Selenoproteins
Selenoproteins are selenium-containing proteins that are present in all three domains of life: bacteria, archaea and eukarya8. Although selenoproteins represent diverse molecular pathways and biological functions, all these proteins contain at least one selenocysteine (Sec), a selenium-containing amino acid, and most serve oxidoreductase functions. Selenoproteins have biological functions in oxidoreductions, redox signaling, antioxidant defense, thyroid hormone metabolism, and immune responses. They thus possess a strong correlation with human diseases such as cancer, Keshan disease, virus infections, male infertility, and abnormalities in immune responses and thyroid hormone function12.
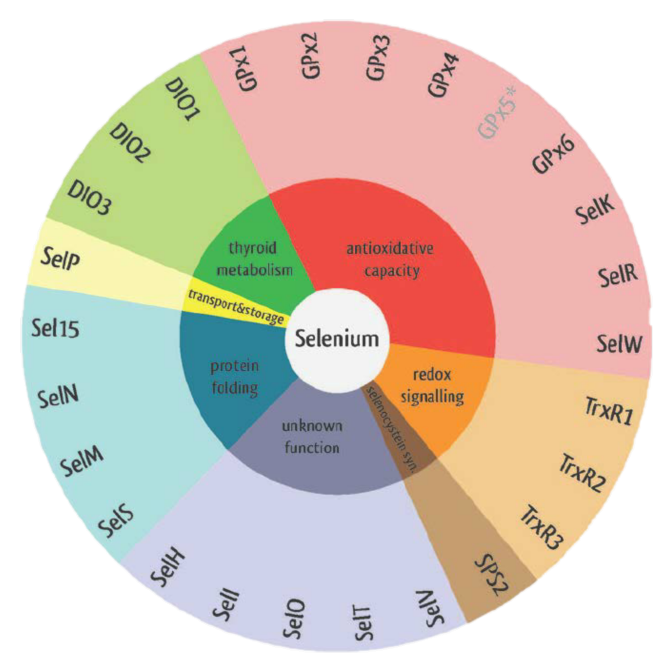
Figure 1. Eukaryotic selenoproteins' functions.
SELENOCYSTEINE
Selenocysteine is recognized as the 21 amino acid in ribosome-mediated protein synthesis and its specific incorporation is directed by the UGA codon. Selenocysteine has a structure similar to that of cysteine, but with an atom of selenium taking the place of the usual sulfur, forming a selenol group2.
Therefore, proteins that contain one or more selenocysteine are called selenoproteins.
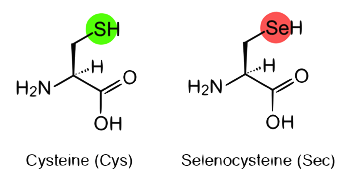
Figure 2. Cysteine and Selenocysteine residues.
SELENOPROTEINS SYNTHESIS
Selenoprotein synthesis is an evolutionary conserved process. Nevertheless, some major differences are found in the mechanisms of selenoprotein synthesis between prokaryotes, eukaryotes, and archaea. The common feature to all organisms is the UGA-Sec codon, the specific tRNA, the SECIS element, and several protein factors10.
Selenocysteine is encoded by a UGA codon in the selenoprotein mRNA. The decoding of UGA as selenocysteine requires the reprogramming of translation because UGA i normally read as a stop codon9. This process requires multiple features such as the selenocysteine insertion sequence (SECIS) element and several protein factors including specific elongation factor EFSec and SECIS binding protein 2 (SBP2).
The SECIS element is essential in order to sintetize selenoproteins. It is located in the 3′ untranslated region (3′ UTR) of the mRNA. Eukaryotic SECIS elements are not highly conserved at the nucleotide level but they all form a similar stem-loop structure, which is composed of two helices separated by an internal loop. Thus, the SECIS element is a structure that provides the platform for the protein factors required for Sec incorporation. One or more of these elements may therefore be directly interacting with SBP2 and thus anchoring it to the ribosome4.
SECIS binding protein 2 (SBP2) is a novel protein that contains an RNA-binding motif, which is found in eukaryotic release factor 1 (eRF1) and ribosomal proteins. Mutagenesis studies demonstrated that this domain is required for the SECIS-binding activity of SBP2. This protein possesses a unique N-terminal domain and a C-terminal domain that is sufficient for all of the known functions of SBP2: SECIS binding, ribosome binding, and in vitro Sec incorporation.
In the selenoprotein synthesis procedure, SECIS element will be recognized by the protein SBP2. When joining, SBP2 recruits a specific elongation factor called eEFsec. This elongation factor is responsible for recruiting the specific tRNA that is linked to Selenocysteine (tRNAsec). This way, the UGA codon will be coded by a selenocysteine and, therefore, the translation will continue until the next STOP codon.
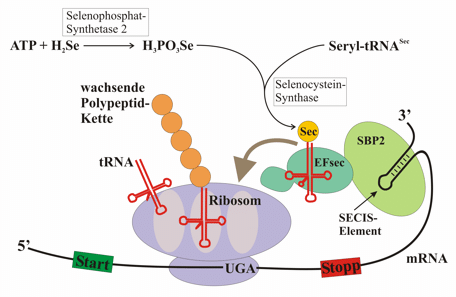
Figure 3. Selenoprotein syntheis procedure.
However, selenoprotein synthesis also depends on other genes that encode for other elements or that have a role in the synthesis itself or participate in the incorporation of selenocysteine. These elements are: Selenophosphate Synthetase (SPS1 and SPS2), Selenocysteine synthase (SLA/LP), Phosphoseryl tRNA Kinase (PSTK) and Selenocysteine associated protein (SECPP43).
SELENOPROTEINS EVOLUTION
While the genome sequence and gene content are available for an increasing number of organisms, eukaryotic selenoproteins remain poorly characterized. The dual role of the UGA codon confounds the identification of novel selenoprotein genes5.
Because of the dual role of the UGA codon, identification of novel selenoproteins in eukaryotes is very difficult. The more direct approach is to search for occurrences of the SECIS structural pattern. Although this approach has been successfully applied in expressed sequence tag (EST) and other cDNA sequences, the low specificity of SECIS searches produces a large number of predictions when applied to eukaryotic genomes.
It should be mentioned that at larger evolutionary distances some selenoproteins are lost and a few evolve and duplicate. In addition, the selenoproteins are irregularly distributed among different species.The highest content of selenoproteins is observed in aquatic organisms, whether they are animals or plants7.
Furthermore, some species have lost Sec, replacing them by Cys-containing homologues, which do not always have the same function9.
The change from aquatic to terrestrial habitats posed a challenge to plants and animals, as the availability of some trace elements greatly diminished, and these organisms were now exposed to higher oxygen levels. As a result, many terrestrial organisms might have lost selenoproteins or replaced them with cysteine-containing homologs.
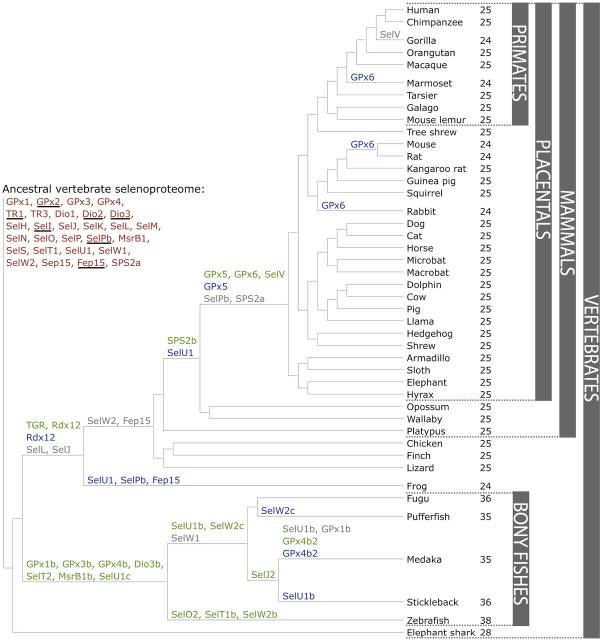
Figure 4. Evolution of the vertebrate selenoproteome.
Nevertheless, considerable progress has been made in characterizing the mechanisms and regulation of selenoprotein synthesis in eukaryotes. Significant discoveries were made recently that helped us better understand Se metabolic pathways, mechanism of Sec biosynthesis in eukaryotes, identities of selenoproteins, and functions of some of these proteins. However, the biological functions of most Sec-containing proteins remain unknown7.