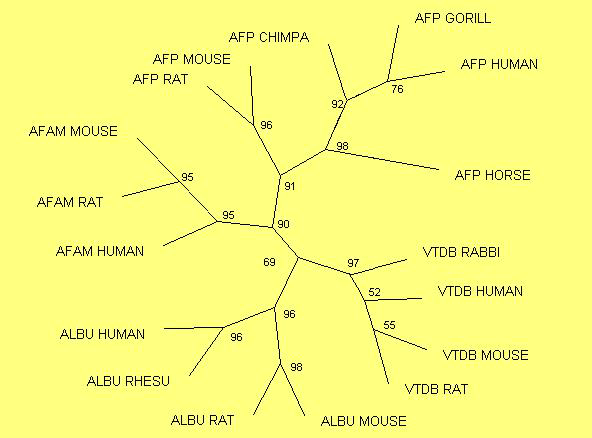
Figure 8. Neighbor-joining tree.
To study the evolutionary relationships among the members of the serum albumin family, a phylogenetic tree has been reconstructed. The multiple alignment used contains sequences of all the proteins of the family (table 1) from different species. The phylogenetic tree has been reconstructed using the PHYLIP 3.5 package program (SeqBoot, Protdist, Neighbor and Consense) and Treeview program has been used to visualize it. The method chosen for the reconstruction of the tree was Neighbor-Joining. The data have been supported by a bootstrap analysis.

Figure 8. Neighbor-joining tree.
All the members of the family derive from a common ancestor which itself arose by triplication (Brown, 1976), vestiges of which are detectable in the modern sequences and the exon structures of the genes themselves.
The first duplication of the progenitor happened 580 Myr ago. From this duplication arose the gene that encodes for VTDB and a second linage. This linage suffered another duplication 295 Myr ago. In this second duplication the ALB gene and a common precursor to ALF and AFP could be observed. The last duplication in this family occurred 250 Myr ago, giving origin to ALF and AFP (Gibbs et al., 1998).
It is possible to correlate
the history of this gene family with the distribution of the members among
different species. The triplication of the ancestral gene must have occurred
before the divergence of vertebrates, since the triplicated structure is
present in mammals, amphibians and bony fish, but not in jawless fish (Gibbs
& Dugaiczyk, 1994). All the members of the family are present
in mammals, while in amphibia ALF and AFP do not exist . Amphibia diverged
from reptiles about 360 Myr ago, about 65 Myr before the divergence of
the albumin and the ALF and AFP progenitor (Gibbs et al.
1998).
Home | Introduction | Gene analysis | Protein analysis | Evolutionary study | References