

Selenoproteins are proteins that contain selenocysteine (Sec, U), known as the 21st amino acid, which has an important role in redox homeostasis. That amino acid is encoded by the UGA codon, which normally signals translation termination. Thus, for the recognition of this codon as a selenocysteine codon, a SECIS element is required, a cis-acting stem-loop located in the 3′-UTR of selenoprotein mRNAs, immediately downstream of the UGA codon.
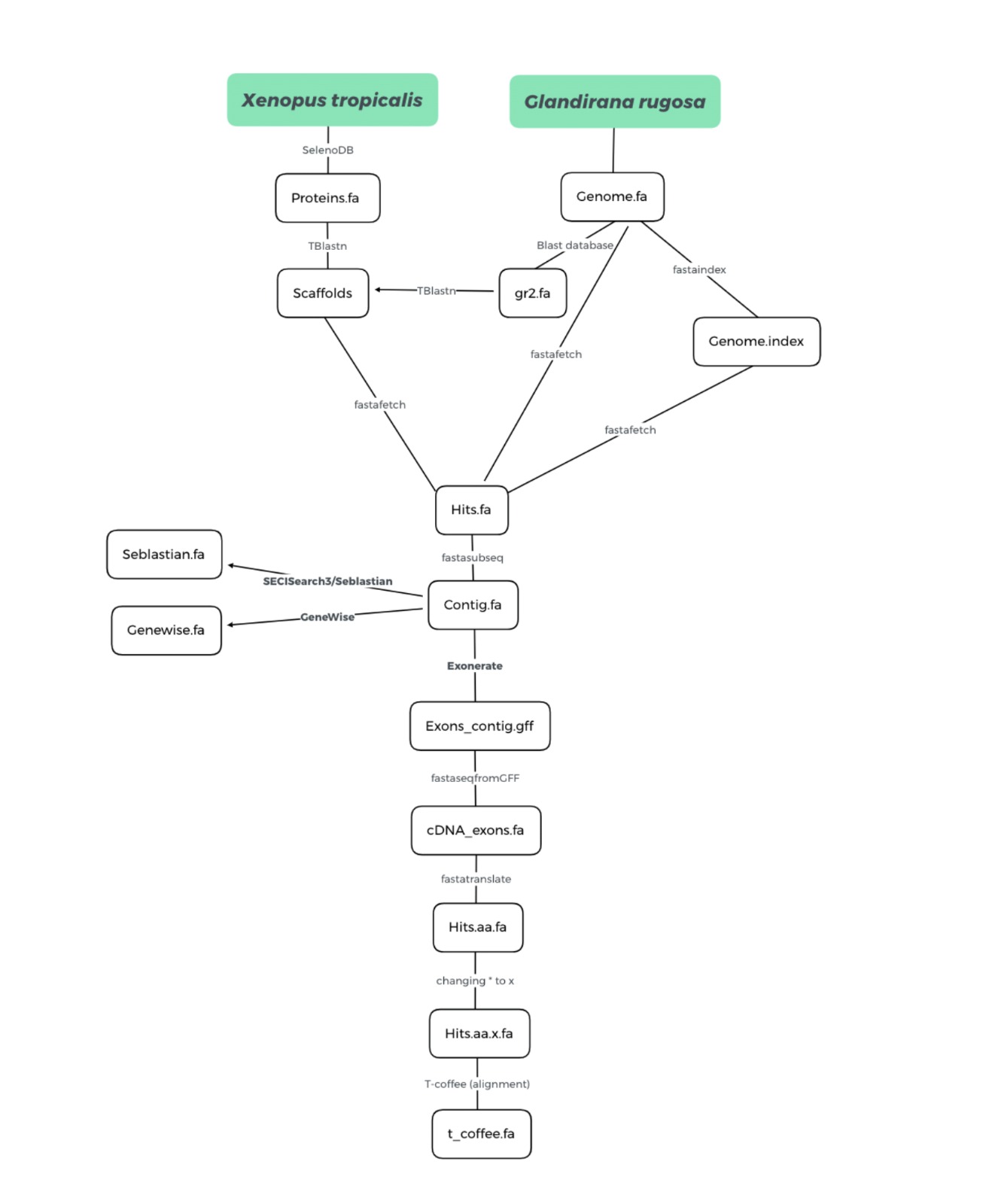
This study aims to annotate the selenoproteome and the machinery necessary for its synthesis of Glandirana rugosa, which is an endemic frog from japan. To archive that, a homology-based approach was used by making a comparison between Glandirana rugosa’s genome and Xenopus tropicalis’ selenoproteins found in SelenoDB 2.0. A program in Bash was created to automate the process of analysis, including different bioinformatic tools such as tblastn, exonerate and t-coffee. Additionally, programs such as Seblastian and GeneWise were also used to identify SECIS elements and to check the result.
As a result, a global alignment of Xenopus’ selenoproteins and predicted G. rugosa’s proteins was obtained. Therefore, it can be concluded that the proteins which have selenocysteines are SecS80, PSTK, Sel15, SelT, SelI, GPx72, GPx73, TR02, GPx69 and DI62. Whereas, proteins which do not have selenocysteine are Fep15, TR04, SelK, GPx67, GPx68, GPx70, GPx71, TR01, TR03, TR04, DI63, DI64, MsrA74, MsrA75, MsrA76, SelR91, SelR92, SelR93, SelI, SelK, SelK, SelS, SelM, SelN, SelO_86, SelO_87, SelO_88, SelP_89, SelP_90, SelW00, SelW99, SelU96, SelU97, SelU98, Fep15, eEFsec05, eEFsec06, SBP2_77, SBP2_78 and SecS79. Finally, there are a group of proteins which do not exist in Glandirana rugosa: TR04, FrnE and SelK.
Selenium (Se) is a trace essential element for humans, plants, and microorganisms. Inorganic
selenium is present in nature in four oxidation states, and these forms are converted by biological
systems into more bioavailable organic forms, mainly as the two seleno-amino acids selenocysteine and
selenomethionine (1). Interest in selenium has considerably grown over the last decades due to the
association of selenium deficiencies with an increased risk of several human diseases, including cancers,
cardiovascular disorders and infectious diseases (2).
The biosynthesis of selenoproteins is unique, since the incorporation of Sec occurs co-translationally
by the ribosome and not post-translationally.
Sec introduction requires special trans-acting protein factors, Sec-tRNA[Ser]Sec and a
cis-acting Sec insertion sequence (SECIS) element.
It is the own tRNA of Sec, which is the longest tRNA sequenced, and the key molecule and central
component of selenoprotein biosynthesis. The gene for tRNA[Ser]Sec is Trsp, it is
found in all three evolutionary lines of descent: eukarya, archaea, and eubacterya.
Its transcription is regulated by three upstream regions: a TATA box motif, a proximal sequence element
and a distal sequence element.

Figure 1. Synthesis of Sec-tRNA[Ser]Sec (5).
The SECIS elements are cis-acting stem-loop located in the 3′-UTR of selenoprotein mRNAs,
immediately downstream of the UGA that encodes for Sec. When a ribosome encounters the UGA codon, which
normally signals translation termination, Sec machinery interacts with the canonical translation machinery
to augment the coding potential of UGA codons and prevent premature termination. SECIS elements serve as
the factors that dictate recoding of UGA as Sec. In response to the SECIS element in selenoprotein mRNA,
Sec-tRNA[Ser]Sec, which has an anticodon complimentary to the UGA, translates UGA as Sec.

Figure 2. Mechanism of Sec insertion (5).
Proteins containing Sec are present in all three evolutionary lines of descent: eukarya,
archaea, and eubacteria, and they were also observed in viruses.
However, selenoproteins were completely lost in fungi, higher plants, and some animal species,
including beetles, silkworms, and several other insects. Interestingly, more than half of the
identified selenoprotein families are present in both single-cell eukaryotes and vertebrates,
indicating that they have an ancient origin (5).
Figure 3. Evolution of the vertebrate selenoproteome (6).
The ancestral vertebrate selenoproteome is indicated in red. The ancestral selenoproteins found
uniquely in vertebrates are underlined. The creation of a new selenoprotein (here always by
duplication of an existing one) is indicated by its name in green. Loss is indicated in grey.
Replacement of Sec with Cys is indicated in blue (apart from SelW2c in pufferfish, which is
with arginine). Events of conversion of Cys to Sec were not found. On the right, the number
of selenoproteins predicted in each species is shown.
Sec is the main element in charge of the physiological functions of selenoproteins. It is
found on the active site of the enzyme and perform catalytic redox reactions. The
classification of selenoproteins bases on their function is the following:
GPxs are well known to be the major components of antioxidant defence. Moreover,
they are involved in hydrogen peroxide (H2O2) signalling, detoxification
of hydroperoxides, and maintaining cellular redox homeostasis.
TR is the only enzyme able to reduce oxidized Thioredoxin (Trx). In addition, the Trx
system participates in many cellular signalling pathways by controlling the activity of
transcription factors containing critical cysteines in their DNA-binding domains, such as
NF-κB, AP-1, p53, and the glucocorticoid receptor.
Three DIs have been identified with a tissue and subcellular localization, which are involved
in regulation of thyroid hormone activity by reductive deodination.
MsrB1 (methionine-R-sulfoxide reductase B1) was initially identified as selenoprotein R
(SelR) and selenoprotein X (SelX) by searching for putative SECIS element structures in EST databases.
Later, this protein was found to function as a stereospecific methionine-R-sulfoxide reductase, which
catalyses repair of the R enantiomer of oxidized methionine residues in proteins. Based on its functional
similarity to methionine-S-sulfoxide reductase A (MsrA), which catalyses reduction of the other
isomer, this selenoprotein was renamed MsrB1.
SPS2 catalyses the synthesis of the active Se donor selenophosphate that is necessary for Sec
biosynthesis.
SelI is found only in vertebrates. It is a transmembrane protein containing a highly conserved
CDP-alcohol phosphatidyltransferase domain, which is present in choline (CHPT1) and choline/ethanolamine
(CEPT1) phosphotransferases. CHPT1 and CEPT1 catalyse the last step in de novo synthesis of the
two major phospholipids through the transfer phosphocholine and phosphoethanolamine groups to
diacylglycerol from CDP-choline and CDP-ethanolamine, respectively.
SelK and SelS have different sequence, but they are studied together because they
similar topology, including a single transmembrane domain in the NH2-terminal sequence;
the presence of a glycine-rich (G-rich) segment and a characteristic location of Sec residues in
the COOH-terminal end of the protein. They are found in ER membrane. Their exact function is
unknown, but it is suggested that both are related to ER-associated degradation (ERAD) of
misfolded proteins.
SelM is an ER-resident thiol-disulphide oxidoreductase that is highly expressed in the
brain and bestows neuroprotective properties, preventing oxidative damage induced by
H2O2. In humans, it seems to be a protector factor in Alzheimer's
disease.
SelN is an ER-resident transmembrane glycoprotein that is highly expressed during
embryonic development and in adult tissues, including skeletal muscle. It plays a role in the
regeneration of skeletal muscle tissue after injury or stress. Mutations in the human SelN
gene (also known as SEPN1) are associated with a group of early-onset muscle disorders
known as SEPN1-related myopathies.
SelO are in mammalian and yeast mitochondria, engaged in redox interaction with an
unknown protein through its CXXU motif. Redox regulation of protein function in mitochondria
may involve kinase functions.
SelP is a selenoprotein with multiple Sec residues per protein subunit, 10 Sec
residues in all, and is present in human plasma. The main role of SelP is the transport and
delivery of selenium to the tissues. An additional role may be to serve as a heavy metal
chelator or antioxidant.
The members of this protein family possess a thioredoxin-like fold and are characterized
by the presence of a conserved Cys-x-x-Sec motif, and a conserved stretch of amino acids in
the COOH-terminal portion of the protein with the tGxFEI(V) consensus sequence.
SelW is a small 9-kDa selenoprotein localized in the cytosol and is expressed at
high levels in muscles and brain. It belongs to the stress-related group of selenoproteins
as its expression is highly regulated by the availability of Se in the diet.
SelT is predominantly localized to the ER and Golgi and is ubiquitously expressed
both during embryonic development and in adult tissues. Knockdown of SelT in mouse fibroblasts
leads to decreased expression of extracellular matrix genes involved in cell structure
organization and alters cell adhesion properties. In addition, the loss of SelT resulted in the
upregulation of SelW.
SelH has a unique subcellular localization pattern and was found to localize specifically
to the nucleoli. Expression of SelH is relatively low in adult mouse tissues, but is elevated during
embryonic development. Similar to SelW, SelH is sensitive to dietary Se intake.
SelV is one of the least characterized selenoproteins. It recently evolved, most
likely by duplication from SelW, and is found only in placental mammals. SelV expression is
detected only in testes, and thus may be involved in male reproduction, but its specific
function is not known.
SelU was firstly found in fish and also reported in birds and unicellular eukaryotes.
In high mammalian species, such as humans and mice, all SelU proteins exist in Cys form. The
function of SelU remains unclear.
Sel15 is known in mammals as Sep15, which is together with Selenoprotein M
(SelM) thioredoxin-like fold ER-resident protein. Sel15 involved in calnexin regulation
cycle, which is essential for the folding process of some glycoproteins in the ER, and its
expression is induced by misfolded proteins in ER. There is a protein called Fep15 (15-kDa
selenoprotein-like protein) which it as a Cys-homologue in frog.
This protein phosphorylates Ser-tRNA[Ser]Sec to create then the
Sec-tRNA[Ser]Sec. The function and homology of this protein are conserved across
archaea and eukaryotes that synthesize selenoproteins.
eEFSec recruits Sec-tRNA[Ser]Sec and includes a Sec amino acid in a protein.
SBP2 promotes Sec incorporation by associating with SECIS elements and recruiting the
eEFSec-selenocysteyl-tRNA[Ser]Sec complex to the ribosome.
This enzyme incorporates the active form of Se to the Ser-tRNA[Ser]Sec phosphorylated to create the final Sec-tRNA[Ser]Sec.
The Japanese rugose frog, which has the scientific name Glandirana rugosa (Temminck & Schlegel, 1838), is an endemic species of Japan, belonging to the family Ranidae.
| Taxonomy | |
|---|---|
| Kingdom | Animalia |
| Phylum | Chordata |
| Class | Amphibia |
| Order | Anura |
| Family | Ranidae |
| Genus | Glandirana |
| Species | G. rugosa |
The species is distributed in Japan (north, central and southern Honshu Island, as well as in the Shikoku,
Kyushu, Sado, Oki, Goto, Yakushima, Tsushima and Tanegashima Islands), north and southwest Korea and northeast
China (Liaoning Provinces, Jilin and Heilongjiang Provinces).
It was also introduced to Hawaii at the end of the 19th century (10).
The dorsal colouration of Glandirana rugosa is predominantly muddy brown, with many short ridges
protruding from its back. The ventral colouration is pale yellow or greyish yellow. The size of males varies
from 30 to 47 mm and females from 45 to 60 mm.
Sexual maturity is reached between 1 and 2 years after metamorphosis in males and 2 or 3 years later in females,
with variations of at most one year after metamorphosis. Males can live for up to four years and females five
years after metamorphosis (11).
In Japan, Glandirana rugosa is distributed mainly in lowland areas, with a wide habitat
preference. It can be found in rice paddies, aqueducts, water reservoirs, ponds (natural and artificial) and
along small streams of rapid flow.
In the northern province of Hokkaido, Japan, the species was accidentally introduced through
the aquaculture production of carp in 1985. It has been reported to predate the terrestrial
insects of Hokkaido, where it could pose a threat to the trophic networks and compete with
native frog species.
The sex determination of frogs is complicated. In some species, males have two distinct
sexual chromosomes (a system of XX-XY sex chromosomes, such as mammals), while females of
other species have two distinct sexual chromosomes (a system of ZZ-ZW sex chromosomes, such
as birds). The fact that a species has a system XX-XY or ZZ-ZW has changed tens of times
throughout the evolution of frogs.
Glandirana rugosa has sometimes been considered a single species, along with
Glandirana emeljanovi, which is found on the East Asian mainland.
The two species are distinguished from others by their rough and uneven skin.
The aim of our project was to find the selenoproteins and cysteine-containing
homologues or selenium machinery proteins in the organism Glandirana rugosa.
Its genome was obtained from executing the following command in the shell:
$/mnt/NFS_UPF/soft/genomes/2021/Glandirana_rugosa/
The file which contains its genome is named as genome.fa.
Firstly, we have chosen Xenopus tropicalis as our animal of reference, due to
its close phylogenetic relationship with our organism.
$sed s/U/X/g $p > $p
Then, the changed sequence of each selenoprotein was named as each protein’s name
manually by us. The name was the one that was on the Xenopus protein annotation.
A BLAST database of the G. rugosa genome was made from the genome.fa file
using the following command:
$makeblastdb -in genome.fa -dbtype nucl -out gr2.fa
Then, it ran TBLASTN, a program that compared the protein query to our genome of interest and gave us all the scaffolds found with their significant hits. All the scaffolds with an E-value bigger than 0.01 were discarded.
$tblastn -query $f -db $gr2in -outfmt 6 -evalue 0.01 -out $blastfile
Furthermore, we identified the start and the end of each scaffold and calculated
the length.
$cut -f2 $blastfile | sort | uniq | while read scaffold; do
$start= grep $scaffold $blastfile | cut -f9-10 |sed 's/\t/\n/' | sort -n | head -1
$end= grep $scaffold $blastfile | cut -f9-10 |sed 's/\t/\n/' | sort -n | tail -1
$hit=$(echo $scaffold | cut -f 2 -d ' ')
$begin= "$(($start-$hit_offset))"
$length= "$(($begin + $hit_offset ))"
To extract the genomic sequence from the found region, firstly, we have created
an index of the genome of G. rugosa by using:
$fastaindex ./genome.fa gr2.index
Afterwards, with the following command, we extracted all the selected scaffolds
from the G. rugosa genome, and the output was saved in a name_scaffold.fa file.
$fastafetch $genin $indexgr2in "$hit" > $fastfechfile
Once we extracted the scaffold, and we defined the start and length position,
we obtained the region of interest (contig) with the fastasubseq program using the
following command line:
$fastasubseq $fastfechfile $begin $length > $fastagenomfile
From the protein query and the genomic regions that we have extracted above, we will
generate an annotation of the genes by using exonerate. In other words, we generated a
FASTA file containing the exons of the predicted protein. Our command line was:
$exonerate -m p2g --showtargetgff -q $f -t $fastagenomfile | egrep -w exon > $exoneratefile>
FasteseqfromGFF was used to generate a FASTA file with the cDNA of the exons of predicted
protein from the .gff file provided by exonerate.
$fastaseqfromGFF.pl $fastagenomfile $exoneratefile > "${fastaseqdir}/fastaseq_${hit}.fa"
In this step, we have translated the cDNA nucleotide sequence into a protein sequence.
For each cDNA, several possible proteins were generated by different reading frames. We have
used the first option of the program (-F 1). This was done with the following command line:
$fastatranslate "${fastaseqdir}/fastaseq_${hit}.fa" -F 1 > "${fastaseqdir}/fastaseq_${hit}.aa.fa"
In the protein sequences acquired by the previous step, the residue U was represented by
a *. In order to compare it later with the query sequence from Xenopus tropicalis,
the * has changed to an X.
$sed s/*/X/g "${fastaseqdir}/fastaseq_${hit}.aa.fa" > "${fastaseqdir}/fastaseq_${hit}.aa.x.fa"
Finally, we have used T-Coffee to perform a global alignment between the predicted protein
sequence and the query protein from Xenopus tropicalis. The command line used by us was:
$t_coffee $f "${fastaseqdir}/fastaseq_${hit}.aa.x.fa" > "${t_coffeedir}/${queryname}_${hit}.tc.fa" "${t_coffeedir}/${queryname}_${hit}.tc.aln" "${t_coffeedir}/${queryname}_${hit}.tc.html
To check the results obtained by Exonerate, we have also executed the program called GeneWise,
which from the protein query and the genomic region we have extracted generate an annotation of
the gene that gives rise to this protein.
$genewise -pep -pretty -cdna -gff $f $fastagenomfile > "${genwisedir}/${queryname}_${hit}.gw.fa"
Furthermore, we have used the SECISearch3 and Seblastian web server
to predict SECIS elements in the 3’-UTR, and then to search upstream for selenoprotein coding
sequences. As Seblastian does not recognize the other symbols from ambiguity code rather than N
we solved it by substituting them by an N.
$sed s/*/N/g $fastagenomfile > "${seblastdir}/${hit}.fa
Once we have substituted the ambiguous bases for N, we have to manually select the proteins and go to the SECISearch3 website to predict the SECIS elements and the selenoprotein coding sequences.
The following table shows, for each query, the reference species (Xenopus tropicalis), the Sec/Cys-homologue, its Tblastn output, the scaffold or scaffolds selected, the gene prediction (fastasubseq), the exonerate prediction for the protein (.gff), the protein prediction, the sequence alignment (T-coffee) for the prediction, the SECIS-element predictions and the Seblastian prediction for the selenoprotein.
The proteins that we have obtained from SelenoDB 2.0 were numbered by families, but in various families we obtained more than one different protein for each family, which we have named by the name of the family and a number that corresponds to the last two digits of the query name of Xenopus tropicalis. In the case we had more than one scaffold protein that had exons, we chose those that present the minor e-value scaffold since it is statistically the most reliable.
Due to the problems mentioned above with the annotation of the Xenopus tropicalis proteins we will discuss our proteins with our own created annotation. It will be difficult to contrast our proteins with the literature hypothesis but even with this issue we will be able to study the differences between each family and them with Xenopus tropicalis proteins.
Of each SECIS element we have chosen the greater grade, being A the best, that is on 3’ position in the same strand as our exons of our scaffold.
In our study we studied the selenoproteins in Glandirana rugosa studying its homology with Xenopus tropicalis (frog) proteins. Our aim was to focus the analysis principally on Xenopus tropicalis, due to its closer phylogeny to Glandirana rugosa.
All the results obtained for each predicted protein were carefully analysed and discussed, paying special attention to the T-Coffee output and SECIS elements prediction.
The criteria for deciding whether a detected protein in Glandirana rugosa genome was a selenoprotein, a cysteine-containing homologue or neither of them was the following:
There is one protein, FrnE, a disulfide isomerase, which we don't have any information about its phylogeny in Xenopus tropicalis, family and didn’t match on the tblastn with the genome of Glandirana rugosa; so we descarted it.
The glutathione peroxidase family (GPx) is the common name for a family of multiple isozymes that catalyze the reduction of H2O2 or organic hydroperoxides to water or corresponding alcohols using reduced glutathione (GSH) as an electron donor (H2O2 + 2GSH fi GS-SG + 2H2O). Aerobic reactions lead to the accumulation of reactive oxygen species that can be toxic to the cells. In this context, aerobic organisms have developed several n-enzymatic and enzymatic systems to neutralize these compounds.
After our results’ analysis, we could tree GPxs selenoprotein in our organism. Into to the information explained below we will expose if they have or not SECIS elements, their Seblastian prediction and if they present or not Sec residus.
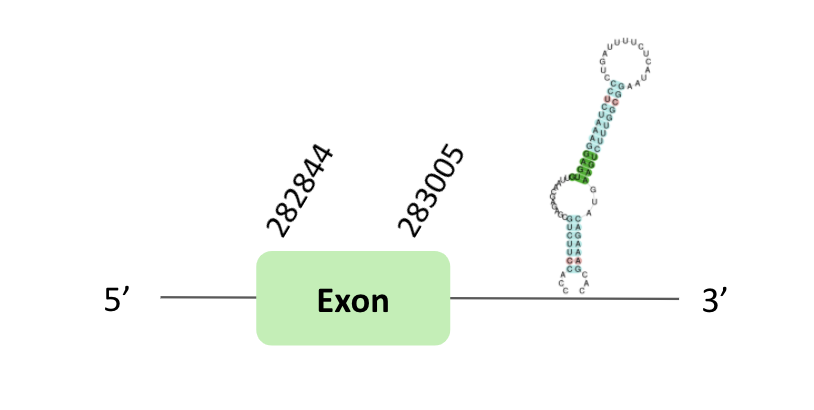
The GPx67 protein location is in the scaffold BLSH010357311.1 between positions 282868 and 283005 in the forward strand. There was no predicted exon.
This protein was predicted blasting Glandirana rugosa genome against the Xenopus tropicalis GPx67 protein found in SelenoDB. To find the gene structure, we analysed the exonerate file.
Regarding the SECIS element, a grade B SECIS was found in the 3'UTR region. Regarding the Seblastian there were no selenoproteins coding sequences. In this case, we did not observe selenocysteine.
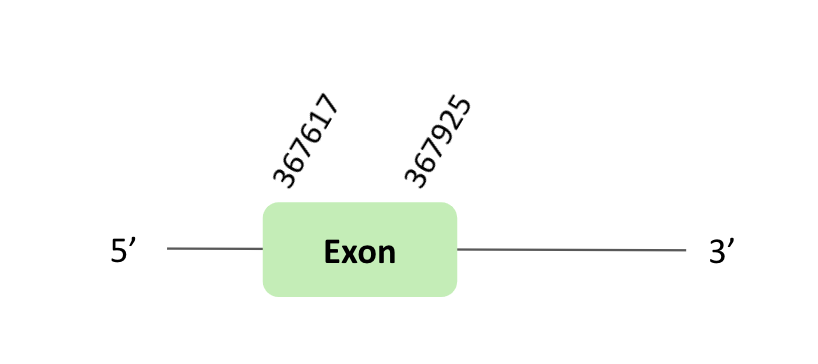
The GPx68 protein location is in the scaffold BLSH010554733.1 between positions 367617 and 367925 in the reverse strand. The gene contains 1 exon.
This protein was predicted blasting Glandirana rugosa genome against the Xenopus tropicalis GPxs protein found in SelenoDB. To find the gene structure, we analysed the exonerate file.
The SECIS element predicted in the 3'UTR region was not useful for this protein, because the coordinates of the SECIS were not larger than the last exon. Regarding the Seblastian there were no selenoproteins coding sequences. In this case, we observed a loss of Xenopus tropicalis selenocysteine.
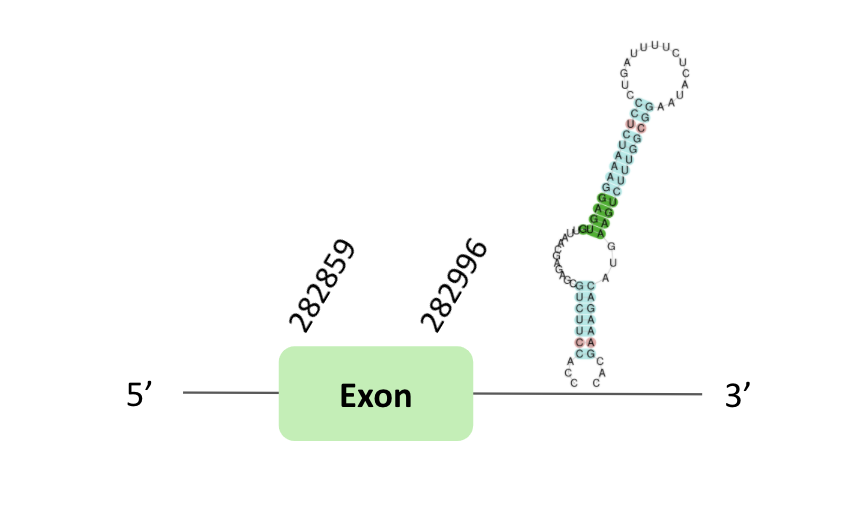
The GPx69 protein location is in the scaffold BLSH010357311.1 between positions 282859 and 408144 in the forward strand. The gene contains 1 exon.
This protein was predicted blasting Glandirana rugosa genome against the Xenopus tropicalis GPxs protein found in SelenoDB. To find the gene structure, we analysed the exonerate file.
Sequentially, SECIS element of grade B, predicted in the 3'UTR region, was found but in the case of Seblastian we did not have any result. Nevertheless, in the t-coffee result we observed a sec gain.
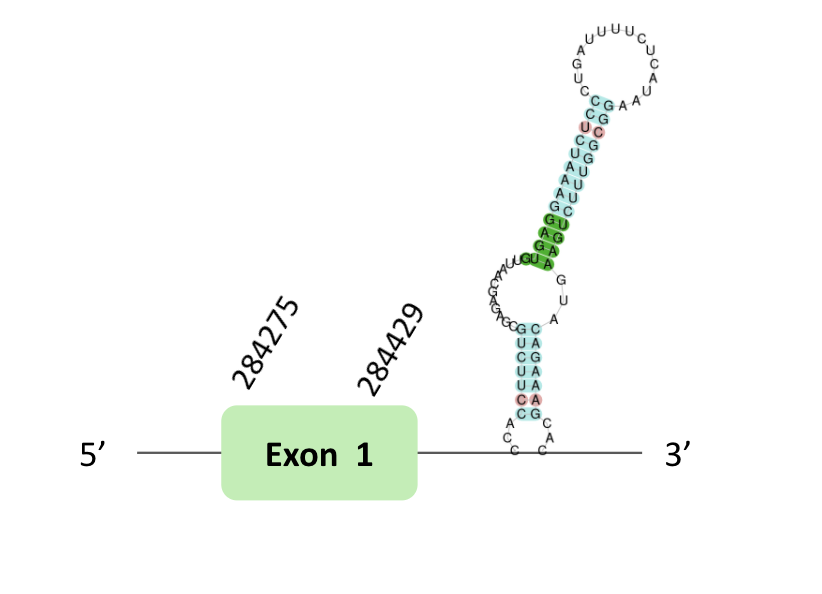
The GPx70 protein location is in the scaffold BLSH010357311.1 between positions 284272 and 284421 in the forward strand. The gene contains 1 exon.
This protein was predicted blasting Glandirana rugosa genome against the Xenopus tropicalis GPxs protein found in SelenoDB. To find the gene structure we analyzed the exonerate file.
Sequentially, SECIS element of grade B, predicted in the 3'UTR region, was found. Finally, we did not have a Seblastian result and in t-coffee results we did not have a selenocysteines.
The GPx71 protein location is in the scaffold BLSH010085861.1 between positions 1079646 and 1079795 in the reverse strand. The gene does not contain exons.
This protein was predicted blasting Glandirana rugosa genome against the Xenopus tropicalis GPx71 protein found in SelenoDB. To find the gene structure, we analyzed the exonerate file.
Finally, neither the SECIS element nor Seblastian nor t-coffee results were obtained.
The GPx72 protein location is in the scaffold BLSH010357311.1 between positions 282850 and 283005 in the forward strand; in this scaffold one exon was predicted.
Another scaffold BLSH010554733.1 is located between positions 367623 and 374034 in the reverse strand, in which two exons were predicted.
These proteins were predicted blasting Glandirana rugosa genome against the Xenopus tropicalis GPx72 protein found in SelenoDB. To find the gene structure, we analyzed the exonerate file.
In the scaffold BLSH010357311.1, a B grade SECIS element was predicted in the 3'UTR region.

In the scaffold BLSH010554733.1, a B grade SECIS element was predicted in the 3'UTR region.
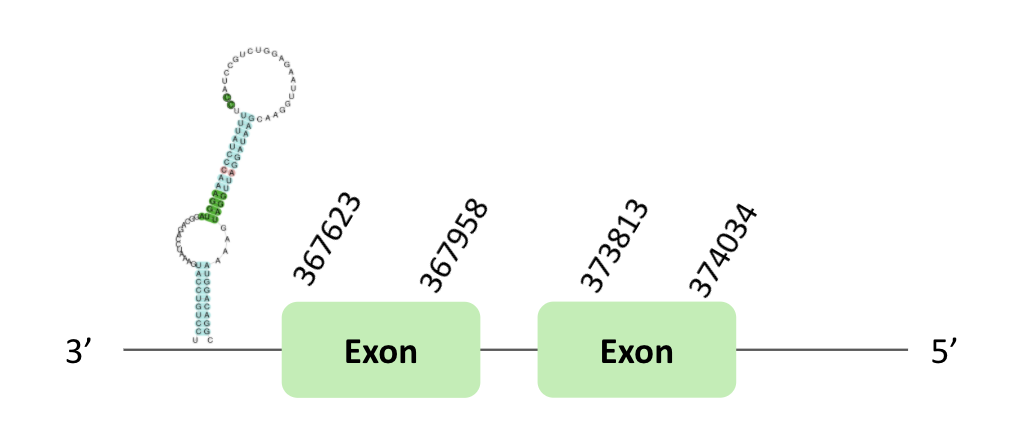
In both cases, regarding the Seblastian, there were no selenoprotein coding sequences. Nevertheless, in both scaffolds there was a sec-homologous selenocysteine.
The GPx73protein location is in the scaffold BLSH010357311.1 between positions 282844 and 283005 in the forward strand. The gene contains 1 exon.
This protein was predicted blasting Glandirana rugosa genome against the Xenopus tropicalis GPx73 protein found in SelenoDB. To find the gene structure we analyzed the exonerate file.
Sequentially, SECIS element of grade B, predicted in the 3'UTR region, was found. Finally, no Seblastian result was obtained. Nevertheless, in t-coffee we predicted 2 selenocysteines, one of them was sec-homologous selenocysteine and the another one was a loss of selenocysteine in G. rugosa.
The thioredoxin reductases family (TR) is a protein family composed by flavoproteins, which function as homodimers, actively involved in redox regulation of cellular processes due to their capacity to control the redox status of thioredoxins. It is the only enzyme known to catalyze the reduction of thioredoxin (Trx) and, hence, is a central component in the thioredoxin system. This system is present in all living cells, and it also has an evolutionary history tied to metabolism and redox signaling.
After our results’ analysis, we could predict one TRs selenoprotein in our organism. According to the information explained below, we didn’t find a Sec residue nor SECIS element nor Seblastian prediction in our two of our four proteins. The other two have SECIS elements but neither have Sec residue, they have experimented a lost on this residue in three of them. The same two that have SECIS elements they also have Seblastian prediction.
The TR01 protein location is in the scaffold BLSH010481670.1 between positions 1521727 and 1595605 in the forward strand, in which 12 exons were predicted. This protein was predicted blasting Glandirana rugosa genome against the Xenopus tropicalis TR01 protein found in SelenoDB. To find the gene structure we analyzed the exonerate file. There was no SECIS element useful for our strand, because the coordinates of the SECIS were not larger than the last exon, and we did not detect any selenocysteine through Seblastian, and in the t-coffee we could observe a loss of X. tropicalis selenocysteine.
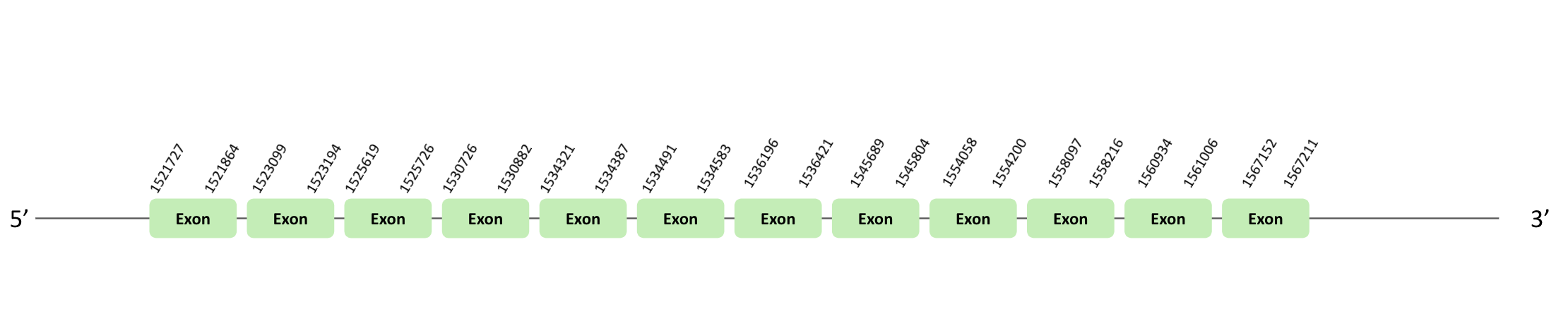
The TR02 protein location is in the scaffold BLSH010481670.1 between positions 1521727 and 1595605 in the reverse strand. The gene contains 14 exons. This protein was predicted blasting Glandirana rugosa genome against the Xenopus tropicalis TR02 protein found in SelenoDB. To find the gene structure we analyzed the exonerate file. A SECIS element of grade A was predicted in the 3'UTR region. Finally, the Seblastian was positive and we found a sec-homologous selenocysteine in t-coffee.
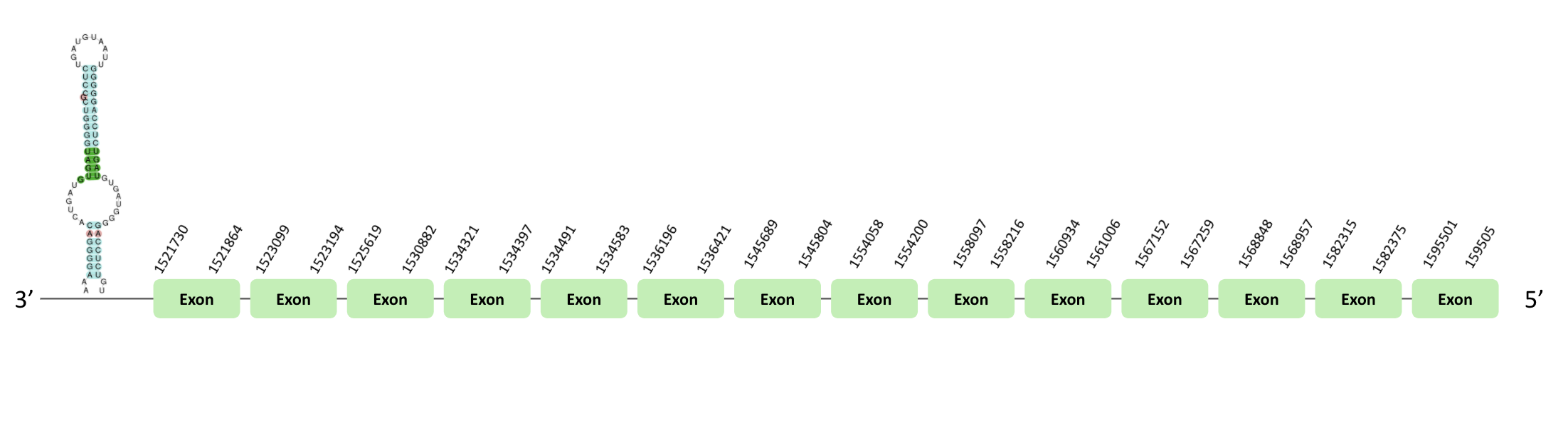
The TR03 protein location is in the scaffold BLSH010481670.1 between positions 1521730 and 1558217 in the reverse strand. The gene contains 5 exons. This protein was predicted blasting Glandirana rugosa genome against the Xenopus tropicalis TR03 protein found in SelenoDB. To find the gene structure we analyzed the exonerate file. Sequentially, a SECIS element of grade A predicted in the 3'UTR region was found. Moreover, the Seblastian result indicated the presence of selenoprotein coding sequences. In the t-coffee result, we found a loss of a X. tropicalis selenocysteine.
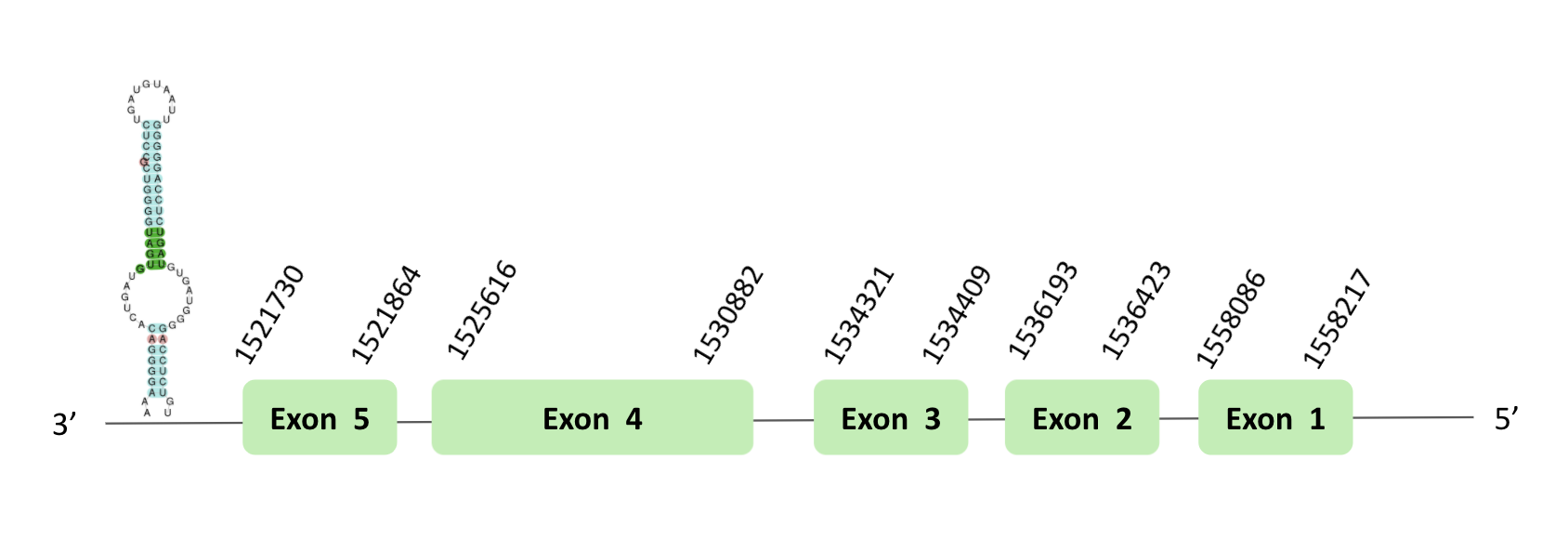
This protein was predicted blasting Glandirana rugosa genome against the i>Xenopus tropicalis TR04 protein found in SelenoDB. But we could not do the tblastn, so we assumed that this protein is not found in Glandirana rugosa.
The iodothyronine deiodinases family (DI) is the general name for a family constituted of enzymes that catalyze the removal of iodine atoms from various thyroid hormones (Ths) in the thyroid gland and extrathyroidal tissues. They are responsible for both the activation and inactivation of these compounds, and are thus important regulators of TH actions.
After our results’ analysis, we could predict one DI selenoprotein in our organism. According to the information explained below, we didn’t find a Sec residue nor SECIS element nor Seblastian prediction in our two of our four proteins. The other two have SECIS elements but neither have Sec residue nor Seblastian prediction.
The DI62 protein location is in the scaffold BLSH010015975.1 between positions 251100 and 251343 in the reverse strand. The gene contains 1 exon. Another scaffold BLSH010535000.1 is located between positions 614022 and 614355 in the reverse strand, which contains 1 exon.
These proteins were predicted blasting Glandirana rugosa genome against the Xenopus tropicalis DI62 protein found in SelenoDB. To find the gene structure, we analyzed the exonerate file.
A SECIS element of grade B was predicted in the 3'UTR region of BLSH010015975.1. In this case, we observed a loss of Xenopus tropicalis selenocysteine.
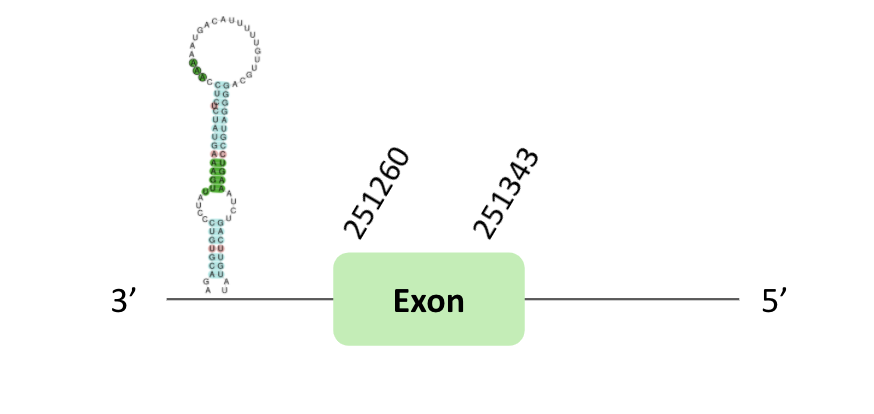
A SECIS element of grade B was also predicted in the 3'UTR region of BLSH010535000.1. In t-coffee results we observed 2 selenocysteines, one of them there was a Xenopus tropicalis selenocysteine which is lost in Glandirana rugosa.
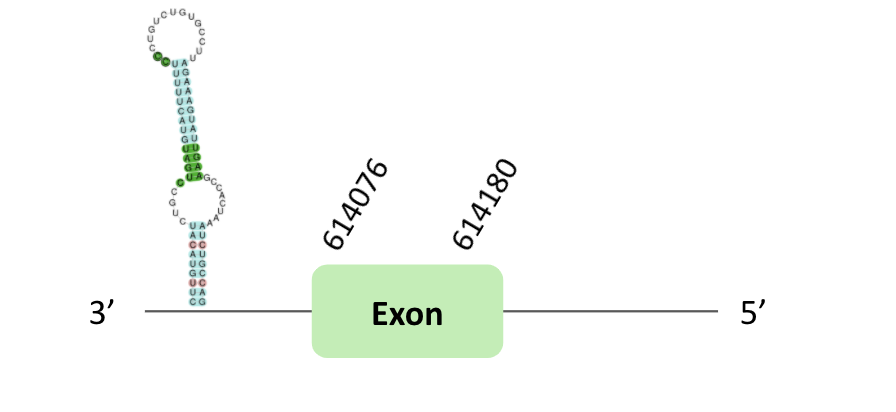
However, non-one of them have a positive Seblastian result.
The DI63 protein location is in the scaffold BLSH010226958.1 between positions 2016293 and 2093027 in the forward strand. The gene does not contain exons.
This protein was predicted blasting Glandirana rugosa genome against the Xenopus tropicalis DI63 protein found in SelenoDB. To find the gene structure, we analyzed the exonerate file.
Moreover, neither the SECIS element nor Seblastian nor t-coffee results were obtained.
The DI64 protein location is in the scaffold BLSH010172101.1 between positions 13299 and 35919 in the forward strand. The gene does not contain exons.
This protein was predicted blasting Glandirana rugosa genome against the Xenopus tropicalis DI64 protein found in SelenoDB. To find the gene structure, we analyzed the exonerate file.
Moreover, neither the SECIS element nor Seblastian nor t-coffee results were obtained.
MsrA exists like a selenoprotein in some lower organisms, as bacteria or green algae, using a Sec-catalytic residue instead of Cys residue. They are also absent in many hyperthermophylic organisms because at higher temperatures, methionine sulfoxide reduction may not require catalysis. Apart from that, it has been seen that Msrs have roles in protecting cellular proteins from oxidative stress and through this function they may regulate lifespan in several model organisms.
Our results showed that the MsrA protein we predicted had no Sec residue, which concords to the literature.
After our results’ analysis, we could not predict MsrA selenoprotein in our organism. According to the information explained below, we didn’t find a Sec residue nor SECIS element nor Seblastian prediction in our 3 proteins. This means a loss of one of the most conserved families of selenoproteins in vertebrates in our organism.
The MsrA74 protein location is in the scaffold BLSH010095144.1 between positions 656655 and 690927 in the forward strand. The gene contains 4 exons.
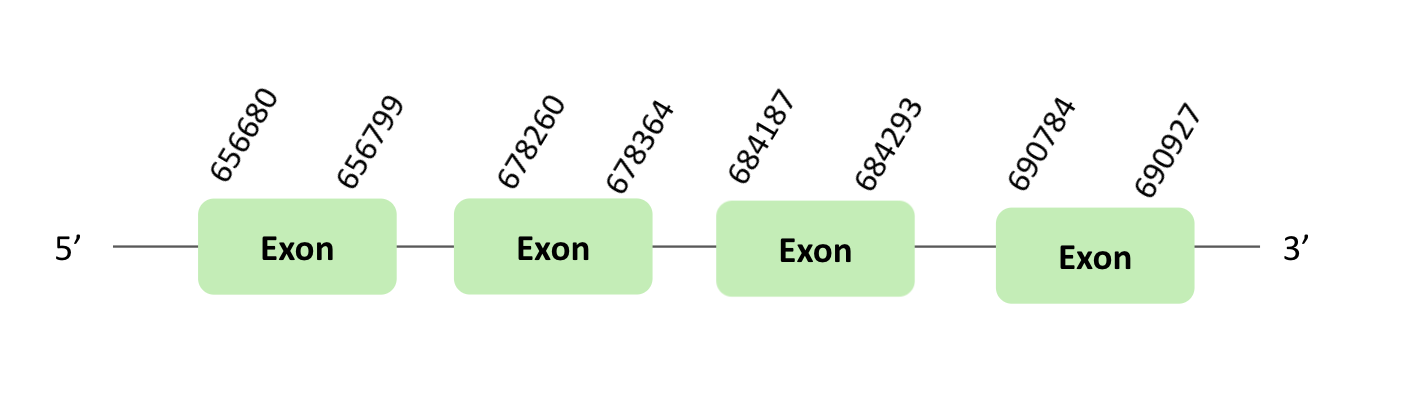
This protein was predicted blasting Glandirana rugosa genome against the Xenopus tropicalis MsrA74 protein found in SelenoDB. To find the gene structure, we analyzed the exonerate file.
SECIS element of grade A, predicted in the 3'UTR region, was found. Regarding the Seblastian there were no selenoproteins coding sequences. Finally, when we analyzed the t-coffee there was any selenocysteine.
The MsrA75 protein location is in the scaffold BLSH010095144.1 between positions 656655 and 690933 in the forward strand. The gene contains 3 exons.
This protein was predicted blasting Glandirana rugosa genome against the Xenopus tropicalis MsrA75 protein found in SelenoDB. To find the gene structure, we analyzed the exonerate file.
There was no SECIS element useful for our strand, because the coordinates of the SECIS were not larger than the last exon. Regarding the Seblastian there were no selenoproteins coding sequences. Finally, when we analyzed the t-coffee, there was any selenocysteine.
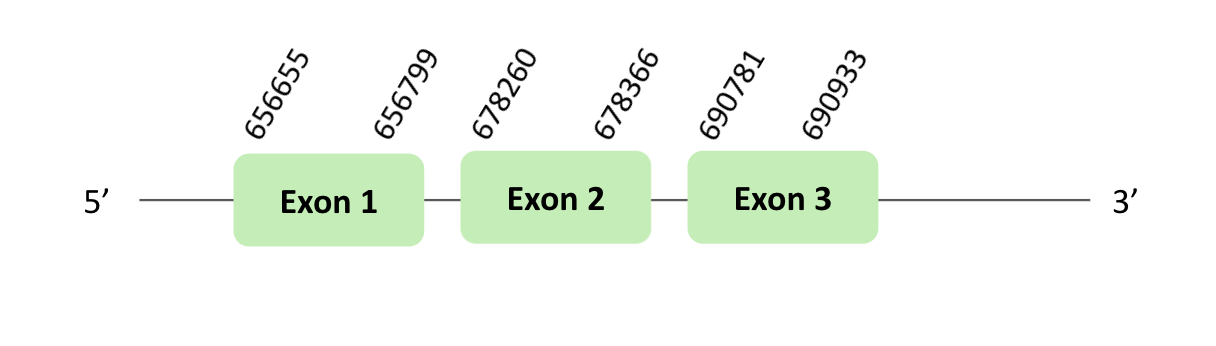
The MsrA76 protein location is in the scaffold BLSH010095144.1 between positions 656676 and 690933 in the forward strand.
This protein was predicted blasting Glandirana rugosa genome against the Xenopus tropicalis MsrA76 protein found in SelenoDB. To find the gene structure we analyzed the exonerate file.
A SECIS element of grade A, predicted in the 3'UTR region, was found. In addition, we did not find a Seblastian result. Finally, when we analyzed the t-coffee there was any selenocysteine.
MrsB1 (SelR) is responsible for the reduction of methionine- R-sulfoxide residues in proteins, is a major MsrB in cytosol and nucleus in mammalian cells.
After our results’ analysis, we could not predict SelR selenoprotein in our organism. According to the information explained below, we didn’t find a Sec residue nor SECIS element nor Seblastian prediction in our 3 scaffolds. This means a loss of two of the most conserved families of selenoproteins in vertebrates in our organism.
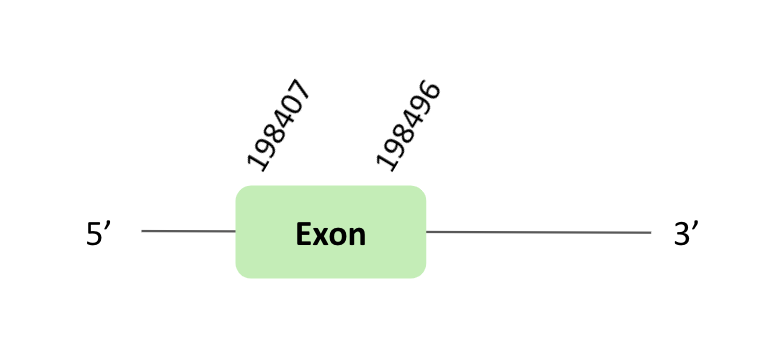
The SelR_91 protein location is in the scaffold BLSH010417220.1 between positions 198407 and 198496 in the forward strand. The gene contains 1 exon.
This protein was predicted blasting Glandirana rugosa genome against the Xenopus tropicalis SelR_91 protein found in SelenoDB. To find the gene structure, we analyzed the exonerate file.
Then, there was no SECIS element useful for our strand. Regarding the Seblastian there were no selenoproteins coding sequences.
Finally, when we analyzed the t-coffee there was any selenocysteine.
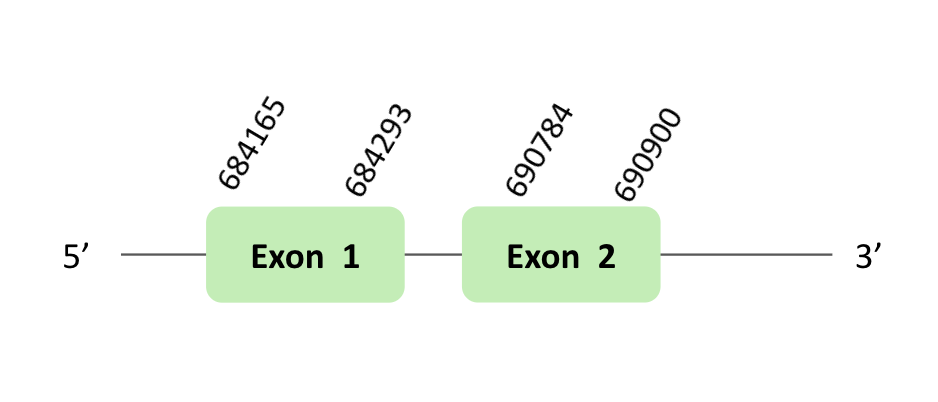
The SelR_92 protein location is in the scaffold BLSH010417220.1 between positions 196901 and 198505 in the forward strand. The gene contains 2 exons.
This protein was predicted blasting Glandirana rugosa genome against the Xenopus tropicalis SelR92 protein found in SelenoDB. To find the gene structure we analyzed the exonerate file.
A SECIS element of grade B was predicted in the 3'UTR region of BLSH010331723.1, that is for a reverse strand. In our case, this SECIS element was not useful, because we had a forward strand. Regarding the Seblastian there were no selenoproteins coding sequences.
Finally, when we analyzed the t-coffee there was any selenocysteine.
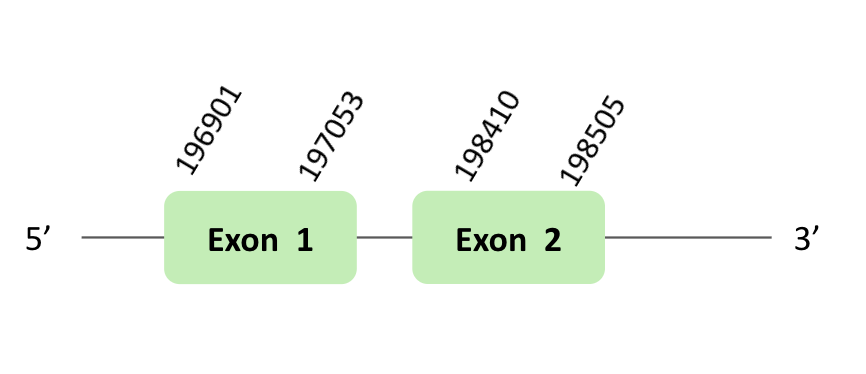
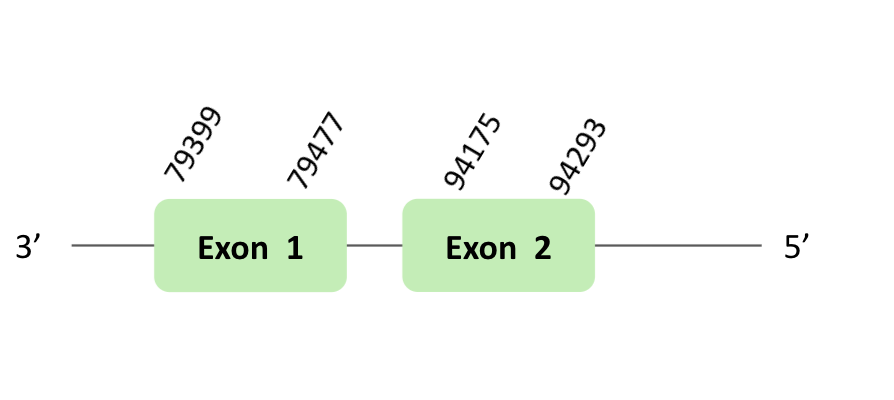
SelR_93
The SelR_93 protein location is in the scaffold BLSH010331723.1 between positions 79399 and 185591 in the reverse strand. The gene contains 2 exons.
This protein was predicted blasting Glandirana rugosa genome against the Xenopus tropicalis SelR93 protein found in SelenoDB. To find the gene structure, we analyzed the exonerate file.
A SECIS element of grade B was predicted in the 3'UTR region of BLSH010331723.1, that is for a forward strand. In our case, this SECIS element was not useful, because we had a reverse strand. Regarding the Seblastian there were no selenoproteins coding sequences. Finally, when we analyzed the t-coffee there was any selenocysteine.
This protein is one of the more recently discovered selenoproteins. It is characterized by a highly conserved CPD-alcohol phosphatidyltransferase domain that is commonly encountered in choline phosphotransferases (CHPT1) and choline/ethanolamine phosphotransferases (CEPT1). Moreover, no Cys forms with homology to the SelI C-terminal extension were found.
After our results’ analysis, we could predict one posible SelI selenoprotein in our organism. According to the information explained below, we didn’t find a Sec residue nor Seblastian prediction in both of our scaffols. But we have found an SECIS element for one of them. We can assume that this is a mistake of our prediction of SECIS elements, so we can discard the possible selenoprotein. This means a loss of one of the most conserved families of selenoproteins in vertebrates in our organism.
The SelI protein location are in two scaffolds, on one hand the scaffold BLSH010070464.1 between positions 1113034 and 1148690, and on the other hand the scaffold BLSH010528819.1 between positions 554328 and 595850, in the reverse and forward strand respectively. The two scaffolds contain 7 exons.
These proteins were predicted blasting Glandirana rugosa genome against the Xenopus tropicalis SelI protein found in SelenoDB. To find the gene structure we analyzed the exonerate file.
A SECIS element of grade A was predicted in the 3'UTR region of scaffold BLSH010070464.1.
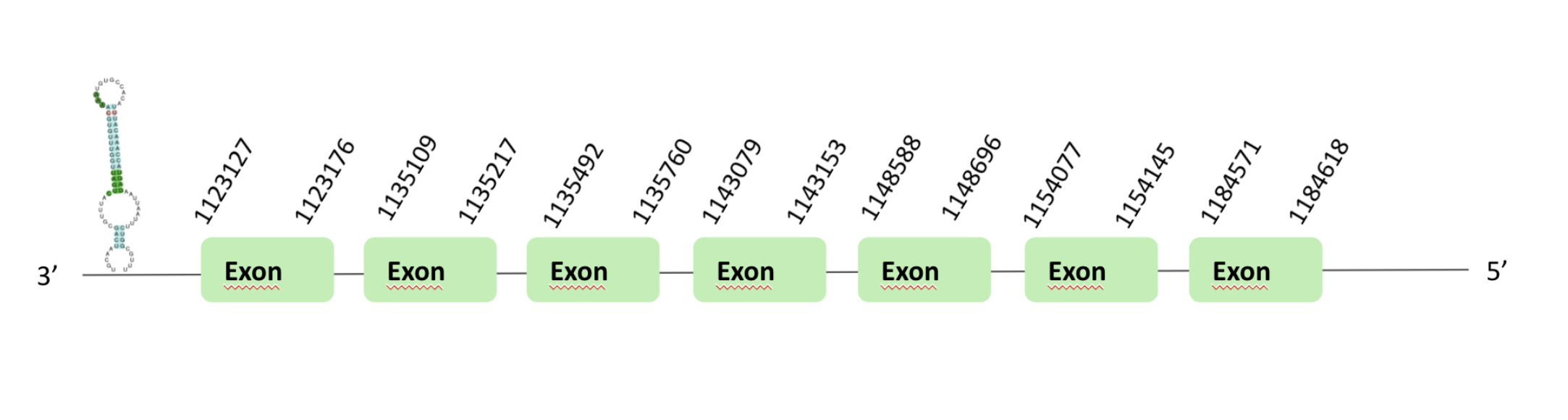
Several SECIS elements were predicted in the scaffold BLSH010528819.1. However, non-one SECIS element in the positive strand was correctly predicted in the 3’ sequence of the protein, because the coordinates of the SECIS were not larger than the last exon.
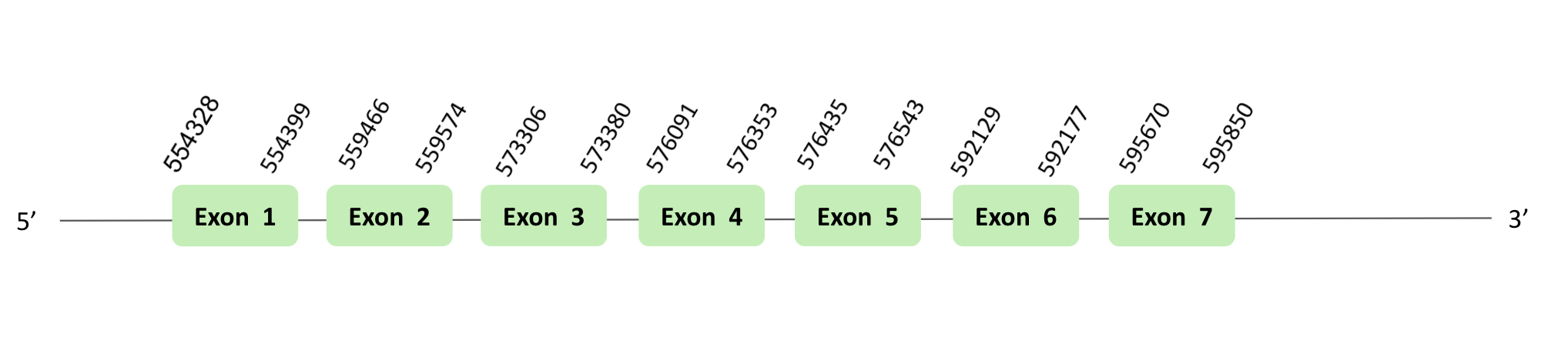
In both cases, there were no positive Seblastian results, t-coffee results did not show any selenocysteine.
SelK and SelS have different sequence, but they are studied together because they similar topology, including a single transmembrane domain in the NH2-terminal sequence; the presence of a glycine-rich (G-rich) segment and a characteristic location of Sec residues in the COOH-terminal end of the protein. They are found in ER membrane. Their exact function is unknown, but it is suggested that both are related to ER-associated degradation (ERAD) of misfolded proteins.
After our results’ analysis, we could not predict SelK SelS selenoprotein in our organism. According to the information explained below, we didn’t find a Sec residue nor SECIS element nor Seblastian prediction in two of our proteins. This means a loss of two of the most conservated families of selenoproteins in vertebrates in our organism.
SelM is an ER-resident thiol-disulfide oxidoreductase that is highly expressed in the brain and bestows neuroprotective properties, preventing oxidative damage induced by H2O2. In humans, it seems to be a protector factor in Alzheimer's diseases.
After our results’ analysis, we could not predict SelM selenoprotein in our organism. According to the information explained below, we didn’t find a Sec residue nor SECIS element nor Seblastian prediction in two of our 3 proteins. This means a loss of one of the most conserved families of selenoproteins in vertebrates in our organism.
The SelM protein location is in the scaffold BLSH010143868.1 between positions 27817 and 31294 in the reverse strand. The gene does not contain exon.
This protein was predicted blasting Glandirana rugosa genome against the Xenopus tropicalis SelM protein found in SelenoDB. To find the gene structure we analyzed the exonerate file.
Finally, neither the SECIS element nor Seblastian nor t-coffee results were obtained.
The selenoprotein N (SelN) is an ancestral selenoprotein found in all the vertebrates. This eukaryotic selenoprotein is located basically in the ER membrane. It has a high expression in fetal and growing muscular tissue, skeletal muscle, heart, lung and placenta.
After our results’ analysis, we could not predict SelN selenoprotein in our organism. According to the information explained below, we didn’t find a Sec residue nor SECIS element nor Seblastian prediction in two of our 3 proteins. This means a los of one of the most conserved families of selenoproteins in vertebrates in our organism.
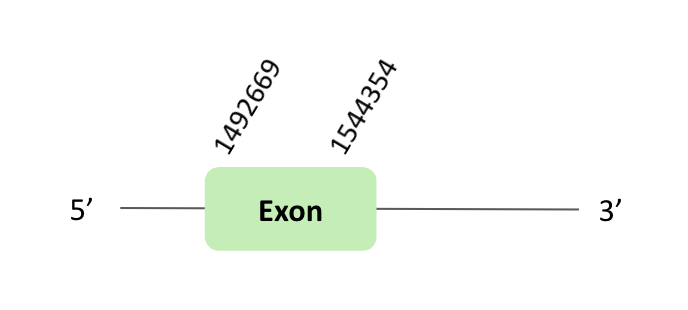
The SelN protein location is in the scaffold BLSH010384004.1 between positions 1492669 and 1494354 in the forward strand. The gene contains 1 exon.
This protein was predicted blasting Glandirana rugosa genome against the Xenopus tropicalis SelN protein found in SelenoDB. To find the gene structure, we analyzed the exonerate file.
Then, several SECIS elements of grade B predicted in the 3'UTR region were found. But non-one SECIS element in the positive strand was correctly predicted in the 3’ sequence of the protein, because the coordinates of the SECIS were not larger than the last exon.
Finally, in t-coffee results, there was any selenocysteine.
The SelO family is the common name for a family of ancestral selenoproteins found in all the vertebrates. These isozymes are localized to mitochondria and expressed in different tissues. This expression is affected by the deficiency of selenium, suggesting that it has a high priority for selenium supply. Although, there is a majority of eukaryotes and bacteria that have a single-copy protein of SelO, many metazoans have duplicated it.
Additionally, this phenomenon is also observed in specific lineages of bony fish such as zebrafish.
After our results’ analysis, we could predict one possible SelO selenoprotein in our organism. According to the information explained below we didn’t find a Sec residue nor SECIS element nor Seblastian prediction in two of our 3 proteins. But we can find one with a SECIS element, but it not has a Seblastian prediction nor the presence of Sec residue. This means that the SECIS element may be wrong and therefore we do not have any selenoprotein in this family.
The SelO_86 protein location is in the scaffold BLSH010113357.1 between positions 425632 and 458181 in the forward strand. The gene contains 1 exon.
This protein was predicted blasting Glandirana rugosa genome against the Xenopus tropicalis SelO86 protein found in SelenoDB. To find the gene structure we analyzed the exonerate file.
Finally, a SECIS element of grade A was predicted in the 3'UTR region. However, regarding the Seblastian there were no selenoproteins coding sequences, we also didn’t have t-coffee results.
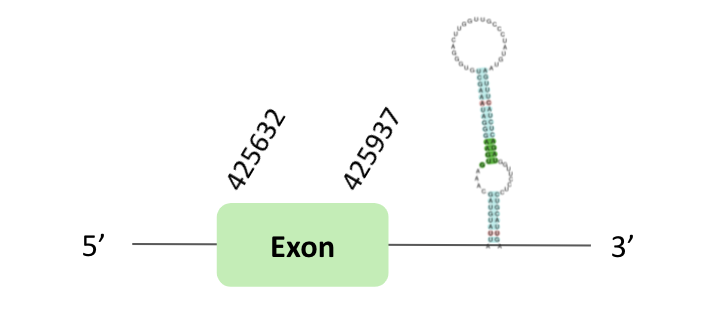
The SelO_87 protein location is in the scaffold BLSH010391438.1 between positions 365955 and 397742 in the forward strand. The gene does not contain exons.
This protein was predicted blasting Glandirana rugosa genome against the Xenopus tropicalis SelO87 protein found in SelenoDB. To find the gene structure we analyzed the exonerate file.
Finally, neither the SECIS element nor Seblastian nor t-coffee results were obtained.
The SelO_88 protein location is in the scaffold BLSH010113357.1 between positions 425710 and 425883 in the forward strand. The gene does not contain exons.
This protein was predicted blasting Glandirana rugosa genome against the Xenopus tropicalis SelO88 protein found in SelenoDB. To find the gene structure we analyzed the exonerate file.
Finally, neither the SECIS element nor Seblastian nor t-coffee results were obtained.
SelP is a selenoprotein with multiple Sec residues per protein subunit, 10 Sec residues in all, and is present in human plasma. The main role of SelP is the transport and delivery of selenium to the tissues. An additional role may be to serve as a heavy metal chelator or antioxidant.
After our results’ analysis, we could not predict SelP selenoprotein in our organism. According to the information explained below we didn’t find a Sec residue nor SECIS element nor Seblastian prediction. This means a lost of one of the most conserved families of selenoproteins in our organism.
The SelP_90 protein location is in the scaffold BLSH010213157.1 between positions 19583 and 20122 in the forward strand. The gene does not contain exons.
This protein was predicted blasting Glandirana rugosa genome against the Xenopus tropicalis SelP90 protein found in SelenoDB. To find the gene structure, we analyzed the exonerate file.
Finally, we cannot search for the SECIS element because there was no comparison between protein and indexed genome. Also, we didn’t obtain t-coffee results.
SelW is a small 9-kDa selenoprotein localized in the cytosol and is expressed at high levels in muscles and brain. It belongs to the stress-related group of selenoproteins as its expression is highly regulated by the availability of Se in the diet. Several SelW homologues were observed across non-mammalian vertebrates. Phylogenetic analysis revealed a distinct group of proteins, SelW2. It was described SelW2 as a selenoprotein in bony fishes, but also in frog and in elephant and shark, which suggests that it was part of the ancestral vertebrate selenoproteome.
After our results' analysis, we could not predict SelW selenoprotein in our organism. According to the information explained below, we didn’t find a Sec residue nor SECIS element nor Seblastian prediction. This means a lost of one of the most conserved families of selenoproteins in our organism.
The SelW00 protein location is in the scaffold BLSH010113654.1 between positions 875118 and 875198 in the reverse strand. The gene does not contain exons.
This protein was predicted blasting Glandirana rugosa genome against the Xenopus tropicalis SelW00 protein found in SelenoDB. To find the gene structure, we analyzed the exonerate file.
Finally, we cannot search for the SECIS element because there was no comparison between protein and indexed genome. Also, we didn’t obtain t-coffee results.
SelT is predominantly localized to the ER and Golgi and is ubiquitously expressed both during embryonic development and in adult tissues. Knockdown of SelT in mouse fibroblasts leads to decreased expression of extracellular matrix genes involved in cell structure organization and alters cell adhesion properties. In addition, the loss of SelT resulted in the upregulation of SelW. This protein is also important in neuroendocrine processes, as it contributes to the homeostasis of the intracellular calcium and secretion of hormones.
After our results’ analysis, we could predict two diferent SelT selenoproteins in our organism. According to the information explained below we found Sec residues on both of our scaffolds, this indicated with the present SECIS element and the prediction of the Seblastian that SelT is a preserved selenoprotein in our organism. Furthermore we can postulated that in our organisme it has suffered a duplication because we can find to different scaffolds from the same inicial protein that was present on Xenopus tropicalis.
The SelT protein location is in two scaffolds, first in the scaffold BLSH010368923.1 between positions 61138 and 61662, and second BLSH010283353.1 between positions 692580 and 692314, in the forward and reverse strand, respectively. The two scaffolds contained 1 exon.
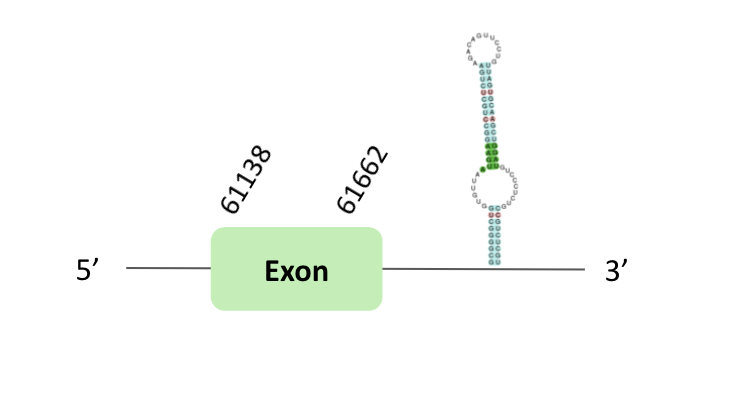
In the case of BLSH010283353.1, a SECIS element of grade B was predicted in the 3'UTR region. However, regarding the Seblastian there are no selenoprotein coding sequences.
Finally, in t-coffee results we observed 4 selenocysteines, one of them was a sec-homologous selenocysteine and another one was a cysteine conversion into selenocysteine.
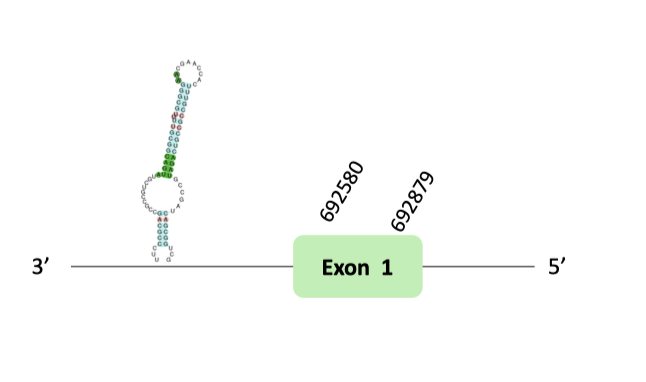
The selenoprotein U family (SelU family) is composed of three members (SelU1, SelU2 and SelU3). The SelU1 selenoprotein is the only member of the family that takes part of the ancestral selenoproteome. A relevant finding in prior phylogenetic analysis of Sec- and Cys-containing forms of the SelU family, suggested that all Sec-containing SelU sequences belong to the SelU1 group. Concretely, in mammals there are three Cys-containing SelU proteins (SelU1-3), while in some fishes there are three Sec-containing SelU proteins.
After our results analysis we could not predict SelU selenoprotein in our organism. According to the information explained below we didn’t find a Sec residue, even though there is a different grade of SECIS element.
The SelU96 protein location is in the scaffold BLSH010317410.1 between positions 95126 and 107060 in the forward strand. The gene doesn't contain exons.
This protein was predicted blasting Glandirana rugosa genome against the Xenopus tropicalis SelU96 protein found in SelenoDB. To find the gene structure, we analyzed the exonerate file.
Finally, neither the SECIS element nor Seblastian nor t-coffee results were obtained.
The SelU97 protein location is in the scaffold BLSH010509110.1 between positions 1433442 and 1438867 in the forward strand. The gene contains 3 exons.
This protein was predicted blasting Glandirana rugosa genome against the Xenopus tropicalis SelU97 protein found in SelenoDB. To find the gene structure we analyzed the exonerate file.
Then, several SECIS elements of grade B y C were predicted in the 3'UTR region. However, non-one SECIS element in the positive strand was correctly predicted in the 3’ sequence of the protein, because the coordinates of the SECIS were not larger than the last exon.
Finally, t-coffee results showed there was any selenocysteine.
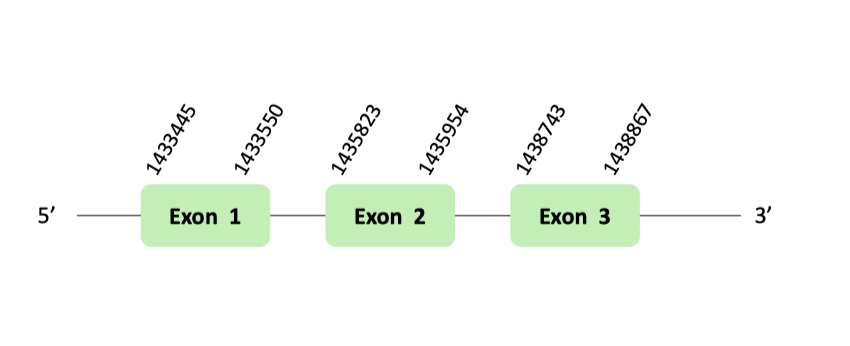
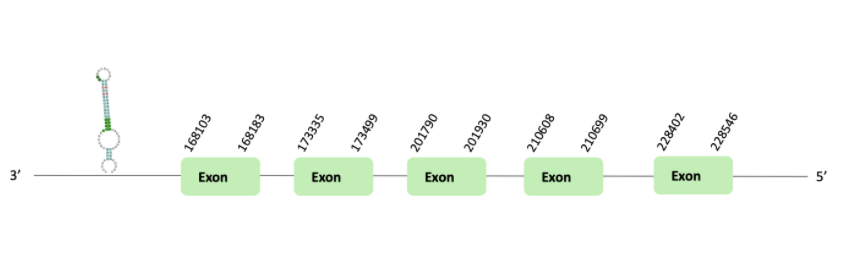
The SelU98 protein location is in the scaffold BLSH010465673.1 between positions 168103 and 228546 in the reverse strand. The gene contains 5 exons.
This protein was predicted blasting Glandirana rugosa genome against the Xenopus tropicalis SelU98 protein found in SelenoDB. To find the gene structure, we analyzed the exonerate file.
Then, a SECIS element of grade A predicted in the 3'UTR region was found. Regarding the Seblastian, there were no selenoproteins coding sequences.
Finally, in t-coffee results showed there was any selenocysteine.
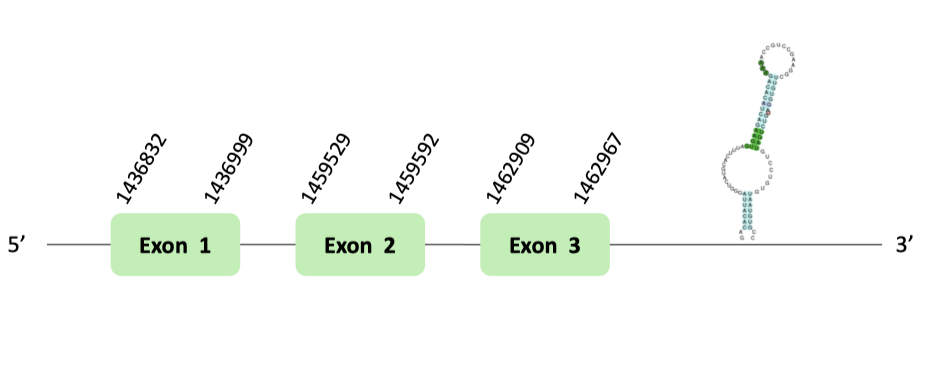
The selenoprotein 15 (Sel15) protein is an ancestral selenoprotein found in all the vertebrates. In 1998 the protein was identified in humans, by Gladyshev et al. But, its specific function remains unknown. It has been shown that Sel15 levels specially respond to selenium addition.
After our results' analysis, we could predict the Sel15 selenoprotein in Glandirana rugosa genome. The protein we predicted contains a Sec residue, and a SECIS element was found in the 3'UTR region of the gene, which concord with the Seblastian output. As it takes part of the common vertebrate selenoproteins, and it was previously well annotated in other databases, this protein was clearly predicted using Xenopus tropicalis as our protein source.
The Sel15 protein location is in the scaffold BLSH010355584.1 between positions 1428416 and 1458525 in the forward strand. The gene contains 3 exons.
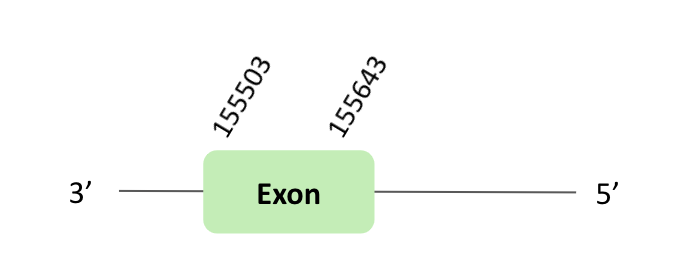
Fep 15 is a 15 kDa selenoprotein-like protein in frog.
The Fep15 protein location is in the scaffold BLSH010059689.1 between positions 155503 and 182936 in the reverse strand. The gene contains 1 exon.
This protein was predicted blasting Glandirana rugosa genome against the Xenopus tropicalis Fep15 protein found in SelenoDB. To find the gene structure, we analyzed the exonerate file.
Then, a SECIS element of grade B was predicted in the 3'UTR region. But it was not correctly predicted in the 3’ sequence of the protein, because the coordinates of the SECIS were not smaller than the exon.
Finally, when we analyzed the t-coffee there was any selenocysteine.
This protein phosphorylates Ser-tRNA[Ser]Sec to create the Sec-tRNA[Ser]Sec. The function and homology of this protein are conserved across archaea and eukaryotes that synthesize selenoproteins. For years, identification of the phosphoseryl kinase remained unknown; however, recent in silico analyses of the archeal and eukaryotic genomes for novel kinase-like genes that are present within genomes containing the Sec incorporation genes revealed a candidate, phosphoseryl-tRNA[Ser]Sec kinase gene. Phosphoseryl-tRNA[Ser]Sec kinase was subsequently cloned and characterized as a protein that phosphorylates the seryl moiety on seryl-tRNA[Ser]Sec in the presence of ATP and Mg2+. Moreover, the function and homology of this protein is conserved across archaea and eukaryotes that sinthetise selenoproteins, a fact that suggests that it plays an important role in selenoprotein biosynthesis and/or regulation.
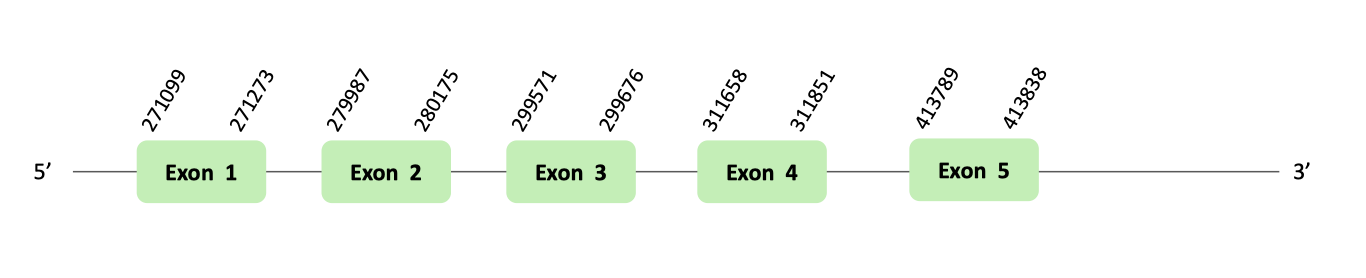
After our results' analysis, we could predict PSTK machinery protein in Glandirana rugosa genome. The protein we predicted contains a Sec residue, but no logic SECIS elements were found in the 3'UTR region of the gene, which concord with the negative Seblastian output.
The PSTK protein location is in the scaffold BLSH010239871.1 between positions 271099 and 1458525 in the forward strand. The gene contains 5 exons.
This protein was predicted blasting Glandirana rugosa genome against the Xenopus tropicalis PSTK protein found in SelenoDB. To find the gene structure we analyzed the exonerate file.
We have found 9 SECIS elements of grade B and C. But non-one SECIS element in the positive strand was correctly predicted in the 3’ sequence of the protein, because the coordinates of the SECIS were not larger than the last exon.
In t-coffee results we observed 4 selenocysteines, one of them was a cysteine conversion into selenocysteine.
eEFSec recruits Sec-tRNA[Ser]Sec and includes a Sec amino acid in a protein. Including a Sec amino-acid in a protein requires a selenocysteyl-tRNA[Ser]Sec specific elongation factor in eukaryotes (eEFSec) and in prokaryotes (SELB or Efsec) that suppresses UGA codons that are upstream of Sec insertion sequence (SECIS) elements bound by SECIS-binding protein 2 (SBP2).
After our results’ analysis, we could predict eEFsec machinery protein in Glandirana rugosa genome. The proteins we predicted do not contain any Sec residue, and any SECIS elements were found in the 3'UTR region of both genes. The negative Seblastian output on both of our proteins might suggest a false positive on the SECIS prediction.
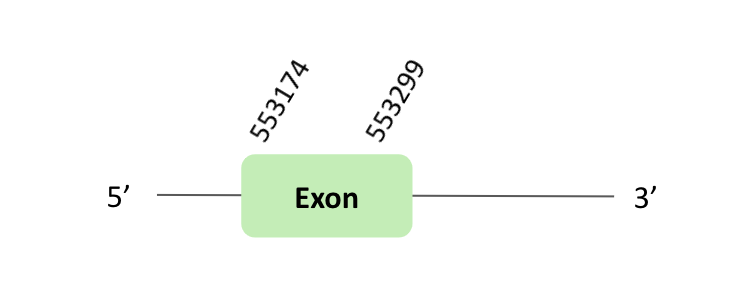
The eEFsec05 protein location is in the scaffold BLSH010363128.1 between positions 553174 and 553299 in the forward strand. The gene contains 1 exon.
This protein was predicted blasting Glandirana rugosa genome against the Xenopus tropicalis eEFsec05 protein found in SelenoDB. To find the gene structure, we analyzed the exonerate file.
Finally, 5 SECIS elements have been predicted, but non-one SECIS element in the positive strand was correctly predicted in the 3 ’sequence of the protein, because the coordinates of the SECIS were not larger than the exon.
In this case, we observed a loss of Xenopus tropicalis selenocysteine since there was an alanine instead of selenocysteine.
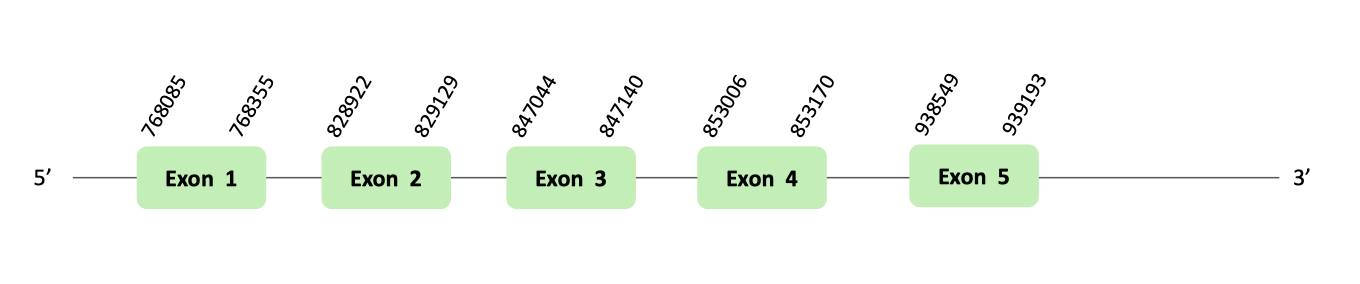
The eEFsec06 protein location is in the scaffold BLSH010494947.1 between positions 768085 and 939193 in the forward strand. The gene contains 5 exons.
This protein was predicted blasting Glandirana rugosa genome against the Xenopus tropicalis eEFsec06 protein found in SelenoDB. To find the gene structure we analyzed the exonerate file.
Finally, 11 SECIS elements have been predicted, where there were 5 for the forward strand. Nevertheless, non-one SECIS element in the positive strand was correctly predicted in the 3’ sequence of the protein, because the coordinates of the SECIS were not larger than the last exon.
When we analyzed the t-coffee there was any selenocysteine.
Therefore, it is not a selenoprotein.
Selenocysteine synthase (SecS), it is an enzyme that incorporates the active form of Sec to the Ser-tRNA[Ser]Sec phosphorylated to create the final Sec-tRNA[Ser]Sec.
Mutations in SecS genes are associated with the development of autosomal-recessive progressive cerebellocerebral atrophy, and that phenotypes could be partially reproduced in the corresponding KO animal models.
After our results' analysis, we could predict two SecS machinery proteins in Glandirana rugosa genome. One of the proteins that we predicted contained a Sec residue, and a SECIS element found in the 3'UTR region, which did not concord with the negative Seblastian output. Both proteins have the same scaffold, and they are in the same positions. We deduce that they are the same protein that was duplicated in Xenopus tropicalis, but in our organism there is only one protein, so maybe it has succeeded a deletion of one of the copies of the gene of Xenopus tropicalis.
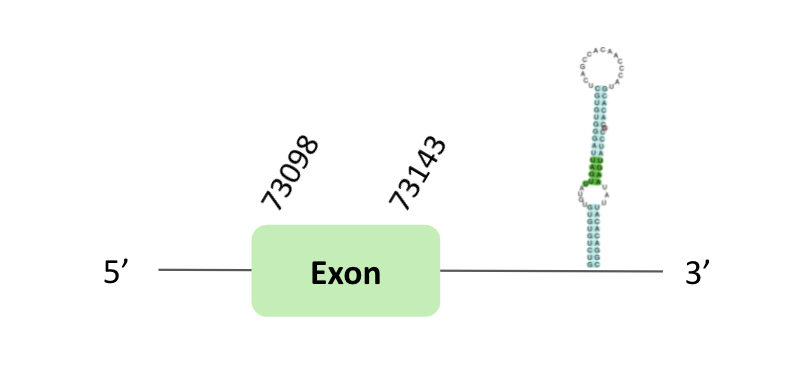
The SecS79 protein location is in the scaffold BLSH010085618.1 between positions 73039 and 73184 in the forward strand, in which one exon was predicted.
This protein was predicted blasting Glandirana rugosa genome against the Xenopus tropicalis SecS79 protein found in SelenoDB. To find the gene structure we analyzed the exonerate file.
A SECIS element of grade A was found in the 3'UTR region. Regarding the Seblastian, there were no selenoprotein coding sequences.
When we analysed the t-coffee, we observed that BLSH010085618.1 did not have a selenocysteine protein.
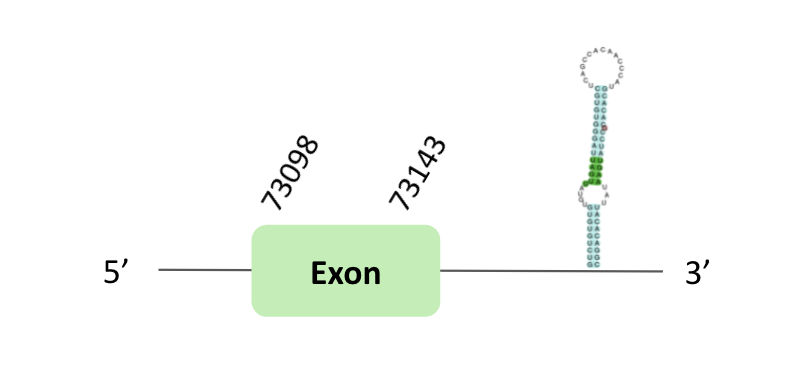
The SecS80 protein location is in the scaffold BLSH010085618.1 between positions 73039 and 73184 in the forward strand. One exon was predicted in this case.
This protein was predicted blasting Glandirana rugosa genome against the Xenopus tropicalis SecS80 protein found in SelenoDB. To find the gene structure, we analyzed the exonerate file.
Finally, a SECIS element of grade B was found in the 3'UTR region. Regarding the Seblastian, there were no selenoprotein coding sequences.
When we analyzed the t-coffee, we observed that BLSH010085618.1 had a selenocysteine protein which it was not homologous with Xenopus tropicalis protein.
In conclusion, this protein contains a selenocysteine and a SECIS element, so we think that Sec80 is a selenoprotein.
Selenoproteins have an important function, acting against oxidation. They are characterized because they have a selenocysteine residue (Sec) in its sequence, a 21st amino acid that contains a selenium atom. However, the selenoproteome differs between species and is not easily identified because the Sec residue is codified by UGA, a stop codon, leading to a low recognition rate by some bioinformatic tools.
For this reason, the objective of this project was to identify the selenoproteins and selenoprotein machinery required for their synthesis present in Glandirana rugosa's genome. To do that, selenoproteins of Xenopus tropicalis are used to do a homology-based prediction, which is the closest relative of our organism.
After having analysed the alignment of each query protein and predicted protein, we concluded that in Glandirana rugosa, there are:
gr2in="/mnt/NFS_UPF/soft/genomes/2021/Glandirana_rugosa/genome.fa"
genin="/mnt/NFS_UPF/soft/genomes/2021/Glandirana_rugosa/genome.fa"
indexgr2in="/mnt/NFS_UPF/soft/genomes/2021/Glandirana_rugosa/genome.index"
mkdir -p results
#estamos en $HOME
for p in ./Proteins/*.fa ; do
sed s/U/X/g $p > $p
#done
for f in ./Proteins.fa/*.fa ; do
#f -> ./Proteins.fa/query.fa
#${string##substring} Deletes longest match of $substring from front of $string.
queryname=${f##*/}
#${string%%substring} Deletes longest match of $substring from back of $string.
queryname=${queryname%%".fa"}
tblastdir="./tblast"
scaffdir="./results/$queryname/scaffolds"
exodir="./results/$queryname/exonerate"
fastaseqdir="./results/$queryname/fastaseq"
t_coffeedir="./results/$queryname/coffee"
genwisedir="./results/$queryname/genwise"
seblastdir="./results/$queryname/seblastian"
mkdir "./results/$queryname"
mkdir $tblastdir
mkdir $scaffdir
mkdir $exodir
mkdir $fastaseqdir
mkdir $t_coffeedir
mkdir $genwisedir
mkdir $seblastdir
#queryname -> query
blastfile="$tblastdir/$queryname.blast"
tblastn -query $f -db $gr2in -outfmt 6 -evalue 0.01 -out $blastfile
#query acc.verc 1
#query acc.verc 1
#subject acc.ver 2
#% identity 3
#alignment length 4
#mismatches 5
#gap opens 6
#q. start 7
#q. end 8
#s. start 9
#s. end 10
#evalue 11
#bit score 12
cut -f2 $blastfile | sort | uniq | while read scaffold; do
#SPP00002862_2.0 BLSH010355584.1:subseq(1420000,35000) 47.059 17 7 1 244 260 9288 9244 7.4 18
start=$(grep $scaffold $blastfile | cut -f9-10 |sed 's/\t/\n/' | sort -n | head -1 )
end=$(grep $scaffold $blastfile | cut -f9-10 |sed 's/\t/\n/' | sort -n | tail -1 )
hit=$(echo $scaffold | cut -f 2 -d ' ')
#start=$(echo $hit_data | cut -f 9 -d ' ')
#end=$(echo $hit_data | cut -f 10 -d ' ')
if [[ $start -gt $hit_offset ]]; then
begin=$(($start-50000))
else
begin=0
fi
length=$(($begin+100000))
fastfechfile="${scaffdir}/${hit}.fa"
fastagenomfile="${scaffdir}/genomic_${hit}.fa"
exoneratefile="${exodir}/${hit}.exonerate.gff"
fastafetch $genin $indexgr2in "$hit" > $fastfechfile
fastasubseq $fastfechfile $begin $length > $fastagenomfile
sed s/*/N/g $fastagenomfile > "${seblastdir}/${hit}.fa"
exonerate -m p2g --showtargetgff -q $f -t $fastagenomfile | egrep -w exon > $exoneratefile
fastaseqfromGFF.pl $fastagenomfile $exoneratefile > "${fastaseqdir}/fastaseq_${hit}.fa"
fastatranslate "${fastaseqdir}/fastaseq_${hit}.fa" -F 1 > "${fastaseqdir}/fastaseq_${hit}.aa.fa"
sed s/*/X/g "${fastaseqdir}/fastaseq_${hit}.aa.fa" > "${fastaseqdir}/fastaseq_${hit}.aa.x.fa"
t_coffee $f "${fastaseqdir}/fastaseq_${hit}.aa.x.fa" > "${t_coffeedir}/${queryname}_${hit}.tc.fa" | rm *.html | rm *.aln
genewise -pep -pretty -cdna -gff $f $fastagenomfile > "${genwisedir}/${queryname}_${hit}.gw.fa"
done
done
exit
We are a 4th Human Biology students at Pompeu Fabra University. This is our project about Glandirana rugosa selenoproteins of Bioinformatics subject.
Human Biology student
Human Biology student
Human Biology student
Human Biology student
Lets get in touch and talk about your next project.